Notes
Article history
The research reported in this issue of the journal was funded by the HTA programme as project number 03/39/18. The contractual start date was in November 2005. The draft report began editorial review in January 2012 and was accepted for publication in April 2013. The authors have been wholly responsible for all data collection, analysis and interpretation, and for writing up their work. The HTA editors and publisher have tried to ensure the accuracy of the authors’ report and would like to thank the reviewers for their constructive comments on the draft document. However, they do not accept liability for damages or losses arising from material published in this report.
Declared competing interests of authors
Karl G Nicholson has been an ad hoc consultant to GlaxoSmithKline and Novartis. He has received funding to speak at meetings organised by Novartis, Baxter, Berna Biotech, Esteves, and the European Scientific Working Group on Influenza, and H5 vaccines from Novartis to support an MRC-funded research project, and H1N1 vaccines from Baxter AG and GlaxoSmithKline to support an NIHR-funded research project. A colleague in Karl G Nicholson’s Department has received research funding from Roche. Maria Zambon has been an investigator of clinical trials sponsored by Novartis, Baxter, Sanofi Pasteur and CSL Australia Ltd. Keith R Abrams has acted as a paid consultant to the health-care industry generally (for the provision of advice and short courses), but specifically has not advised any organisation as regards diagnostics. Tristan W Clark has been an investigator of clinical trials sponsored by Novartis and Roche.
Permissions
Copyright statement
© Queen’s Printer and Controller of HMSO 2014. This work was produced by Nicholson et al. under the terms of a commissioning contract issued by the Secretary of State for Health. This issue may be freely reproduced for the purposes of private research and study and extracts (or indeed, the full report) may be included in professional journals provided that suitable acknowledgement is made and the reproduction is not associated with any form of advertising. Applications for commercial reproduction should be addressed to: NIHR Journals Library, National Institute for Health Research, Evaluation, Trials and Studies Coordinating Centre, Alpha House, University of Southampton Science Park, Southampton SO16 7NS, UK.
Chapter 1 Background and rationale
Elderly demographics
Western industrialised nations face a large increase in the number of older people. In 2006 there were about 200,000 more children of < 16 years of age than people at state pension age in the UK. However, in 2007, for the first time ever, the population at state pension age exceeded the number of children. And, despite increases to state pension age, the population at state pension age is projected to exceed the number of children of < 16 years by 400,000 in 2016, and by over 2 million in 2031. 1 Annual figures published by the NHS Information Centre reveal that people aged > 60 years accounted for almost half of all of the 16.8 million hospital admissions (finished consultant episodes) in 2009–10. 2,3 During 2009–10, infections of the respiratory tract accounted for about 1 in 30 of all hospital admissions in England and 1 in 20 of the 51.5 million bed-days. 4 Preparation for this population growth, including the prevention and care of illness, is of paramount importance.
Acute respiratory illness
Illness surveys conducted in general practice indicate an overwhelming importance of acute respiratory illness in comparison with other conditions. During the most recent (Fourth) National Morbidity Study, a higher proportion of people (31%) consulted for respiratory conditions at least once during the year than for diseases in any other single International Classification of Diseases (ICD) chapter. 5 Overall, 67% of patients who saw their general medical practitioner (GP) with a respiratory condition did so because of an acute infection, i.e. ≈ 20% of all consultations in primary care occur because of acute respiratory infections (ARIs), which are mostly viral. The rates of acute respiratory illness were highest among small children. They were lowest among subjects aged 45–64 years and then increased with age, and the percentage that was graded as ‘serious’ reached ≈ 25% in those aged > 65 years.
Owing to the increasing severity of acute respiratory illness in older people, the number of hospital admissions in England for influenza, pneumonia and other acute lower respiratory infections (ICD-10 codes J10-J18, J20-J22) is approximately three times higher for people aged ≥ 75 years than in younger people ( Figure 1 ). 4 Annual figures published by the NHS Information Centre reveal that among those aged > 75 years the number of admissions for influenza and pneumonia and other lower respiratory tract infections has doubled to almost 200,000 in England over the last 10 years (see Figure 1 ). 4
FIGURE 1.
Annual number of admissions by age to NHS hospitals in England for ‘Influenza and Pneumonia’ (ICD-10 codes J10-J18), and ‘Other acute lower respiratory infections’ (J20-J22), years 1998–99 to 2009–10. Source: www.hesonline.nhs.uk/Ease/servlet/ContentServer?siteID = 1937&categoryID = 202 (accessed 30 October 2010).
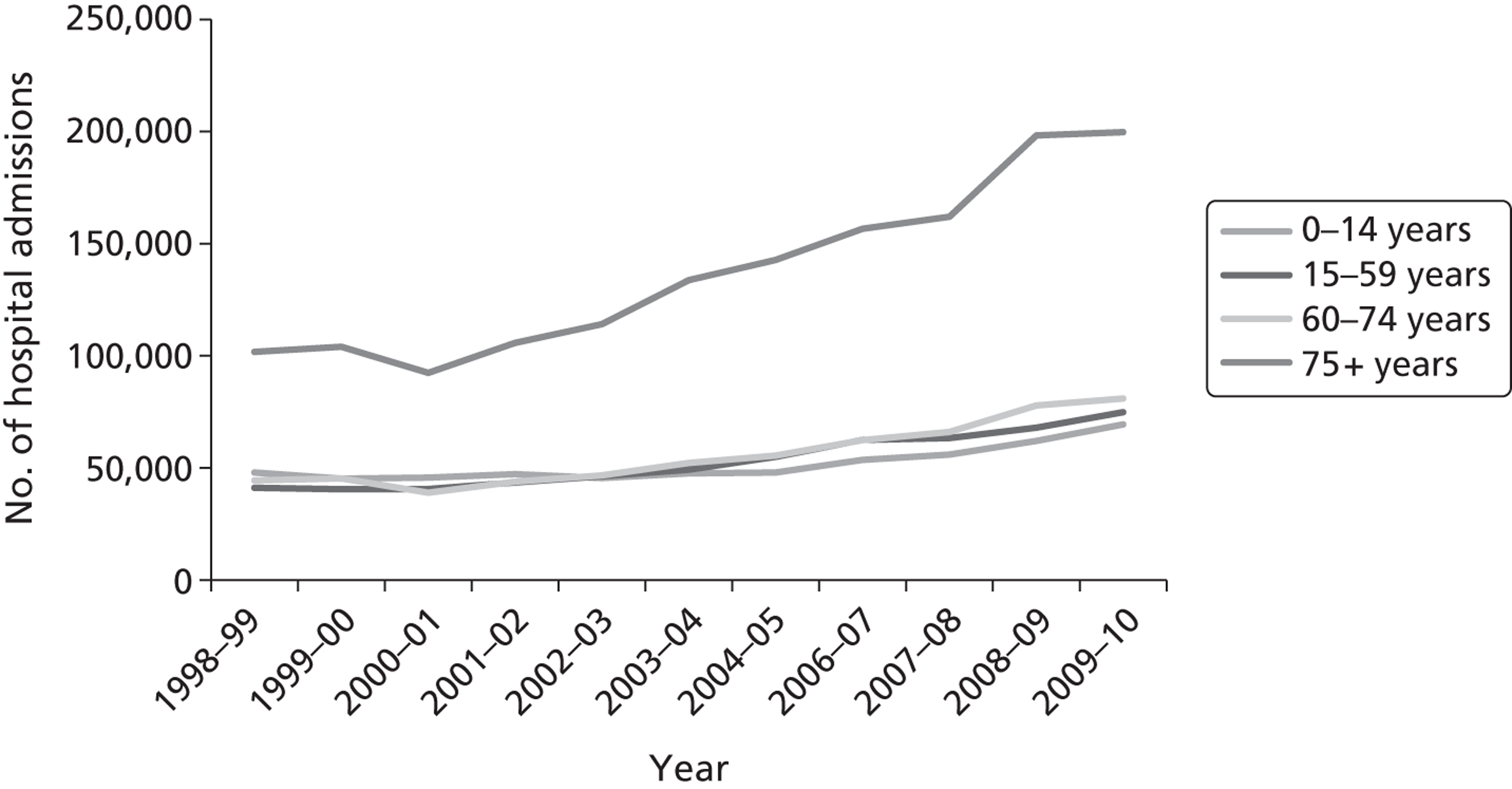
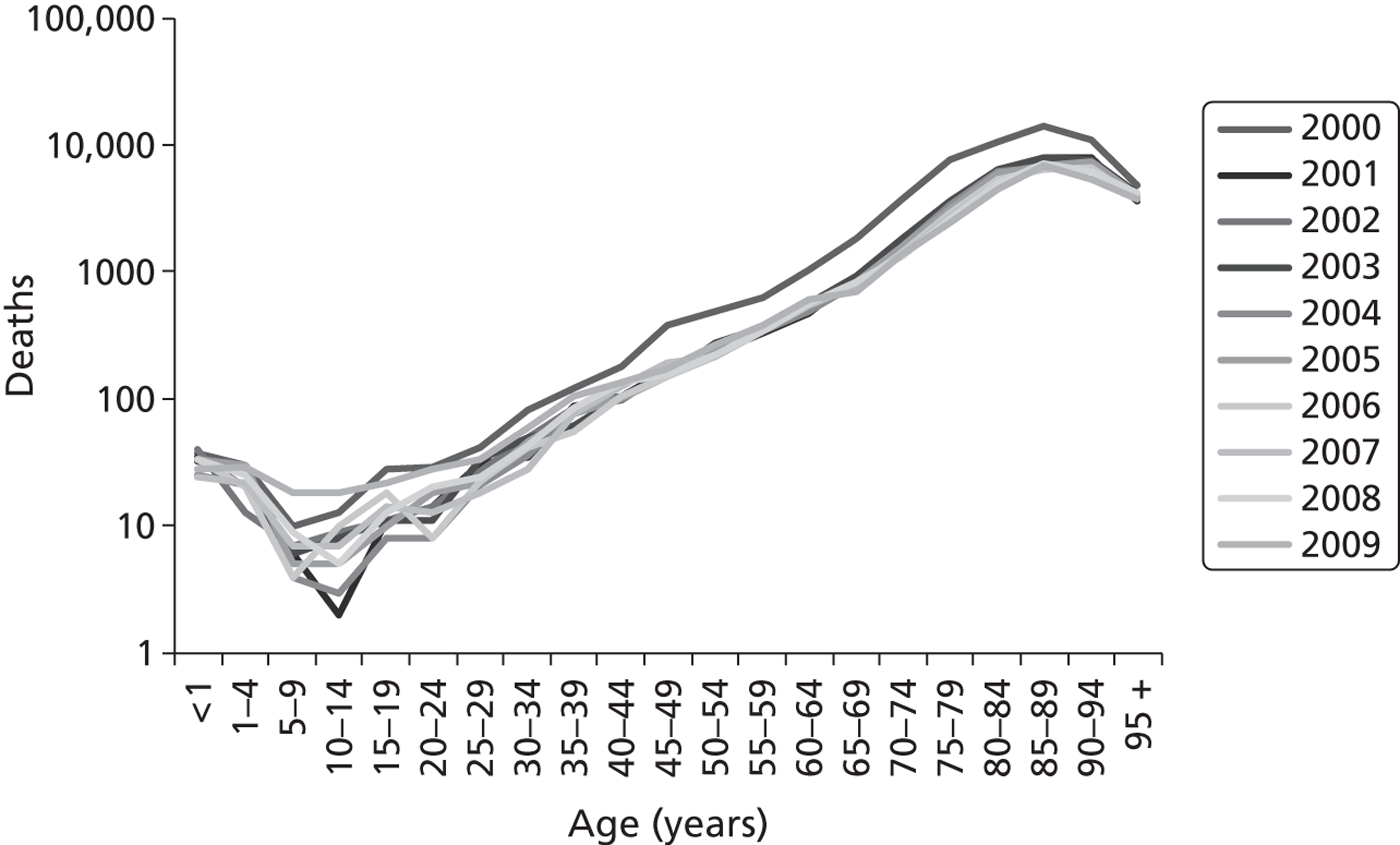
The average length of stay for acute respiratory conditions increases progressively with age. 6 Although the average length of stay for pneumonia has fallen during the last 20 years, it is 10–15% longer in those aged > 65 years than in younger adults ( Figure 2 ). 7 The annual number of pneumonia and influenza deaths (ICD-10 codes J10–J18) in England and Wales increases with increasing age and exceeds 1000 per annum in each of the 5-year age bands in those aged > 70 years ( Figure 3 ). Strategies that prevent acute lower respiratory infections, ameliorate their severity or shorten the average duration of stay will have the greatest benefit in the elderly.
FIGURE 2.
Average length of stay for ‘pneumonia’ in short-stay hospitals, by age: USA, 1990, 2000 and 2006. Source: www.cdc.gov/nchs/data/hus/hus09.pdf#102oryID = 202 (accessed 30 October 2010).
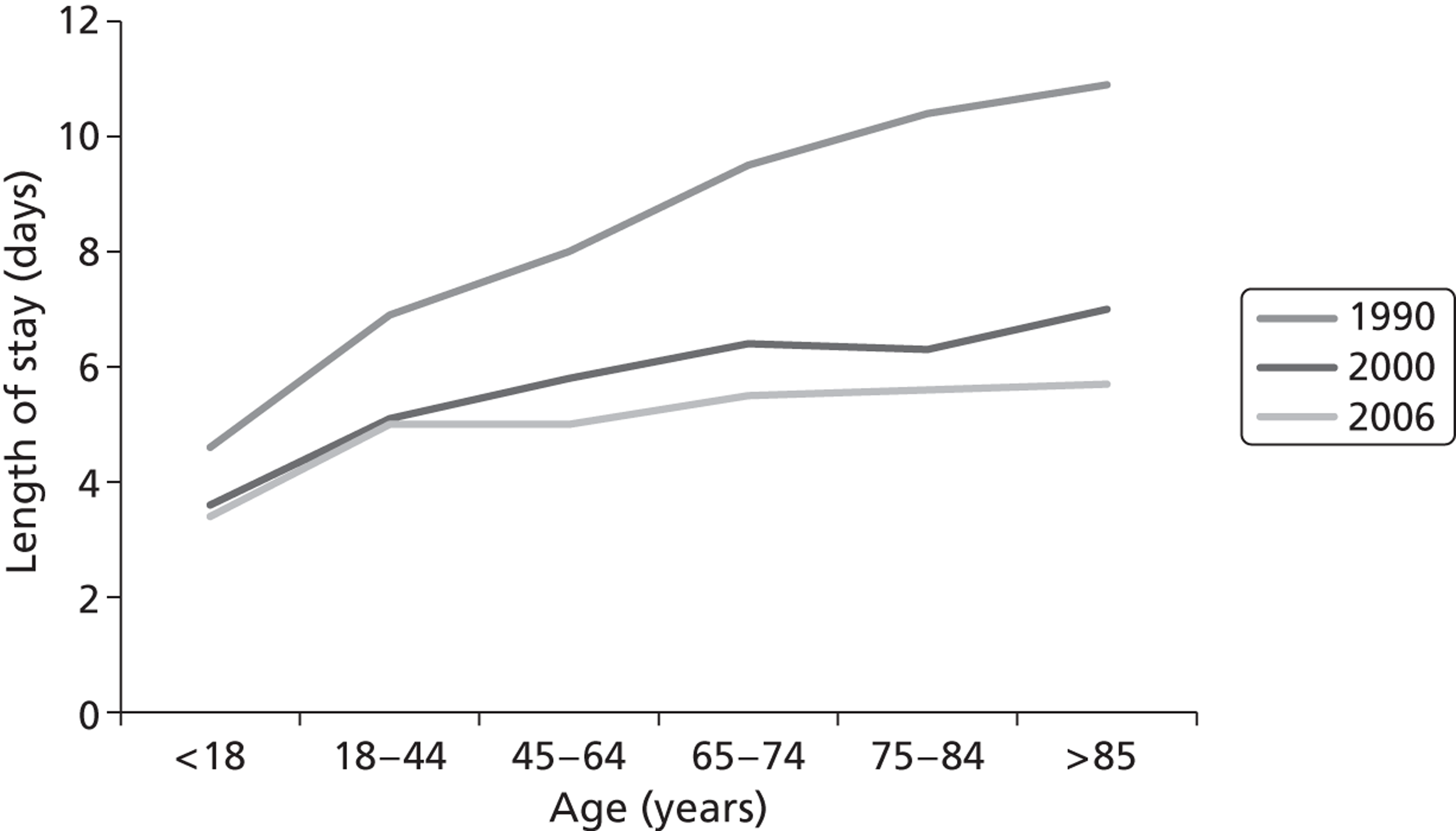
FIGURE 3.
Annual number of pneumonia and influenza deaths (ICD-10 codes J10-J18) in England and Wales, years 2000–9 (compiled from annual ONS mortality statistics).
In this study, we evaluate rapid diagnostic technologies for three target pathogens – influenza, respiratory syncytial virus (RSV) and Streptococcus pneumoniae – which are key aetiological agents of acute respiratory illness and, collectively, are responsible for considerable morbidity and mortality in the elderly.
There is a paucity of information on the relative incidence of influenza, RSV and pneumococcal disease among elderly cardiopulmonary admissions. Falsey et al. 8 evaluated the number of hospitalisations for RSV infection relative to influenza in several thousand elderly people admitted to six hospitals in New York State between November and April 1989–1992. This and other studies suggest that RSV may be found in up to 5% of patients hospitalised with acute respiratory disease,9–13 although with molecular diagnostic tests, the number identified may be higher. Previous studies8–10,12,13 indicate that about 10% of cardiopulmonary admissions have influenza but the number of admissions is influenced by the severity of epidemics, which have been generally mild since 1999/2000, including the recent 2009 H1N1 pandemic. None of these studies was conducted in the UK, and referral and admission practices may differ in the UK from those elsewhere. About one-third of all patients who are hospitalised in Northern Europe with community-acquired pneumonia (CAP) have S. pneumoniae infection. 14 S. pneumoniae is the most common microbiological cause of CAP,15,16 including the UK,17 and is the most commonly identified cause of CAP death. 16
Influenza
About 20% of children and 5% of adults worldwide develop symptomatic influenza A or B each year. 18 Although influenza A and B viruses circulate virtually every winter, quantification of the burden of influenza on consultations, emergency department examinations, hospital admissions and mortality has been difficult because influenza lacks pathognomonic features, it co-circulates with other respiratory pathogens, and it causes a range of non-specific complications, such as exacerbations of chronic cardiopulmonary disease. Indeed, during 2009, many hospital admissions with confirmed influenza A H1N1 infection presented with an exacerbation of asthma. 19 During outbreaks, sentinel schemes, such as the Royal College of General Practitioners network in England, report increased consultation rates for influenza-like illness (ILI) and other respiratory syndromes that are strongly associated with excess mortality. In England and Wales, an estimated 6200–29,600 people died during each of the epidemics between 1975–6 and 1989–90. 20 These estimates are about 10 times the number of death certifications for influenza, suggesting that influenza is responsible for many ‘hidden deaths’. About 90% of influenza-associated excess deaths are among people aged ≥ 65 years. 18 Although there are age-related increases in deaths from seasonal influenzal illness in both ‘at-risk’ and ‘low-risk’ groups,21 most deaths and hospitalisations occur in elderly people with chronic cardiopulmonary disorders.
The burden of influenza on winter admissions is poorly reflected by hospital activity analysis – as shown by our recent study of rapid molecular diagnosis of paediatric admissions in Leicester. We found that very few children with influenza were diagnosed or coded correctly. 22 Moreover, analysis of hospital activity statistics for Leicester for winters 2002–3 and 2003–4 showed that only 2 of 5614 cardiopulmonary admissions among the elderly had a confirmed diagnosis of influenza. These local observations suggest that hospital activity data may grossly underestimate the true burden of influenza in hospitals, and the infrequency with which influenza is diagnosed may explain why hospital doctors consider conventional diagnostic virology for respiratory pathogens to be unhelpful.
Respiratory syncytial virus
Respiratory syncytial virus infection produces incomplete protection and reinfection is common. Like influenza, RSV infection in the elderly has no pathognomonic features, and cannot be distinguished from other respiratory virus infections clinically. Evidence indicates that RSV may be severe in the elderly, causing a spectrum of illness including pneumonia. 8,23,24 Outbreaks in residential care facilities causing severe morbidity and mortality are well documented. Pneumonia occurs in 5–55% of cases and mortality of up to 20% is described. 8,23 Because RSV has traditionally been considered a paediatric infection, evidence of the virus in community-dwelling elderly or admissions with cardiopulmonary disorders is usually not sought.
Community-acquired pneumonia and Streptococcus pneumoniae
Invasive pneumococcal disease and CAP exhibit a distinct winter seasonality that may be attributed to climatic conditions, crowding, air pollution, and respiratory virus activity, including influenza, RSV and rhinoviruses. 25–27 In Leicester, weekly admissions data for 1352 cases of CAP admitted during the winters of 2002–3 and 2003–4 showed two peaks of 5 and 6 weeks’ duration during 2002–3 and a 14-week peak during 2003–4, confirming the seasonal pattern and its possible association with respiratory virus activity. Bacterial pneumonia is a well-recognised complication of influenza, and pneumococcus was the most common microbe associated with life-threatening and fatal 2009 pandemic H1N1 infection. Between 9.5% and 48% of CAP may involve co-infection of typical and atypical organisms. 28 Of the 148 cases of S. pneumoniae infection identified by Porath et al. ,29 100 had co-pathogens identified, usually ‘atypicals’. Although the clinical importance of polymicrobial infection is uncertain, mixed infection may be associated with a more complicated course. 30 The overall mortality from CAP can be substantial – in one meta-analysis involving 33,148 patients with CAP, it was 12.3% for patients with S. pneumoniae, 9% for influenza, and 5% for RSV. 16 Treatment cannot await the results of conventional microbiological tests, so an empiric regimen is necessary, which in the UK typically includes a β-lactam antibiotic, with or without a macrolide. 17
It is possible that a positive point-of-care (POC) pneumococcal antigen test result could lead to the prescription of a single antimicrobial agent, placing patients with polymicrobial infection that includes S. pneumoniae at increased risk of death. Oosterheert et al. 31 undertook a systematic review to assess whether treatment with a β-lactam plus macrolide or quinolone monotherapy is truly superior to β-lactam treatment alone. Eight relevant studies were selected. In six, significant reductions in mortality were found; in one, a reduction in hospital length of stay was found; and in another no beneficial effects could be demonstrated for treatment regimens with fluoroquinolone monotherapy or combinations of β-lactams and macrolides. The studies supporting the recommended treatment regimen were designed as non-experimental cohort studies and confounding may have influenced the results. The authors concluded that a randomised controlled trial (RCT) is warranted to circumvent the methodological flaws in the designs of the available studies.
Diagnostic tests
Diagnostic tests for influenza
Viral isolation and haemagglutination inhibition antibody testing are standard methods for influenza diagnosis but have drawbacks. Virus isolation by culture from respiratory secretions may take a week or more – for example, a median of 8 days in one recent study;21 it requires specialised laboratory facilities, and the results cannot be provided soon enough to influence treatment decisions or infection control. Serology provides a retrospective diagnosis. 32 Neither test alone is considered a reference standard for influenza diagnosis, as each lacks sensitivity, but culture and serology have been used together as the reference standard in assessment of molecular tests. 33
Tests for rapid diagnosis of influenza A and B virus by immunofluorescence (IF) of exfoliated nasopharyngeal cells have shown variable sensitivity (40–100%) and specificity (86–99%);34 they require specialist equipment and expertise, and are labour intensive. Rapid, near-patient tests (NPTs) for influenza vary in complexity, sensitivity and specificity. 18 They can potentially aid clinical management, but their value in the hospital setting in influencing prescribing and infection control of adults is unclear. We used the Quidel® QuickVue Influenza A + B test (Quidel, San Diego, CA, USA) due to its apparent ease and speed of use, and reports of its sensitivity and specificity. 18 However, its diagnostic accuracy in the elderly is unclear. Molecular diagnosis of influenza by reverse-transcriptase [reverse transcriptase-polymerase chain reaction (RT-PCR)] provides improved sensitivity and specificity, allows accurate detection, and facilitates the subtyping of influenza. 35 Like virus culture, multiplex polymerase chain reaction (PCR) offers the potential to identify several pathogens (e.g. influenza subtypes A/H1N1, A/H3N2 and B; RSV types A and B) in one sample and in one reaction. 35,36 The technique is used routinely within the specialist diagnostic facilities of the Centre for Infections, Colindale, London, where it has a sensitivity of 92% and specificity of 84%. 33
Diagnostic tests for respiratory syncytial virus
Factors contributing to underestimations of the incidence and burden of RSV in the elderly include virus lability; the brief period of virus shedding and low titre of virus in nasal specimens during reinfection; the relative insensitivity of standard diagnostic tests – including the complement fixation test (CFT), virus culture (even when performed under rigorous conditions including bedside inoculation),37 and rapid antigen detection tests [IF and Directigen® enzyme immunoassay (Becton Dickinson, Franklin Lakes, NJ, USA)] in the elderly,37,38 and the frequent co-circulation of RSV with influenza. 22,24 Multiplex RT-PCR has emerged as a sensitive and specific method of detecting RSV infection. 36 Examination of nose and throat swabs by multiplex RT-PCR from 167 elderly subjects (age ≥ 65 years) who presented to their GP with ILI during the winters of 1995–6, 1996–7 and 1997–8 showed that 15% had RSV. 39 These investigators detected one RSV infection for every two influenza infections, suggesting that the previously unrecognised burden of RSV in the elderly may be substantial.
Diagnostic tests for Streptococcus pneumoniae
Diagnosis of pneumococcal pneumonia is complicated by the lack of a diagnostic reference standard that is highly sensitive and specific. Despite being the single most important pathogen causing CAP, S. pneumoniae is undoubtedly underdiagnosed owing to limitations of conventional tests. Limitations of Gram stain and culture of sputum include failure to obtain sputum for culture – a fraction of patients produce sputum;28 the overall diagnostic yield of sputum examination is very low (< 25%),40 and isolation of S. pneumoniae from sputum may represent colonisation. Blood cultures have been considered as a standard in patients with CAP,41 but positive cultures are found in < 10% of patients with CAP, particularly those with low-severity CAP or have started antibiotics already. 17,41,42 The test is often unhelpful, as positivity becomes evident no earlier than 24 hours after obtaining the specimen, and results typically have little influence on therapeutic decisions and outcomes. 41,43–45 However, a review of patients with confirmed pneumococcal pneumonia found that 42% of patients with positive blood culture results had their treatment changed as a result. 46 As the overall prevalence of β-lactam resistance remains low in the UK, rapid near-patient testing for pneumococcal infection could influence therapeutic decisions.
Measurement of pneumococcal antibodies has not proven reliable for diagnosing pneumococcal pneumonia. 47 PCR appears to be more sensitive than blood culture but most studies have tested only a small number of samples, and have not compared different sample types from the same patients. 48 Murdoch et al. 48 used a nested PCR to target the pneumolysin gene in multiple sample types from 474 adults with CAP. The authors concluded that the pneumolysin PCR adds little to existing diagnostic tests and that it was less sensitive than the rapid urine antigen test. Other investigators have evaluated different PCR methods on sputum – in general they have had good sensitivity but poor specificity, whereas PCRs that have been developed to evaluate blood have had poor sensitivity but good specificity. 49 The detection of S. pneumoniae antigens in the urine of patients with pneumonia has been extensively studied using a variety of techniques. Although the performance of most tests has been somewhat disappointing, the BinaxNOW® urinary antigen test (Binax, Portland, ME, USA) that we used is simple to perform; it can detect the C polysaccharide cell wall antigen common to all S. pneumoniae strains and it provides results within 15 minutes. It has sensitivity of 80% or more in adults and children when positive blood cultures are used as reference standard. 50–54
Rationale for the study
The three respiratory pathogens, influenza, RSV and S. pneumoniae, are responsible for considerable morbidity and mortality, and become increasingly important as pathogens with advancing age and comorbidity. The elderly population of the UK is rapidly increasing in life expectancy and size. People aged > 60 years now account for almost half of the annual number of all hospital admissions, placing huge pressure on the health-care system. RSV and influenza can exacerbate chronic cardiopulmonary disease in adults of working age, adding to the demand for hospital beds and pressure on health-care providers to discharge patients at the earliest opportunity, preferably before admission to a hospital ward.
Vaccines and drugs to prevent RSV transmission and illness are unlikely to be available for use within the next decade. Vaccines against influenza and pneumococcal infection provide incomplete protection. Many pathogens can cause CAP, and the appreciable risk of death from CAP demands that treatment is given empirically at the earliest opportunity. Early treatment with neuraminidase inhibitors (NIs) – within 48 hours of onset of illness – is considered essential.
Conventional diagnostic tests, especially viral culture, provide information too late to influence care and containment decisions. They are expensive and require specialised facilities and expertise. Blood cultures are often negative in patients with CAP caused by the pneumococcus; the result is also influenced by antecedent antimicrobial treatment. Treatment of CAP is usually empirical.
Compared with other diagnostic tests, the rapid point-of-care tests (POCTs) offer the greatest potential to influence antibiotic prescribing, ameliorate illness and prevent nosocomial transmission – but only if the tests are sufficiently sensitive and specific. PCR could provide comparable benefits if its longer turnaround time is compensated for by better test performance.
This study was designed to evaluate the ease and speed of use of the different tests, assess their costs, and identify whether they provide clinical and health-economic benefits, and rationalise the use of single-roomed accommodation.
The three diagnostic strategies assessed in this study were (1) POCTs for influenza A and B and pneumococcal infection; (2) RT-PCR tests for influenza A and B, and RSV A and B; and (3) conventional culture for these pathogens.
Chapter 2 Methods
Study design
We undertook a prospective RCT and economic evaluation of rapid POC, molecular and conventional diagnostic tests for influenza, RSV and S. pneumoniae in the management and outcome of acute cardiopulmonary admissions in the elderly (age ≥ 65 years) and ‘high-risk’ individuals with underlying chronic heart or lung disease, including asthma, who were 18–64-years of age at the time of presentation in two teaching hospitals in the UK. A summary of the protocol for the study is provided in Appendix 1 . A copy of the full protocol is available from the Principal Investigator, Karl Nicholson.
Setting
The participating hospitals were Glenfield General Hospital and Leicester Royal Infirmary in the University Hospitals of Leicester (UHL) NHS Trust, Leicester, a city in the English East Midlands. The UHL NHS Trust serves a population of approximately one million subjects of all ages. It is the only facility within the county of Leicestershire that provides inpatient emergency medical care to the population of Leicestershire. The laboratory tests were carried out in the Department of Microbiology, Leicester Royal Infirmary.
Participants
We recruited people presenting to medical admissions units, or any ward accepting acute medical admissions, with an acute exacerbation of chronic cardiopulmonary illness of ≤ 168 hours’ (7 days’) duration or an acute cardiopulmonary illness of ≤ 7 days’ duration [including pneumonia, ‘influenza’/ILI, exacerbations of chronic obstructive pulmonary disease (COPD), bronchitis, asthma, congestive heart failure or cardiac arrhythmia], who satisfied the study inclusion and exclusion criteria and could be recruited to the study within a 16-hour period of initial assessment by the patient’s medical team.
The inclusion and exclusion criteria for participants are shown in Box 1 .
-
Written informed consent, or written informed assent by a relative or carer.
-
Men or women aged ≥ 65 years, or 18–64 years, with underlying chronic heart or lung disease including asthma.
-
Acute exacerbation of chronic cardiopulmonary illness,* or acute cardiopulmonary illness or ILI of ≤ 168 hours’ duration, including pneumonia, influenza or ILI, exacerbations of COPD, bronchitis, asthma, congestive heart failure and cardiac arrhythmia.
-
Recruitment within 16 hours of initial medical assessment.
-
Able to comply with the study protocol.
-
Access to a telephone.
-
Angina or suspected myocardial infarction.
-
Previously recruited within 28 days of the current admission.
-
Enrolment in a trial of antimicrobial therapy.
-
Sore throat and/or hoarseness.
-
Nasal symptoms (stuffiness, and/or runny nose, and/or thick nasal discharge, or sneezing).
-
Cough (new or increased).
-
Sputum (new or increased).
-
Wheezing (new or increased).
-
Difficulty breathing/shortness of breath (new or increased).
-
Chest pain with breathing.
-
Feverishness/sweating.
-
Chills, shivers or rigors.
-
Tiredness or fatigue.
-
Decrease or loss of appetite.
-
Headache.
-
Muscle or body aches.
-
Generally feel unwell.
Recruitment
Medical and nursing staff on medical admissions units and wards providing acute medical care to patients with acute cardiopulmonary conditions identified eligible patients. Research nurses provided trial information and obtained signed informed consent from the patient or signed informed assent from a relative or carer.
Randomisation
Participants were then randomly allocated to one of three diagnostic study groups: (1) NPTs for pneumococcal infection and influenza; (2) rapid molecular tests for influenza and RSV; or (3) conventional laboratory diagnostic tests. Their investigations, medical care and discharge planning was provided as usual by the medical and nursing teams on the medical admissions units and other wards, not by the investigators. The randomisation process enabled the investigators to evaluate the role of the diagnostic tests on clinical outcomes. Ultimately, all diagnostic tests were performed on specimens from all subjects, providing the means to compare diagnostic accuracy.
The trial statistician generated randomisation codes, stratified by centre and using randomly permuted block sizes of 9, 12 or 15, which were not revealed to any person before randomisation. The randomisation code allocated participants in the ratio 1 : 1 : 1 to one of the three study groups. It was provided in sequentially numbered sealed study envelopes, which were stored securely (within a locked filing cabinet, within a locked office, within a locked department). The randomisation code for an individual patient became known to the research nurse only when signed informed consent or assent was obtained. The randomisation codes were then checked by the trial statistician against the master copy to ensure that the sequences concurred. It was not revealed to the participants or to the medical and nursing team providing care.
Planned interventions
Participants were randomised to receive:
-
diagnostic assessment using rapid near-patient diagnostic tests (Quidel for influenza, and BinaxNOW for the pneumococcal antigen), or
-
rapid molecular tests (for influenza A and B and RSV A and B), plus laboratory pneumococcal antigen testing, or
-
conventional laboratory diagnostic assessment, notably culture for influenza A and B, RSV A and B, and S. pneumoniae, and serology for influenza A and B.
Although all tests were eventually carried out on all participants, clinicians were provided with rapid test results relating only to their randomisation group.
Collection of samples for microbiological analyses
Identical samples were taken from each person but were processed differently depending on the randomisation ( Box 2 ).
-
Blood culture Undertaken by the admitting medical team or research nurse, and transported to/processed in the laboratory according to local protocols.
-
Paired (acute and convalescent) sera An ‘acute’ venous blood sample was collected and transported to the laboratory, where the serum was separated and stored. A convalescent sample was collected, where possible, up to 90 days after admission. Acute and convalescent sera from participants were batched and tested at the HPA Centre for Infections for antibodies to seasonal strains of influenza A and B.
-
Freshly expectorated sputum Sputum was collected from participants with a productive cough and cultured in the laboratory using standard operating procedures.
-
Freshly voided urine Collected and tested on the ward, by a research nurse, for pneumococcal soluble antigen using the near-patient BinaxNOW S. pneumoniae test, according to the manufacturer’s instructions (see Appendix 2 ).
-
Nasopharyngeal swabs A nasal swab sample was collected from the nostril that presented the most secretion (if any) under visual inspection. This specimen was analysed on the ward by a research nurse for the presence of influenza A and B antigen using the near-patient QuickVue Influenza A + B test (www.cliawaived.com/web/items/pdf/QDL-20183-Quidel_Influenza_Tests_Insert∼619file1.pdf). A nasopharyngeal specimen was collected for deferred molecular testing and conventional virus culture using the opposite nostril to that used previously. The protocol for collecting and transporting nasopharyngeal specimens is provided in Appendix 3 . The nasopharyngeal specimen was transported to laboratory (and stored at 4 °C if received after hours), where one aliquot was cultured for influenza A and B and RSV using standard operating procedures, and another was stored at –80 °C and analysed by deferred molecular diagnostic tests for influenza A and B and RSV A and B.
-
Blood culture Undertaken and processed as above.
-
Paired (acute and convalescent) sera Collected and processed as above.
-
Freshly expectorated sputum Collected from participants with a productive cough and processed as for the ‘near patient test group’.
-
Freshly voided urine Collected and transported to laboratory, where it was tested promptly for the presence of pneumococcal soluble antigen using the near-patient BinaxNOW S. pneumoniae test.
-
Nasopharyngeal swabs Nasopharyngeal swabs were collected as for the ‘near-patient test group’. They were transported to the laboratory and stored at 4 °C if they were received after hours. The nasal swab sample was stored at –20 °C for deferred testing for the presence of influenza A and B antigen using the near-patient QuickVue Influenza A + B test. An aliquot of the nasopharyngeal specimen was analysed promptly by molecular diagnostic tests for influenza A and B and RSV A and B. Another aliquot was cultured for influenza A and B and RSV using standard operating procedures.
-
Blood culture Undertaken and processed as for ‘near-patient test group’.
-
Paired (acute and convalescent) sera Collected and processed as for ‘near-patient test group’.
-
Freshly expectorated sputum Collected from participants with a productive cough and processed as for the ‘near-patient test group’.
-
Freshly voided urine Collected and transported to laboratory where it was stored at –20 °C for deferred testing for the presence of pneumococcal soluble antigen using the near-patient BinaxNOW S. pneumoniae test.
-
Nasopharyngeal swabs Nasopharyngeal swabs were collected as ‘near-patient test group’ and transported to the laboratory as above. One aliquot was cultured for influenza A and B and RSV using standard operating procedures, and another was stored and analysed by deferred molecular diagnostic tests for influenza A and B and RSV A and B. The nasal swab sample was stored at –20 °C for deferred testing for the presence of influenza A and B antigen using the near-patient QuickVue Influenza A + B test.
Participants in each group gave a venous blood sample on entry to the study for blood culture if it had not Partcipants in each group gave a venous blood sample on entry to the study for blood culture if it had not been collected already by the medical team. Blood cultures were processed in the laboratory according to local protocols. Serum from an ‘acute’ blood sample was collected from participants in each group on entry to the study and stored for titration of antibodies against influenza A and B in paired ‘acute’ and ‘convalescent’ sera. It was originally planned that the ‘convalescent’ sample would be collected 10 days after admission. Because many people were discharged within 10 days of admission, we amended the protocol to collect ‘convalescent’ sera up to 30 days, and subsequently up to 90 days, after admission. Acute and convalescent sera were processed in the Influenza Laboratory at the Health Protection Agency (HPA) Centre for Infections, Colindale, London.
On entry to the study we collected freshly expectorated sputum for Gram stain and culture from participants in each study group with a productive cough. Freshly voided urine was collected from all participants on entry to the study and processed in the BinaxNOW S. pneumoniae test, as shown in Box 2 , and according to the manufacturer’s instructions (see Appendix 2 ). One nasal swab (collected from one nostril only), one nasal swab (collected from the opposite nostril) and one throat swab were collected on entry to the study (see Appendix 3 ), and were processed as shown in Box 2 .
EuroQol quality-of-life assessment (European Quality of Life-5 Dimensions)
We assessed patients using the European quality of life (EuroQol) assessment at baseline on admission, and 7 and 28 days later. The EuroQoL European Quality of Life-5 Dimensions tool (EQ-5D)55 has been used in many cost-effectiveness studies56,57 and is recommended for use in the economic evaluation of health-care technologies within the UK in guidance issued by the National Institute for Health and Care Excellence (NICE). 58 It defines health in five dimensions: morbidity, self-care, usual activities, pain or discomfort, anxiety or depression. Each dimension has three levels: no problems, a moderate problem, or a severe problem. Health states defined by the level chosen for each dimension can be scored using utility weights reflecting the values from a representative sample of the UK population. 59 These utilities are scaled so that full health = 1 and death = 0, and they allow for severe health states for which health-related quality of life (HRQoL) is valued lower than death. The EQ-5D was self-completed by participants or was done by proxy if the participant was incapable of completing the self-administered questionnaire or providing a verbal response. Where patients were discharged, the EQ-5D was assessed by telephone interview of the patient, by postal questionnaire or by proxy.
Record-keeping
The mainstay of the record-keeping for this study was the case report form (CRF) (see Appendix 5 ). Individual CRFs were stored in locked filing cabinets in a secure University of Leicester research laboratory within Leicester Royal Infirmary.
The CRFs captured basic demographic data (including details of residential status, smoking habits, alcohol consumption, household contacts, influenza and pneumococcus immunisation status during the previous 3 years, and hospital admissions during the period 1 September to 30 April of the previous winter), the date and time of admission, the randomisation group, GP details, symptoms of the presenting illness, examination findings on admission, past medical history, and provisional diagnosis information on recruitment. The CRFs also recorded information on prescribed medication, oxygen and intravenous (i.v.) fluids, investigations, isolation status, complications, transfer to the intensive care unit, duration of stay, deaths, quality of life (QoL), and the timing of specimen collection and test results. Information was collected from the participant by research staff on recruitment and during follow-up visits, from medical case notes, computerised hospital records, and by post, and was entered into the CRF. Clinical and QoL data, together with laboratory findings, were entered into a database written in Microsoft Access 2000 (Microsoft Corporation, Redmond, WA, USA), which was maintained on University of Leicester mainframe computers, behind electronic firewalls allowing limited access with passwords. This proved a secure and confidential way of maintaining the records.
Near-patient tests
BinaxNOW Streptococcus pneumoniae test
Urine was tested for S. pneumoniae antigens using the BinaxNOW S. pneumoniae urinary antigen tests according to the manufacturer’s instructions. The principles of the test and the test procedure are described in Appendix 2 . Freshly voided urine samples were collected from participants in each study group, as described in Box 2 . Briefly, the test swab provided by the manufacturer was dipped into the urine specimen at room temperature, so that the specimen completely covered the swab head. The swab was removed from the urine and then placed into the bottom hole (the swab well) of the test device, and pushed upwards so that the swab tip was visible in the top hole. Three drops of the reagent was added to the bottom hole and the adhesive cover was immediately removed from the right edge of the test device, which was then closed and sealed. A positive-control swab (containing heat-inactivated S. pneumoniae) and an S. pneumoniae negative-control swab were also provided. The results were read 15 minutes later. A positive test result was indicated by the appearance of a pink-to-purple coloured line for both the specimen and positive control. A negative test result was indicated by a colour reaction to only the positive control.
Quidel QuickVue Influenza A + B test
Nasopharyngeal specimens collected as described in Box 2 were tested for influenza type A and type B antigens using the Quidel QuickVue Influenza A + B test, according to the manufacturer’s instructions. The principles of the test and the test procedure are described at www.cliawaived.com/web/items/pdf/QDL-20183-Quidel_Influenza_Tests_Insert∼619file1.pdf.
Molecular diagnostic tests
We used molecular diagnostic tests that were developed at the Centre for Infections, HPA (London, UK), and the HPA Laboratory, Cambridge.
Ribonucleic acid extraction from clinical and control samples
Swab samples in virus transport medium (VTM) were vortexed vigorously for 1 minute to dislodge material attached to the swab. Each sample was mixed with 2 ml of amphotericin B (Fungizone®, Gibco®, New York, NY, USA) (2.5 µg/ml concentration) then vortexed to mix before removal of an aliquot for nucleic acid extraction. Viral ribonucleic acid (RNA) was extracted from nasopharyngeal specimens that were collected, transported and stored, as described in Box 2 . During the first study season, viral RNA was manually extracted from 150-µl aliquots of nasopharyngeal specimen using the guanidium isothiocyanate method described by Boom et al. ,60 with a final elution volume of 30 µl. Although this method provided high-quality RNA for PCR, subsequent work was performed using an automated nucleic acid extraction platform (X-tractorGene, Corbett Robotics, Australia) with a guanidine isothiocyanate reagent pack (Sigma-Aldrich, St Louis, MO, USA) according to the manufacturer’s specifications. Briefly, manual cell lysis was performed by adding 100 µl of sample digest buffer and 360 µl of lysis buffer to a 180-µl nasopharyngeal sample aliquot. The entire 640-µl volume was then transferred into a lysis block, which was placed in X-tractorGene for automated processing, providing a final elution volume of 50 µl.
Positive-control viruses and two negative controls were included with each extraction. The control viruses were influenza strains A/Taiwan/1/86 (H1N1), A/Moscow/10/99 (H3N2) and B/Panama/45/90; RSV strains Long (RSV A) and N2 (RSV B); and human metapneumovirus (hMPV). The no-template controls were VTM and deoxyribonuclease (DNAse)/ribonuclease (RNAse)-free water. The controls were made as quality-controlled batches, stored at –80 °C and then undergoing the same lysis procedure and nucleic acid extraction described above.
Reverse transcription
Synthesis of complementary deoxyribonucleic acid (cDNA) from extracted RNA was primed using random hexamers and Moloney murine leukaemia virus (MMLV) reverse transcriptase (RT) as described by Stockton et al. ,36 prior to amplification by either the multiplex semi-nested conventional PCR or real-time PCR methods. In a 40-µl reaction, 22.2 µl of RNA was added to a RT mix containing 20 mM Tris-HCl, 50 mM KCl, 7.5 mM MgCl2, 100 mM of each deoxynucleotide triphosphate (dNTP), 5.3 nM of a random hexamer (Promega, Madison, WI, USA), 1.6 U of ribonuclease inhibitor (RNasin) (Promega) and 200 U of MMLV transcriptase (Invitrogen, Carlsbad, CA, USA). The mixture was incubated at room temperature for 10 minutes, at 37 °C for at least 45 minutes, at 95 °C for 5 minutes and then quenched on ice for at least 1 minute.
Polymerase chain reaction tests
Different PCR assays were used to detect influenza A and B and RSV A and B during the 3-year study that took into account ongoing technological advances and availability. The method applied during the first year of the study used conventional PCR. Subsequently, real-time PCR methods were applied to facilitate rapid, high throughput of specimens.
Semi-nested multiplex conventional PCR During the first year of the study, detection and subtyping of influenza A H1 and H3, influenza B, and RSV A and RSV B were carried out by conventional, semi-nested PCR targeting the haemagglutinin (HA) region of the influenza genome, and the N and P regions of RSV in a multiplex reaction as described by Stockton et al. 36 For primary PCR, 20 µl of cDNA was added to the master mix to make up a final reaction volume of 100 µl. For secondary PCR, 2 µl of the primary PCR product was added to 48 µl of reaction master mix for a final volume of 50 µl. Secondary PCR amplicons were visualised using ethidium bromide following gel electrophoresis on 1.2% agarose (Roche, Mannheim, Germany). Band patterns were compared against positive-control viruses for influenza and RSV. Negative controls included DNAse/RNAse-free water and VTM.
HPA Cambridge one-step quadriplex real-time RT-PCR During the second year of the study, we used a one-step quadriplex PCR to detect influenza A and B according to the National Standard Method virology standard operating procedure (VSOP) 25. 61 This assay amplified 5 µl of extracted RNA template in a 25-µl reaction volume with generic primers targeting the conserved region of the matrix gene for influenza A/B detecting all known influenza subtypes (H1–H15) and influenza B, and includes a bacteriophage MS2 internal control. The specificity of this assay was evaluated against a panel of influenza A (subtypes H1–H15), influenza B strains and a respiratory panel of pathogens to ensure specificity of the assay for influenza type A, influenza A subtype H5 and influenza type B. All samples were analysed in duplicate, and both results had to be concordant for a definitive positive or negative diagnosis. Data analysis was performed using the Rotor Gene software versions 6.0.41 and 6.1.71 (Corbett Research, Australia). This method and all other real-time PCR methods in the study used a Rotor-Gene real-time PCR machine (Corbett Research, Australia), and master mix and enzyme kits purchased from Invitrogen (Carlsbad, CA, USA). Forward and reverse primers were obtained from Eurofins (London, UK); probes tagged with minor groove binders (MGBs) were synthesised by Applied Biosystems (ABI, Warrington, UK) and probes labelled with Cy5 and Rox were purchased from Metabion (Martinsried, Germany).
HPA Colindale, one-step influenza multiplex real-time RT-PCR During the third year of the study, we detected H1 and H3 subtypes of influenza A and influenza B using a one-step multiplex PCR according to the National Standard Method VSOP 50 as described by Stephenson et al. 62 but with minor modifications. This assay amplified 7.5 µl of extracted RNA template in a 25-µl reaction volume, with primers and probes targeting conserved regions of the HA gene of H1, H3 and B viruses, and includes a soil-borne cereal mosaic virus (SBCMV) internal control. All samples were analysed in duplicate, and both results had to be concordant for a definitive positive or negative diagnosis. Primers and probes tagged with black hole quencher (BHQ) were obtained from Eurofins (London, UK): other probes tagged with MGB were purchased from ABI. Modifications included substitution of the fluorophores VIC to JOE and NED to Cy5. Data analysis was performed using the Rotor-Gene software version 6.1.71.
HPA Colindale, One-step real-time RT-PCR for RSVA and RSVB and hMPV, and two-step real-time, multiplex PCR for RSVA, RSVB, and hMPV We used one-step real-time RT-PCR for RSVA, RSVB, and hMPV during the second year of the study, and a two-step RT-PCR for these pathogens, according to the HPA SOP V-5381/01–06. The same primers and probes were used in both the one-step and two-step assays and targeted conserved regions of the respective nucleocapsid genes in RSV A, RSV B and hMPV63 and both assays included an SBCMV internal control. In the one-step method, 7.5 µl of extracted RNA template was used in a 25 µl total reaction volume. In the two-step assay, 22.2 µl of extracted RNA was added to 17.8 µl RT mix and then 2.5 µl of the cDNA product was added to 22.5 µl reaction mix for a total volume of 25 µl. All samples were analysed in duplicate with results validated as positive or negative where both replicates were concordant. Probes tagged with BHQs and primers were purchased from Eurofins (London, UK).
Conventional diagnostic tests
Virus culture
Nasopharyngeal samples were processed in the laboratory according to local protocols. Two cell lines were used for viral culture: primary liver cells (PLC/PRF5) (a continuous primary liver carcinoma line)64 and Medical Research Council 5 cells (MRC-5) from human fetal lung. 65
Approximately 0.25 ml of the nasopharyngeal specimen in VTM was inoculated into monolayered PLC/PRF5 and MRC-5 cells in culture tubes and incubated, stationary, at 33 °C overnight. Tubes negative for cytopathic effect (CPE) after 24 hours were reincubated, with rolling at two to three revolutions per minute, at 33 °C in fresh maintenance medium.
MRC-5 cell culture tubes were observed until 28 days after inoculation. Those that were CPE positive were confirmed and subtyped by IF using group-specific antibodies, whereas those that were CPE negative were reincubated and observed up to 28 days after initial inoculation. If contamination occurred during this period, the original samples were filtered and reinoculated into fresh culture tubes. If the cell monolayer sheet appeared to be of poor quality then cells were scraped off the tubes, transferred into a fresh culture tube and reincubated and analysed for up to another 28 days.
PLC/PRF5 cell culture tubes were observed for CPE twice weekly and haemadsorption (HAd) was concurrently performed using human red blood cells type O. CPE- and/or HAd-positive tubes were confirmed and subtyped by IF, whereas those that were negative were reincubated and observed for up to 14 days post initial inoculation. As with MRC-5 tubes, contaminated tubes were filtered and reincubated, whereas those with poor cell sheets were scraped off and transferred into fresh culture tubes for analysis up to another 14 days.
Haemagglutination inhibition
Antibody responses were titrated by the Respiratory Virus Unit, Centre for Infections, HPA (London, UK), by haemagglutination inhibition (HAI) assay using established protocols. Sera were tested at an initial dilution of 1/8 and were given serial two-times dilutions to establish end point titres. Acute and convalescent sera were tested in parallel, in duplicate, under masked conditions using A/New Caledonia/20/99 (H1N1), A/California/7/2004 (H3N2), and B/Malaysia/2506/04 (a descendant of the B/Victoria/2/87 lineage) as test antigens during the 2005–6 season; A/New Caledonia/20/99 (H1N1), A/Wisconsin/67/2005 (H3N2), B/Malaysia/2506/04 during the 2006–7 season; and A/Solomon Islands/03/2006 (H1N1), A/Brisbane/10/2007 (H3N2), and B/Florida/4/2006 (a descendant of the B/Yamagata/16/88 lineage) during the 2007–8 season. A fourfold or greater rise in antibody titre was considered a significant rise.
Blood cultures
Blood cultures bottles were incubated using the BacT/ALERT 3D automated blood culture system (bioMérieux, Durham, NC, USA) according to local protocols. Bottles flagging positive growth were removed and a drop of the blood culture broth was Gram stained and examined under the microscope. Blood culture broth with Gram-positive cocci in chains were inoculated onto blood agar plates and incubated aerobically and anaerobically at 37 °C. Primary antibiotic susceptibility for streptococci to include optochin were also set up on blood agar and Iso-sensitest agar (Oxoid, Basingstoke, UK), and incubated aerobically at 37 °C. The presence of S. pneumoniae was confirmed by demonstrating streptococci in the Gram stain of colonies, negative catalase test and susceptibility to optochin.
Gram stain and microscopy of sputum
Sputum was examined by Gram stain and microscopy according to local protocols. Briefly, sputum smears on slides were flame heated to fix, and then stained in the following order: Crystal violet, Gram’s iodine, ethanol and carbol fuchsin. Microscopic analysis was performed using immersion oil and bright-field microscopy. Samples positive for Gram-positive cocci, Gram-positive bacilli, Gram-negative cocci and Gram-negative bacilli were rated, subjectively, using a ‘+’ system, whereby the lowest copy number receives one + and confluent samples are given +++. The presence of epithelial cells and white blood cells was also observed and reported.
Sputum culture
Sputum was cultured on blood and chocolate agar according to local protocols. Samples were diluted with equal volume of sterile saline, homogenised and inoculated onto chocolate agar with bacitracin, and blood agar with optochin. Plates were incubated aerobically in CO2 at 37 °C, were read at 24 hours and 48 hours. S. pneumoniae were identified by Gram stain showing streptococci, negative catalase test and susceptibility to optochin.
Outcome measures
Clinical
Impact of test result on prescribing:
-
Time, from admission to first administration of ‘narrow-spectrum’ antibiotics.
-
Time, from admission to first administration of oral antibiotics.
-
Time, from admission to prescription of ‘no antibiotics’ (oral or i.v.) administered to patients with influenza or RSV.
-
Proportion of patients in each group who are prescribed NIs.
Impact of test result on duration of hospitalisation:
-
For all patients in each diagnostic group.
-
For patients in each diagnostic group with:
-
Influenza.
-
RSV.
-
S. pneumoniae infection.
-
Fever duration during first 10 days of hospitalisation
-
Time, from admission until patients became apyrexial, for:
-
All patients in each group.
-
Patients with S. pneumoniae infection.
-
Supplemental oxygen dependence and continuous positive airway pressure dependence during first 10 days of hospitalisation
Admissions to intensive care and ventilator support during first 10 days of hospitalisation
Deaths within 28 days of hospitalisation
-
For all patients in each diagnostic group.
-
Overall, for patients with:
-
Influenza.
-
RSV.
-
S. pneumoniae infection.
-
Quality of life:
-
For all patients in each diagnostic group.
-
For patients in each diagnostic group with influenza, RSV and S. pneumoniae infection.
Use of isolation facilities by patients with influenza and RSV, and inappropriate use by those with S. pneumoniae infection
Financial
Costs of the diagnostic tests
Care costs
Cost savings arising from the use of diagnostic tests
Total NHS costs
Incremental cost per case detected and cost per QALY
Laboratory
Diagnostic accuracy: sensitivity, specificity, and positive and negative predictive values
Ease of use of diagnostic tests
Speed of use of diagnostic tests
Cost per case detected
Antimicrobial spectrum of activity
Broad-spectrum antibiotics
The following antibiotics, or classes of antibiotics, for the purposes of this study, were considered to be broad-spectrum antibiotics, i.e. having activity against Gram-positive and Gram-negative pathogens, with or without anaerobic organisms:
-
Cephalosporins.
-
Co-amoxiclav (Augmentin®, GSK).
-
Piperacillin with tazobactam (Tazocin®, Pfizer).
-
Carbapenems [imepenem with cilastatin (Primaxin®, MSD), meropenem (Meronem®, AstraZeneca), ertapenem (Invanz®, MSD)].
-
Quinolones [ciprofloxacin (Ciproxin®, Bayer), levofloxacin (Tavanic®, Sanofi-aventis), moxifloxacin (Avelox®, Bayer), etc.].
-
Tetracyclines [doxycycline (non-proprietary), oxytetracycline (non-proprietary), minocycline (non-proprietary)].
-
Cotrimoxazole (non-proprietary).
-
Clarithromycin and azithromycin (non-proprietary).
-
Clindamycin (non-proprietary).
Narrow-spectrum antibiotics
The following antibiotics, or classes of antibiotics, are considered to be narrow-spectrum antibiotics, i.e. predominantly having activity against either Gram-positive or Gram-negative organisms, or anaerobic organisms:
Gram-positive antibiotics
-
Benzylpenicillin (and penicillin V).
-
Flucloxacillin.
-
Amoxicillin (and ampicillin).
-
Erythromycin.
-
Vancomycin.
-
Rifampicin.
-
Fusidic acid.
-
Linezolid.
-
Daptomycin.
Gram-negative antibiotics
-
Gentamicin and other aminoglycosides.
-
Aztreonam.
-
Trimethoprim.
-
Nitrofurantoin.
Anaerobic antibiotics
-
Metronidazole.
Where multiple agents are used together, they are classified as broad spectrum if either is a broad-spectrum agent, or if together they cover Gram-negative and Gram-positive organisms, with or without anaerobes.
Sample size
Statistical power of the Three Winters Study (3WS) was estimated for one laboratory end point (sensitivity/specificity), the primary clinical end point (i.e. length of stay), and one secondary end point (i.e. appropriate isolation levels).
The initial sample size calculation proposed that if the average sensitivity/specificity of the tests is assumed to be 80% and a 20% dropout rate was assumed then 2752 patients in total would enable the sensitivity/specificity to be estimated to within two standard errors (SEs), i.e. 7.6% if the disease prevalence was 5% and 5.4% if the prevalence was 10%.
In terms of clinical end points, 2752 patients in total would also enable a minimum clinically significant difference (MCSD), between diagnostic policies, of 1 day in the mean length of stay [assuming standard deviation (SD) = 6 days] to be detected at the 5% significance level with over 80% power, assuming a 20% dropout rate and adjusting for the fact that there are three groups. In total 2752 patients would also enable a MCSD, between diagnostic policies, of an improvement in appropriate use of isolation facilities from 5% to 15% to be detected at the 1% significance level with over 95% power, assuming a 20% dropout rate and adjusting for the fact that there are three groups.
Clearly, the initial sample size calculation was driven by the desire to estimate the sensitivity/specificity with a sufficient level of precision but that this depended crucially upon the disease prevalence. Consequently, the sample size calculation was revisited in 2007 after the first two winters’ data were available. During 2005–6, the overall disease prevalence was 18.7%, and during 2006–7 it was 10.3%, giving a combined disease prevalence across the first two winters of 13.2%. Consequently, if the average sensitivity/specificity was 80% and the dropout rate continued to be 20% then 1200 patients in total would enable the sensitivity/specificity to be estimated to within, i.e. two SE, 7.8% assuming a prevalence of 13%. Hence, the required sample size was revised to 1200 patients in total, as this still enabled both a MCSD of 1 day in mean length of stay (SD = 6 days) to be detected at the 5% significance level with over 80% power, and a MCSD of an improvement in appropriate use of isolation facilities from 5% to 15% to be detected at the 1% significance level with over 95% power, both assuming a 20% dropout rate and allowing for the fact that there are three groups.
Statistical methods
Statistical analyses were carried out using Stata version 11 (StataCorp LP, College Station, TX, USA). The study was analysed as an intention-to-treat (ITT) study; effectively all patients were analysed according to the group to which they were randomly allocated, regardless of whether they were in fact managed according to the results of their designated diagnosis method.
For continuous, non-time-to-event, outcomes the three intervention groups were compared using either parametric [analysis of variance (ANOVA)] or non-parametric (Kruskal–Wallis) test as appropriate, and summary statistics were reported as means (SD) and medians [interquartile range (IQR)], respectively. For categorical outcomes, the three interventions groups were compared using either Pearson’s chi-squared test or Fisher’s exact test, as appropriate. For time-to-event outcomes, these were reported as median (IQR) for each of three intervention groups, and compared formally using Cox proportional hazards regression models to allow for censoring/death and reported as hazard ratios (HRs) [and associated 95% confidence intervals (CIs)]. For the primary outcome, length of stay, the three intervention groups were also compared graphically using cumulative probability plots, i.e. one minus the Kaplan–Meier survivor function. EQ-5D data – collected at baseline, 7 days and 28 days – was initially summarised at each of the three time points and compared using parametric/non-parametric methods as for other continuous outcomes. Further analysis using linear mixed-effect regression models to allow for within-patient correlation enabled an assessment of the change in EQ-5D over time to be made and whether there was evidence of an intervention group interaction with time.
A number of patients were randomised more than once owing to the nature of their age and clinical condition. The primary analyses used their first admission/randomisation. However, to assess the impact that this assumption had on the results, two sensitivity analyses were undertaken – the first using all admissions but assuming that they were independent, i.e. ignoring the fact that some patients appeared more than once, and the second, perhaps more appropriately, including a random effect in the analyses to account for the inherent correlation induced by patients being rerandomised.
All statistical tests were reported at the 5% significance level, and 95% CIs or credible intervals (CrIs) were reported throughout as the sample size calculations explicitly made allowance for the fact that there were three rather two groups and inflated the sample size accordingly to maintain an overall 5% significance level.
Ethical arrangements
This study was sponsored by the University Hospitals of Leicester NHS Trust. It was approved by Leicestershire, Northamptonshire and Rutland Research Ethics Committee on 20 July 2005, reference no. 05/Q2502/76.
Revisions to the protocol
We requested protocol amendments as outlined below. All requests were approved by the Local Research Ethics Committee. Protocol amendments that were implemented are outlined below:
-
On 13 January 2006 we requested the following amendments to the protocol in response to poor recruitment:
-
Approve a one-page ‘Synopsis’ of the six-page Patient Information Sheet to be used by patients who are not known to be suffering from dementia but were too unwell to read the full Patient Information Sheet.
-
Allow recruitment of subjects with an illness of up to 7 days’ duration (i.e. ≤ 168 hours) rather than five days’ duration (i.e. ≤ 120 hours).
-
Change from 6 hours to 8 hours the interval between initial assessment by the admitting medical team and recruitment to the trial.
-
Allow trained medical students to recruit patients to the study during out-of-hours periods.
-
-
On 14 March 2006, we requested the following protocol amendments.
-
Change the timing of the convalescent blood sample from day 10 to ‘between day 10 and day 30’.
-
Approve recruitment of all 18- to 64-year-old subjects with pneumonia and ILI, rather than just those 18- to 64-year-old subjects with pneumonia and ILI who have underlying chronic heart and lung disease, including asthma.
-
Recruit patients until the end of June, rather than the period ‘September to April’.
-
Approve a 20-ml urine collection and amendments to protocol where the volume was stated differently.
-
Change the labelling of the packs to correspond exactly with the trial identification numbers. For patients recruited in the Royal Infirmary, the packs were changed from RI-HTA-0001, RI-HTA-0002, etc., to 1–0001, 1–0002, etc. For patients recruited at Glenfield Hospital, the packs were changed from GH-HTA-0001, GH-HTA-0002, etc., to 2–0001, 2–0002, etc.
-
-
On 21 December 2006 we requested an amendment to aid recruitment:
-
Change, from 8 hours (originally 6 hours) to 16 hours the interval between initial assessment by the admitting medical team and recruitment to the trial.
-
-
On 20 March 2007 we requested an amendment to aid collection of convalescent sera:
-
Extend from day 10 to day 90 (i.e. 90 days after admission) the period when we could collect convalescent blood.
-
-
On 5 November 2007 we requested an amendment to aid recruitment of incapacitated adults:
-
Allow personal and professional representatives to provide consent for incapacitated adults to take part in the study.
-
Chapter 3 Study population
Recruitment and compliance
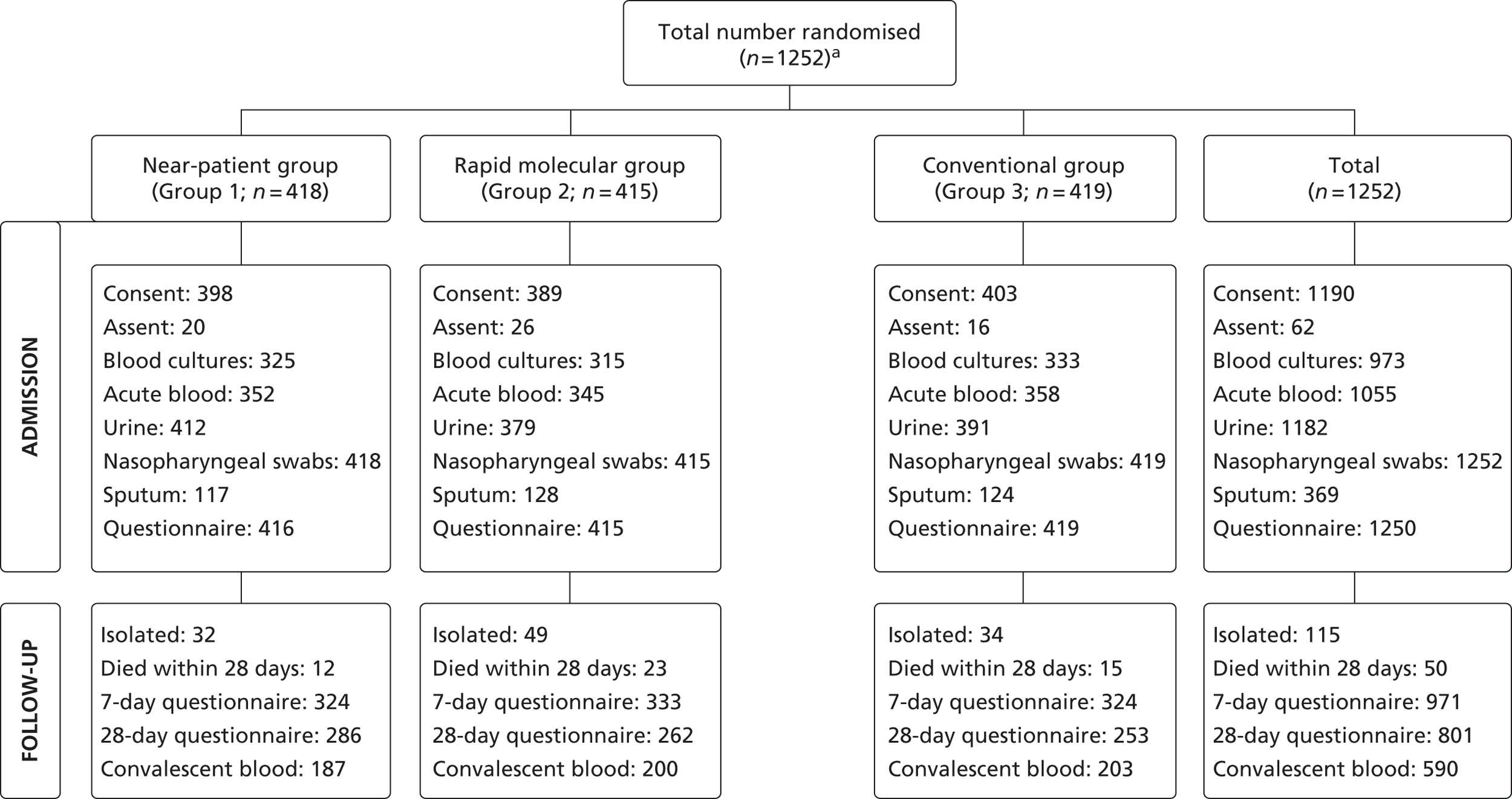
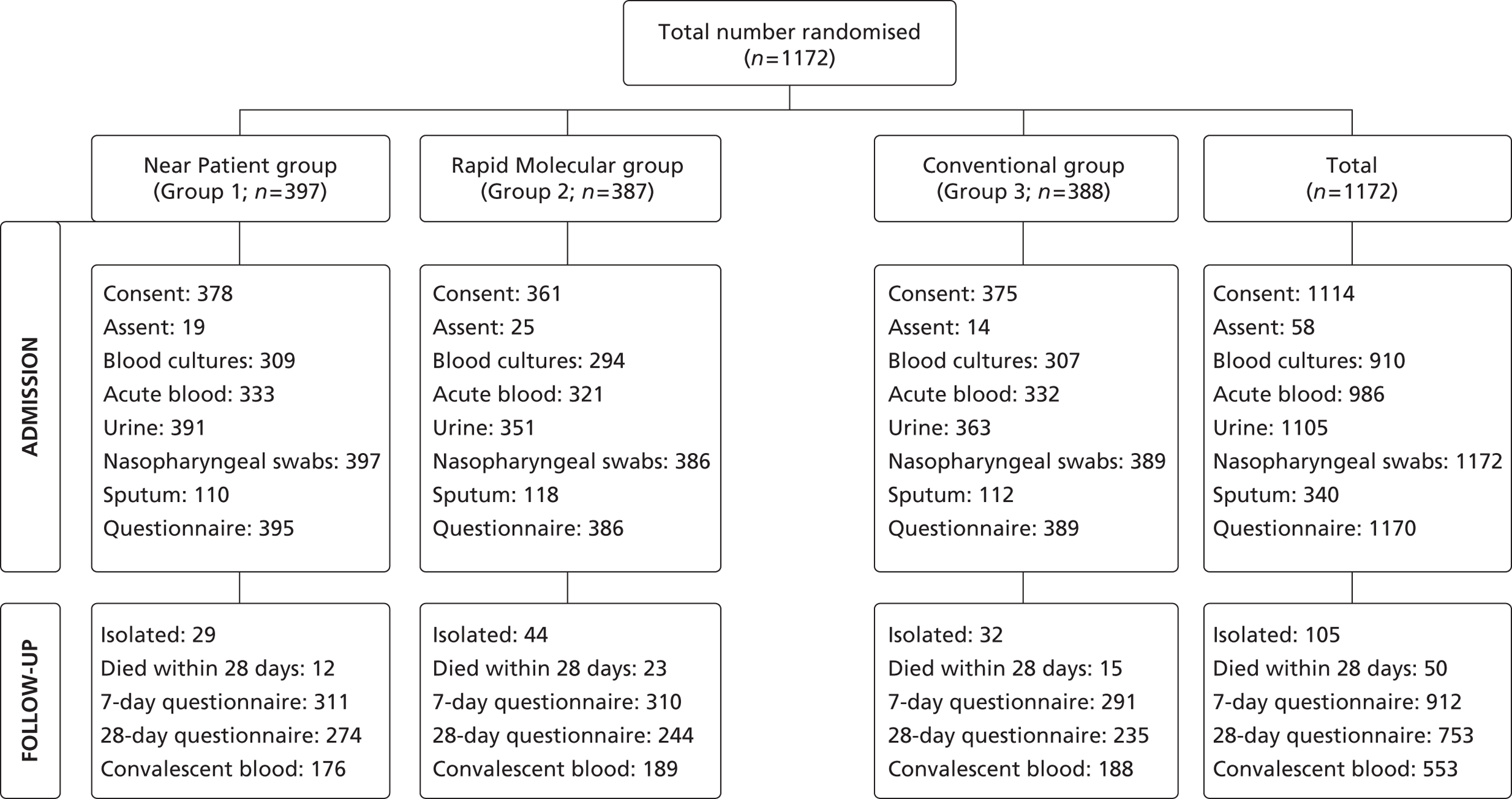
Between 14 December 2005 and 23 May 2008, 1253 admissions were enrolled and randomised to the rapid near-patient test group (n = 418), the molecular diagnostic group (n = 415) and the conventional diagnosis group (n = 420). One individual withdrew from the study. Altogether a total of 1252 admissions were randomised as shown in Figure 4 , and participated in the study.
FIGURE 4.
Randomisation of admissions, according to diagnostic group. a, The figure 1252 refers to admissions or patient admissions (and not unique patients) – there were 1172 patients of whom 67 were admitted/randomised at least twice.
Demography
The data set comprised data relating to 1252 separate hospital inpatient episodes. There were 1172 unique patients in the study, with 67 individuals having up to four separate inpatients episodes each. The main analyses and results use the first admission for the 1172 patients (see Chapter 2 for other sensitivity analysis details). The number of admissions per patient is shown in Table 1 , together with the frequencies of inpatient episodes by patient numbers. The 1172 first admissions were randomised as shown in Figure 5 .
| Admission | No. of patients (% out of 1252 admissionsa) |
|---|---|
| First | 1172 (93.6) |
| Second | 67 (5.4) |
| Third | 10 (0.8) |
| Fourth | 3 (0.2) |
FIGURE 5.
Randomisation of first cases, according to diagnostic group.
The demographic variables are described for first admissions only (1172 patients) and are set out in Table 2 , as well as other patient background information deemed of interest. The baseline demographics are intended to describe the characteristics of the patients participating in the trial and to assess whether the randomisation process had been applied successfully.
| Covariate | Near patient (n = 397) | Rapid molecular (n = 387) | Traditional (n = 388) | p-valuea | Missing, n b |
|---|---|---|---|---|---|
| Gender, n (%) | 0.415 | 0 | |||
| Male | 203 (51.1) | 197 (50.9) | 182 (46.9) | ||
| Female | 194 (48.9) | 190 (49.1) | 206 (53.1) | ||
| Age (years), n (%) | 0.971 | 0 | |||
| 18–29 | 23 (5.8) | 21 (5.4) | 23 (5.9) | ||
| 30–49 | 69 (17.4) | 72 (18.6) | 76 (19.6) | ||
| 50–64 | 96 (24.2) | 83 (21.5) | 81 (20.9) | ||
| 65–74 | 89 (22.4) | 84 (21.7) | 83 (21.4) | ||
| ≥ 75 | 120 (30.3) | 127 (32.8) | 125 (32.2) | ||
| BMI (kg/m2) (mean, SD) | |||||
| 26.6 (6.96) | 26.6 (7.49) | 25.5 (6.22) | 0.091 | 231b | |
| Ethnicity, n (%) | 0.940 | 0 | |||
| White | 361 (90.9) | 347 (89.7) | 354 (91.2) | ||
| Indian | 25 (6.3) | 29 (7.5) | 25 (6.4) | ||
| Other | 11 (2.8) | 11 (2.8) | 9 (2.3) | ||
| Smoking status, n (%) | 0.123 | 0 | |||
| Current | 93 (23.4) | 90 (23.3) | 118 (30.4) | ||
| Previous | 184 (46.4) | 187 (48.3) | 169 (43.6) | ||
| Never | 120 (30.2) | 110 (28.4) | 101 (26.0) | ||
| Flu vaccination this season, n (%) | 0.831 | 0 | |||
| Yes | 235 (59.2) | 221 (57.1) | 224 (57.7) | ||
| No | 162 (40.8) | 166 (42.9) | 164 (42.3) | ||
| Flu vaccination last season, n (%) | 0.435 | 0 | |||
| Yes | 240 (60.5) | 218 (56.3) | 233 (60.1) | ||
| No | 157 (39.6) | 169 (43.7) | 155 (40.0) | ||
| Flu vaccination two seasons ago, n (%) | 0.856 | 0 | |||
| Yes | 220 (55.4) | 209 (54.0) | 217 (55.9) | ||
| No | 177 (44.6) | 178 (46.0) | 171 (44.1) | ||
| Flu vaccination ever, n (%) | 0.866 | 0 | |||
| Yes | 297 (74.8) | 283 (73.1) | 287 (74.0) | ||
| No (never) | 100 (25.2) | 104 (26.9) | 101 (26.0) | ||
| Hospital admissions in previous year IRR (95% CI; p-value) | |||||
| 1 | 1.017 (0.744 to 1.391; 0.915) | 1.037 (0.762 to 1.412; 0.816) | 132b | ||
From Table 2 , the only demographic variable that appears to show weak statistically significant evidence of unbalanced distribution across the three trial arms is body mass index (BMI), with a p-value of 0.091. However, the mean values of BMI across the three groups are very similar, and it is unlikely that any difference across the groups is clinically significant.
Regarding hospital admissions in the previous year, out of the 1140 first admission patients with data available, 727 had zero admissions in the previous year. The maximum number of previous admissions was 20 (one patient).
Comorbidity
Pre-existing medical conditions for the 1172 patients at first admission are set out in Table 3 . As can be seen from Table 3 , there is no evidence of any association between trial arm and comorbidity for any disease.
| Medical condition | Near patient (n = 397) | Rapid molecular (n = 387) | Traditional (n = 388) | p-valuea | Missing, n b |
|---|---|---|---|---|---|
| Myocardial infarction, n (%) | 0.627 | 37b | |||
| Yes | 48 (12.5) | 56 (14.9) | 51 (13.5) | ||
| No | 335 (87.5) | 319 (85.1) | 326 (86.5) | ||
| Heart failure, n (%) | 0.860 | 37b | |||
| Yes | 21 (5.5) | 21 (5.6) | 18 (4.8) | ||
| No | 362 (94.5) | 354 (94.4) | 359 (95.2) | ||
| Angina, n (%) | 0.768 | 37b | |||
| Yes | 44 (11.5) | 37 (9.9) | 41 (10.9) | ||
| No | 339 (88.5) | 338 (90.1) | 336 (89.1) | ||
| Stroke, n (%) | 0.369 | 37b | |||
| Yes | 21 (5.5) | 21 (5.6) | 29 (7.7) | ||
| No | 362 (94.5) | 354 (94.4) | 348 (92.3) | ||
| Chronic bronchitis, n (%) | 0.533 | 37b | |||
| Yes | 42 (11.0) | 51 (13.6) | 45 (11.9) | ||
| No | 341 (89.0) | 324 (86.4) | 332 (88.1) | ||
| Asthma, n (%) | 0.999 | 37b | |||
| Yes | 162 (42.3) | 159 (42.4) | 160 (42.4) | ||
| No | 221 (57.7) | 216 (57.6) | 217 (57.6) | ||
Time to convalescent blood samples
Convalescent blood samples were taken from patients following their admission. Median times to collection of the samples are set out in Table 4 , for first admissions only, and are similar across trial arms.
| Investigation | Near patient (n = 174) | Rapid molecular (n = 185) | Traditional (n = 183) | Total (n = 542) |
|---|---|---|---|---|
| Median days from admission to collection of convalescent samples (IQR) | 28.25 (14.58–59.45) | 27.46 (17.95–55.97) | 27.72 (19.09–61.59) | 28.00 (17.07–59.49) |
Chapter 4 Patient outcomes
Introduction
In this section, we report the findings relating to the patient outcomes. The main results reported are based on the first admission for 1172 patients. The following diagnostic tests contributed to the diagnoses made:
-
rapid molecular tests (PCR): influenza and RSV
-
Quidel (NPT for influenza)
-
viral cultures (influenza, RSV)
-
BinaxNOW test (S. pneumoniae)
-
sputum cultures (S. pneumoniae), and
-
blood cultures (S. pneumoniae).
A patient was diagnosed as having any of the three infectious conditions (influenza, RSV or S. pneumoniae) if any of the diagnostic tests (not necessarily a test to which the patient had been randomised) showed a positive result, regardless of the results of the other tests. The Gram stain test was not used in determining diagnosis as none of the outcomes were positive for S. pneumoniae. In Chapter 5 , sensitivity to diagnostic test results is further explored using serology data for influenza.
Diagnostic test results for first admissions
The results of the diagnostic tests for 1172 first admissions are set out in Table 5 . Note that an outcome of ‘Not diagnosed’ does not indicate a definite negative diagnosis, as for the influenza tests, a diagnostic result was not available for two patients (e.g. due to lack of a suitable sample). From the first admissions, four patients were diagnosed with both RSV and S. pneumoniae; also four patients were diagnosed with both influenza and S. pneumoniae. No patients were diagnosed with both RSV and influenza.
| Diagnosis | Near patient (n = 397) | Rapid molecular (n = 387) | Traditional (n = 388) | p-valuea | n missing |
|---|---|---|---|---|---|
| Influenza, n (%) | 0.590 | 0 | |||
| Diagnosed | 27 (6.8) | 30 (7.8) | 34 (8.8) | ||
| Not diagnosed | 370 (93.2) | 357 (92.3) | 354 (91.2) | ||
| RSV, n (%) | 0.160 | 0 | |||
| Diagnosed | 11 (2.8) | 13 (3.4) | 5 (1.3) | ||
| Not diagnosed | 386 (97.2) | 374 (96.6) | 383 (98.7) | ||
| S. pneumoniae, n (%) | 0.099 | 0 | |||
| Diagnosed | 24 (6.1) | 36 (9.3) | 39 (10.1) | ||
| Not diagnosed | 373 (94.0) | 351 (90.7) | 349 (90.0) | ||
Overall, the diagnoses appeared to be relatively evenly spread across the three trial arms, with no evidence of an association between trial arm and diagnoses. No attempt is made in these analyses to distinguish patients with multiple diagnoses (e.g. if a patient has a diagnosis of both RSV and S. pneumoniae, this patient will be counted in both groups).
Prescribing outcomes
Time from admission to first narrow-spectrum antibiotic
One of the outcomes of interest in this trial is the potential impact of the investigation on the time from admission to the time of prescription of the first narrow-spectrum antibiotic. Of the 1252 admissions, a narrow-spectrum antibiotic was prescribed during 555 admissions. For 161 of these admissions, the duration of time until the prescription was ≤ 0 hours, indicating that the time of prescription of the narrow-spectrum antibiotic was prior to the recorded time of hospital admission, for example in the accident and emergency (A&E) department. For these patients, duration of 0.01 hours was substituted to facilitate the analysis. Of the 1172 first admissions, a narrow-spectrum antibiotic was prescribed during 527 admissions.
For first admissions only, the time to prescription of the first narrow-spectrum antibiotic is shown in Table 6 , which includes the HR with 95% CI based on a Cox proportional hazards model (with the ‘traditional’ group as the reference group) and median survival times for all three diagnostic groups.
| Investigation | Traditional (n = 199) | Near patient (n = 170) | Rapid molecular (n = 158) | Total (n = 527) |
|---|---|---|---|---|
| Median hours to first narrow-spectrum antibiotic (IQR) | 3 (0.01–8.17) | 3.5 (0.01–9.83) | 2.67 (0.01–6.75) | 3 (0.01–8.08) |
| Cox HR (95% CI; p-value) | 1 | 1.023 (0.833 to 1.257; 0.829) | 1.205 (0.976 to 1.487; 0.082) |
Based on Table 6 , there is weak evidence (p-value 0.082 for the Cox HR model) for an association between diagnostic group and time to first narrow-spectrum antibiotic comparing the ‘rapid molecular’ group to the ‘traditional’ group. However, in the light of the width of the 95% CI for the HR (0.976 to 1.487) and wide IQRs for both groups, the association does not appear to be strong. There is no evidence for an association between diagnostic groups comparing the ‘near-patient’ group with the ‘traditional’ group.
Time from admission to first oral antibiotic
All patients who received antibiotics were analysed for time until first oral antibiotic (if prescribed oral antibiotics at all), regardless of whether the patient received antibiotics by another route (e.g. i.v.) prior to receiving oral antibiotics. In total, of the 1252 admissions, 851 received at least one oral antibiotic during the hospital inpatient episode. Of these, 216 admissions had a time to first oral antibiotic that was zero or negative, and duration of 0.01 hours was substituted to facilitate the analysis. Of the 1172 patients with a first admission, 800 received at least one oral antibiotic. The analysis of time to first oral antibiotic, for first admissions only, by diagnostic group, is set out in Table 7 . Table 7 shows no evidence for any association between diagnostic group and time to first oral antibiotic.
| Investigation | Traditional (n = 283) | Near patient (n = 265) | Rapid molecular (n = 252) | Total (n = 800) |
|---|---|---|---|---|
| Median hours to first oral antibiotic (IQR) | 4 (0.1–10.5) | 3.75 (0.25–11.33) | 4 (0.01–11.5) | 4 (0.1–11) |
| Cox HR (95% CI; p-value) | 1 | 0.957 (0.809 to 1.132; 0.605) | 0.905 (0.763 to 1.074; 0.253) |
Time from admission to cessation of antibiotics
For patients with RSV or influenza only, the time to cessation of antibiotics (by all routes of administration) was analysed. For patients with RSV or influenza, 96 received at least one antibiotic during the hospital admission (across all 1252 admissions). Among the 1172 first admissions, 93 of the 120 patients diagnosed with influenza or RSV received at least one antibiotic during their admission. Six of these patients had a time to cessation of antibiotics that was negative, and was replaced by a duration of 0.01 hours. The results of the Cox proportional hazards model, comparing diagnostic groups, and median times to cessation of antibiotics, are shown in Table 8 .
| Investigation | Traditional | Near patient | Rapid molecular | Total |
|---|---|---|---|---|
| All first admissions with RSV or influenza (n = 120) | ||||
| Median hours to cessation of antibiotics (IQR; n) | 79.5 (34–116.75; 32) | 56.42 (22.5–118; 24) | 58.17 (18.98–152.75; 37) | 77 (25.83–131.83; 93) |
| Cox HR (95% CI; p-value) | 1 | 0.887 (0.512 to 1.537; 0.669) | 1.014 (0.628 to 1.637; 0.955) | |
| All first admissions (n = 1172) | ||||
| Median hours to cessation of antibiotics (IQR; n) | 78.5 (27–116.75; 296) | 60.75 (20.25–151; 286) | 77.5 (17–152.75; 275) | 77 (20.33–146.62; 857) |
| Cox HR (95% CI; p-value) | 1 | 1.066 (0.906 to 1.255; 0.440) | 1.030 (0.873 to 1.214; 0.726) | |
Table 8 shows no evidence for an association between time from initial hospital admission to final dose of antibiotics among patients with influenza or RSV. However, the numbers of eligible patients in each diagnostics group are relatively small, leading to wide 95% CIs and IQRs.
A corresponding analysis was also performed for all patients with a first admission (regardless of diagnostic outcomes). Across all 1252 admissions, 911 patient episodes were associated with administration of at least one antibiotic (in 44 cases the duration to cessation of antibiotics was negative and replaced by 0.01 hours). Of the 1172 patients admitted for the first time within the study, 857 received at least one antibiotic (42 of whom had a negative duration to cessation of antibiotics, which was replaced by 0.01 hours). The results are shown in Table 8 . Again, there is no evidence to support an association between diagnostic group and time to cessation of antibiotics.
Neuraminidase prescriptions in patients with influenza
One patient was treated with a NI. This patient was a first admission, in the traditional diagnostic group, and was not diagnosed with S. pneumoniae, influenza or RSV.
Clinical outcomes
Length of hospital stay
Length of hospital stay among survivors was calculated as the time between admission to hospital and discharge for patients who were discharged only (excluding patients who died while in hospital). For the first admissions, only 46 patients died in hospital (39 within 28 days), with the remaining 1126 being discharged.
Using a Cox proportional hazards model to investigate any associations between the duration of hospital stay and diagnostic group, the results are set out in Table 9 , which shows the Cox HR and median hospital stay (days). Two further analyses were performed on the full cohort of all discharged patients (1205), including discharges resulting from admissions subsequent to the first. These analyses used a Cox proportional hazards model, one of which included a frailty (random effect on individual patient across multiple admissions for those patients who had more than one admission, effectively treating patient as a cluster variable).
| Investigation | Traditional | Near patient | Rapid molecular | Total |
|---|---|---|---|---|
| All first admissions discharged (n = 1126) | ||||
| Median days hospital stay (IQR; n) | 3.23 (1.21–7.86; 374) | 2.44 (0.90–7.56; 388) | 2.81 (0.76–7.58; 364) | 2.98 (0.95–7.75; 1126) |
| Cox HR (95% CI; p-value) | 1 | 1.041 (0.903 to 1.201; 0.577) | 1.109 (0.960 to 1.282; 0.160) | |
| All first admissions discharged who were diagnosed with influenza (n = 90) | ||||
| Median days hospital stay (IQR; n) | 3.59 (1.40–7.28; 34) | 2.08 (0.37–4.43; 26) | 1.96 (0.74–6.82; 30) | 2.52 (0.90–6.56; 90) |
| Cox HR (95% CI; p-value) | 1 | 1.274 (0.758 to 2.143; 0.361) | 1.248 (0.762 to 2.043; 0.380) | |
| All first admissions discharged who were diagnosed with RSV (n = 27) | ||||
| Median days hospital stay (IQR; n) | 2.99 (2.98–3.16; 5) | 2.56 (2.05–8.60; 11) | 6.18 (1.27–11.75; 11) | 3.10 (1.32–8.84; 27) |
| Cox HR (95% CI; p-value) | 1 | 0.625 (0.203 to 1.920; 0.412) | 0.559 (0.183 to 1.709; 0.308) | |
| All first admissions discharged who were diagnosed with S. pneumoniae (n = 90) | ||||
| Median days hospital stay (IQR; n) | 4.21 (2.78–8.75; 37) | 3.10 (1.60–6.19; 22) | 4.02 (1.10–8.30; 31) | 4.13 (1.82–8.50; 90) |
| Cox HR (95% CI; p-value) | 1 | 1.131 (0.665 to 1.924; 0.651) | 1.033 (0.637 to 1.676; 0.896) | |
| All patients discharged (all admissions, 1205 discharges; no adjustment for multiple admissions) | ||||
| Median days hospital stay (IQR; n) | 3.23 (1.17–7.63; 404) | 2.43 (0.90–7.48; 409) | 2.81 (0.79–7.58; 392) | 2.99 (0.97–7.52; 1205) |
| Cox HR (95% CI; p-value) | 1 | 1.035 (0.902 to 1.188; 0.622) | 1.098 (0.955 to 1.262; 0.190) | |
| All patients who died or who were discharged (all admissions, 1205 discharges; frailtya on individual patient to adjust for multiple admissions in same patient) | ||||
| Cox HR (95% CI; p-value) | 1 | 1.055 (0.904 to 1.230; 0.499) | 1.118 (0.957 to 1.306; 0.159) | |
As shown in Table 9 , there was little difference in the HR whether including first admissions only or all admissions. Also, there was little difference in the HR when including frailty in the Cox proportional hazards model. Frailty itself did not appear to be significant (p-value = 0.166), indicating no evidence of an effect on hospital stay, independent of diagnostic group.
Comparing the three diagnostic groups, there did not appear to be any evidence to support any association between duration of hospital stay and diagnostic group. Figure 6 shows a Kaplan–Meier estimated cumulative probability plot of time to discharge or death by randomised group. It shows the overall similarity in length of stay by randomisation group.
FIGURE 6.
Kaplan–Meier estimated cumulative probability of time to discharge (all first admissions discharged n = 1126) by randomised group.

Fever duration
Patients were considered to have been pyrexial from admission if any of the following were indicated:
-
temperature on admission (> 37.2 °C),
-
pyrexial (> 37.2 °C) within 48 hours of admission, and
-
end date of pyrexia present.
The time/date of the end of fever (or total duration of fever) was derived as follows (in order of precedence):
-
stated end time/date of fever if present (460 admissions out of 1252), or
-
discharge/death date if patient discharged/died within 10 days of admission (note: there were 148 patient admissions who were pyrexial on admission but with no stated end time/date of fever out of 1252*).
[*1252 refers to admissions or patient admissions (and not unique patients) – there were 1172 patients, of whom 67 were admitted/randomised at least twice.]
Also, there were two admissions with an unknown date of end of pyrexia (and hence substituted with date of discharge or death) who were recorded as being pyrexial within 10 days, but who had a fever duration of more than 10 days when using the substituted date of end of pyrexia. For these patients, a fever duration of 240 hours was used. There were 26 patients with duration of fever calculated as ≤ 0 hours, and for these patients fever duration of 0.01 hours was substituted to facilitate the analysis. Only patients who became apyrexial within ≤ 240 hours of admission to hospital were included in the analyses (583 admissions out of 1252, and 549 patients out of 1172 first admissions).
The results of a Cox proportional hazards model for median fever duration (hours) comparing the ‘near-patient’ and ‘rapid molecular’ diagnostic groups against the ‘traditional’ group are shown in Table 10 . Results are shown for all patients at first admission, and first admissions with S. pneumoniae only. Based on the results in Table 10 , there is no evidence to support an association between fever duration and diagnostic group in all patients (regardless of any diagnosis), or for patients diagnosed with S. pneumoniae only, although the small numbers of patients with S. pneumoniae should be noted.
| Investigation | Traditional | Near patient | Rapid molecular | Total |
|---|---|---|---|---|
| All first admissions (n = 1172) | ||||
| Median fever duration, hours (IQR; n) | 22.75 (11.25–51.33; 190) | 22.5 (8.92–60; 185) | 21.75 (8.17–46; 174) | 22.5 (9.17–51.33; 549) |
| Cox HR (95% CI; p-value) | 1 | 1.003 (0.819 to 1.230; 0.974) | 1.158 (0.941 to 1.424; 0.165) | |
| All first admissions diagnosed with S. pneumoniae (n = 99) | ||||
| Median fever duration, hours (IQR; n) | 28.17 (8.92–71.5; 23) | 28.5 (8.42–70.33; 19) | 24 (11–46; 13) | 28.17 (8.92–66.75; 55) |
| Cox HR (95% CI; p-value) | 1 | 1.136 (0.608 to 2.122; 0.689) | 1.223 (0.610 to 2.450; 0.570) | |
Supplemental oxygen dependence
Of all 1252 admissions, 546 were associated with prescription of oxygen during the admission. Of these 546, 28 had no confirmed end date for cessation of oxygen therapy. For these patients, the cessation date for oxygen therapy was considered to be the date of discharge (n = 25) or death (n = 3). For patients whose duration of oxygen therapy in hours (from admission to cessation of oxygen) was zero or negative (i.e. their time of cessation of oxygen was recorded as being prior to the recorded time of admission to hospital), their duration was replaced by 0.01 hours (41 patients). Patients with duration of oxygen therapy of more than 240 hours (10 days) were excluded from the analyses.
| Investigation | Traditional | Near patient | Rapid molecular | Total |
|---|---|---|---|---|
| All first admissions (n = 1172) | ||||
| Median hours from admission to cessation of oxygen therapy (IQR; n) | 32.33 (11–75; 146) | 23.92 (9.33–74; 146) | 26 (9.25–70.08; 144) | 26 (10–72.5; 436) |
| Cox HR (95% CI; p-value) | 1 | 1.004 (0.798 to 1.265; 0.970) | 1.043 (0.828 to 1.314; 0.718) | |
| All first admissions diagnosed with S. pneumoniae (n = 99) | ||||
| Median hours from admission to cessation of oxygen therapy (IQR; n) | 45.33 (25.75–130; 18) | 40 (17.25–74.5; 9) | 30.17 (5.5–59.5; 13) | 44.17 (12–80.5; 40) |
| Cox HR (95% CI; p-value) | 1 | 1.401 (0.618 to 3.176; 0.420) | 1.791 (0.847 to 3.790; 0.127) | |
Regarding the 1172 first admissions only, 436 patients received oxygen therapy that ceased within 240 hours. The results of the time-to-event analysis by diagnostic group for the 436 patients, and for 40 out of 99 patients who were with diagnosed S. pneumoniae only, are shown in Table 11 .
Based on these results, there is no evidence to indicate an association between diagnostic group and time to cessation of oxygen, in all patients, or in those diagnosed with S. pneumoniae.
Continuous positive airway pressure dependence
Of all 1252 admissions, 17 were associated with the use of continuous positive airway pressure (CPAP) during the admission, with 14 patients receiving CPAP during a first admission. There were three patients with no date of cessation of CPAP; for two of these, the date of CPAP cessation was taken as the date of discharge, and for the third patient the time of death was used. There were three patients with a negative duration of CPAP (from admission to cessation of CPAP, indicating that time of commencement of CPAP was recorded as being before the recorded time of admission to hospital), replaced by a duration of 0.01 hours. All 14 patients who received CPAP during a first admission had duration of CPAP of ≤ 240 hours.
The results of the time-to-event analysis by diagnostic group, for all patients with a first admission, are shown in Table 12 . Owing to the small numbers of patients who received CPAP, the results should be viewed with caution; however, there is no evidence to support any association between diagnostic group and time to cessation of CPAP. Only two patients diagnosed with S. pneumoniae received CPAP during a first admission, one in the ‘near-patient’ diagnostic group and one in the ‘traditional’ group. Owing to the very small numbers of patients, analysis of the data is not feasible.
| Investigation | Traditional | Near patient | Rapid molecular | Total |
|---|---|---|---|---|
| All first admissions (n = 1172) | ||||
| Median hours from admission to cessation of CPAP (IQR; n) | 19.75 (8.5–26; 5) | 15.5 (6.42–22.5; 4) | 43.92 (0.01–48.83; 5) | 19.75 (6.42–3.4; 14) |
| Cox HR (95% CI; p-value) | 1 | 0.849 (0.220 to 3.276; 0.812) | 0.254 (0.045 to 1.440; 0.122) | |
Admissions requiring intensive care and ventilator support
Across the 1252 admissions, six patients were admitted to intensive treatment unit (ITU), of whom five were admitted on a first admission. Of the total of six admissions, three were in the ‘near-patient’ group, one was in the ‘rapid molecular’ group and two were in the ‘traditional’ group. One patient admitted to ITU was diagnosed with S. pneumoniae (in the ‘near-patient’ group and a first admission). None of the patients who were diagnosed with influenza or RSV were admitted to ITU.
All patients who were admitted to ITU required ventilator support (no other patients were ventilated while admitted). Owing to the small numbers of patients requiring ITU admission and ventilation, it is not feasible to make any comparisons across the diagnostic groups for this outcome.
Deaths
A total of 58 deaths were confirmed among the 1252 admissions (57 in first admissions), of which 50 (all in first admissions) occurred within 28 days of admission. Deaths that occurred more than 28 days after admission were not comprehensively recorded, so only deaths that occurred within 28 days are considered in this analysis. All 50 patients who died within 28 days of admission were patients who had been admitted for the first time (of the 1172 patients with first admissions, 50 died within 28 days, 4.3%). Of the 50 deaths, 39 died in hospital and 11 died after discharge. Deaths occurred within 28 days in three (3.3%) admissions with influenza, two (6.9%) with RSV, and ten (10.1%) with S. pneumoniae. Table 13 shows the distribution of deaths within 28 days of admission in each diagnostic group, overall and by diagnosis, for first admissions only. Despite the small number of deaths in the study, we found no evidence to associate any diagnostic group with increased mortality for all patients, or for patients with any specific diagnosis.
| Death within 28 days | Near patient (n = 397) | Rapid molecular (n = 387) | Traditional (n = 388) | p-valuea |
|---|---|---|---|---|
| Total cohort (N = 1172), n (%) | 0.115 | |||
| Yes | 12 (3.0) | 23 (5.9) | 15 (3.9) | |
| No | 385 (97.0) | 364 (94.1) | 373 (96.1) | |
| Influenza (N = 91), n (%) | 0.635b | |||
| Yes | 1 (3.7) | 0 (0) | 2 (5.9) | |
| No | 26 (96.3) | 30 (100) | 32 (94.1) | |
| RSV (N = 29), n (%) | 0.648b | |||
| Yes | 0 (0) | 2 (15.4) | 0 (0) | |
| No | 11 (100) | 11 (84.6) | 5 (100) | |
| S. pneumoniae (N = 99), n (%) | 0.241b | |||
| Yes | 2 (8.3) | 6 (16.7) | 2 (5.1) | |
| No | 22 (91.7) | 30 (83.3) | 37 (94.9) | |
Use of isolation facilities
Across the 1252 admissions, 112 patients were admitted to isolation, and, of the 1172 first admissions, 102 patients were admitted to isolation. Of the 1172 patients at a first admission, 120 were diagnosed with influenza or RSV of whom 14 were admitted to isolation at some time during their admission. In five cases, their time from admission to hospital to admission to isolation was zero or negative, and was replaced by duration of 0.01 hours. Owing to the small numbers of patients with RSV or influenza who were admitted to isolation, a time-to-event analysis is difficult, but the results are set out in Table 14 .
| Investigation | Traditional | Near patient | Rapid molecular |
|---|---|---|---|
| All first admissions (n = 1172), of whom 14 with influenza or RSV were isolated | |||
| Median hours from admission to ‘isolation’ (IQR; n) | 0.01 (0.01–27.37; 2) | 0.01 (0.01–0.01; 4) | 23.33 (5.50–55.08; 8) |
| Cox HR (95% CI; p-value) | 1 | 0.65 (0.10 to 4.20; 0.655) | 0.69 (0.14 to 3.39; 0.650) |
The numbers of patients with influenza or RSV who were admitted to isolation within 120 hours of admission in each group are shown in Table 15 . Owing to the small number of admissions to isolation within 120 hours of the admission, comparisons between the diagnostic groups are considered inappropriate.
| Patients | Near patient (n = 397) | Rapid molecular (n = 387) | Traditional (n = 388) |
|---|---|---|---|
| All patients with influenza or RSV, n (%) | 38 (9.6) | 43 (11.1) | 39 (10.1) |
| No. ‘isolated’ with influenza or RSV, n (%) | 3 (7.9) | 7 (16.3) | 2 (5.1) |
Eight patients who experienced a first admission and were diagnosed with S. pneumoniae (but not concomitant influenza or RSV) were admitted to single-room accommodation. Seven were isolated for > 12 hours, including two of 22 (who were diagnosed with streptococcal infection by any means but did not have concomitant influenza or RSV) in the ‘near-patient’ group (9.1%), four of 34 in the ‘rapid molecular’ group (11.8%) and one of 35 in the ‘traditional’ group (2.9%). Overall, taking data relating to RSV, influenza and S. pneumoniae infections into consideration, we found no evidence to indicate that diagnostic group has any association with use of isolation facilities, or that patients with RSV and influenza, who pose a higher threat of nosocomial transmission than S. pneumoniae, are isolated more often.
Quality of life
Data regarding QoL (EuroQoL EQ-5D scores) are available at three time points in the trial: admission, 7 days and 28 days following admission, and was self-completed by participants. At first admission, all 1172 patients were available for EQ-5D assessment, whereas at day 7, 16 patients (with a first admission) were deceased, and at day 28, 50 patients (with a first admission) were deceased. The numbers of patients alive, by diagnostic group, and with EQ-5D data available at different times, is shown in Table 16 . Numbers of patients across the diagnostic groups (first admissions only) with full EQ-5D data available at different times during the study period are shown in Table 17 , which also provides descriptive analyses of EQ-5D scores as combined into one overall score. 66,67
| Days | Near patient (n = 397) | Rapid molecular (n = 387) | Traditional (n = 388) | Total (n = 1172) |
|---|---|---|---|---|
| Availability of all five EQ-5D scores n (%) | ||||
| Admission only | 57 (14.4) | 64 (16.5) | 78 (20.1) | 199 (17.0) |
| Day 7 only | 1 (0.3) | 3 (0.8) | 1 (0.3) | 5 (0.4) |
| Day 28 only | 1 (0.3) | 0 (0) | 0 (0) | 1 (0.1) |
| Admission and day 7 | 72 (18.1) | 91 (23.5) | 90 (23.2) | 253 (21.6) |
| Admission and day 28 | 29 (7.3) | 22 (5.7) | 26 (6.7) | 77 (6.6) |
| Days 7 and 28 | 5 (1.3) | 4 (1.0) | 4 (1.0) | 13 (1.1) |
| Admission, days 7 and 28 | 228 (57.4) | 202 (52.2) | 188 (48.5) | 618 (52.7) |
| No EQ-5D at any time | 4 (1.0) | 1 (0.3) | 1 (0.3) | 6 (0.5) |
| Analysis | Near patient (n = 397) | Rapid molecular (n = 387) | Traditional (n = 388) |
|---|---|---|---|
| Quality-of-life data on admission | |||
| All five EQ-5D scores, no. (%) | 386 (97.2) | 379 (97.9) | 382 (98.5) |
| EuroQoL score: | |||
| Mean (SD) | 0.463 (0.330) | 0.459 (0.352) | 0.469 (0.330) |
| Median (IQR) | 0.487 (0.201–0.743) | 0.487 (0.189–0.760) | 0.516 (0.260–0714) |
| Quality-of-life data on day 7 | |||
| All five EQ-5D scores, no. (%) | 306/394 (77.7) | 300/378 (79.4) | 283/384 (73.7) |
| EuroQoL score: | |||
| Mean (SD) | 0.563 (0.332) | 0.593 (0.322) | 0.591 (0.287) |
| Median (IQR) | 0.639 (0.378–0.796) | 0.689 (0.325–0.840) | 0.620 (0.433–0.796) |
| Quality-of-life data on day 28 | |||
| All five EQ-5D scores, no. (%) | 263/385 (68.3) | 228/364 (62.6) | 218/373 (58.4) |
| EuroQoL score: | |||
| Mean (SD) | 0.634 (0.308) | 0.636 (0.288) | 0.588 (0.310) |
| Median (IQR) | 0.691 (0.516–0.814) | 0.691 (0.516–0.814) | 0.656 (0.516–0.796) |
There were no statistically significant differences between the three intervention groups in terms of median EQ-5D score at admission (p = 0.931), 7 days (p = 0.466) or 28 days (p = 0.117).
Repeated measures analyses, with the aim of investigating an interaction between diagnostic group and time of EQ-5D measurement on EQ-5D scores, using a linear mixed-effects model with a random effect for individual patient, are reported in Table 18 , for first admissions only. These analyses were performed initially on all data available at each time point, and subsequently on data from patients with EQ-5D scores available at admission, days 7 and 28.
| Covariate | Coefficient (95% CI) | p-value |
|---|---|---|
| All patients included with data available at each individual time (no. observations = 2745; no. patients = 1166) | ||
| Diagnostic group | ||
| Near patient | –0.007 (–0.052 to 0.038) | 0.763 |
| Rapid molecular | –0.008 (–0.054 to 0.037) | 0.723 |
| Time | ||
| Day 7 | 0.124 (0.086 to 0.162) | < 0.001 |
| Day 28 | 0.106 (0.064 to 0.148) | < 0.001 |
| Interactions | ||
| Day 7 × near patient | –0.012 (–0.065 to 0.041) | 0.665 |
| Day 7 × rapid molecular | 0.011 (–0.043 to 0.064) | 0.696 |
| Day 28 × near patient | 0.062 (0.005 to 0.120) | 0.033 |
| Day 28 × rapid molecular | 0.054 (–0.005 to 0.112) | 0.074 |
| Only patients with data at all three time points included (no. observations = 1854; no. patients = 618) | ||
| Diagnostic group | ||
| Near patient | –0.038 (–0.099 to 0.022) | 0.211 |
| Rapid molecular | –0.015 (–0.077 to 0.047) | 0.639 |
| Time | ||
| Day 7 | 0.123 (0.076 to 0.169) | < 0.001 |
| Day 28 | 0.096 (0.050 to 0.143) | < 0.001 |
| Interactions | ||
| Day 7 × near patient | –0.007 (–0.069 to 0.056) | 0.837 |
| Day 7 × rapid molecular | 0.004 (–0.060 to 0.069) | 0.897 |
| Day 28 × near patient | 0.078 (0.015 to 0.141) | 0.015 |
| Day 28 × rapid molecular | 0.057 (–0.008 to 0.122) | 0.084 |
The analysis of all available data indicated that there was a significant interaction between ‘near-patient’ diagnostic group and time of measurement at day 28 (p-value 0.033) and a borderline significant interaction between ‘rapid molecular’ diagnostic group and time of measurement at day 28 (p-value 0.074). In both cases the coefficient of the interaction was positive, indicating that the EQ-5D scores for day 28 were greater for the ‘near-patient’ and ‘rapid molecular’ groups compared with the traditional group. Under both sets of analyses, the effect of changing EQ-5D levels over time was also highly statistically significant (p < 0.001), with increases in EQ-5D at both day 7 (0.123; 95% CI 0.076 to 0.169), and day 28 (0.096; 95% CI 0.050 to 0.143) compared with admission.
Case reviews
Altogether eight patients in the ‘near-patient’ study group had a positive Quidel QuickVue Influenza A + B result. We examined the CRFs of these eight patients to examine at a patient level whether their medical management could have been influenced by the result of diagnostic testing. We also reviewed the CRFs of the 21 patients in the ‘near-patient’ study group whose urine on admission was positive for pneumococcal antigen using the BinaxNOW test to see if there was a temporal association between the test result and changes to case management and isolation.
QuickVUE influenza A + B test
The following results should be treated with caution given the small number of admissions in the ‘near-patient’ study group that tested positive in the QuickVUE Influenza A + B test.
A summary of the findings is shown in Table 19 . None of the patients was admitted within 48 hours of onset of symptoms and none was given NI. All seven patients with data on comorbidity had comorbidities that made them eligible for treatment if their symptoms were of ≤ 48 hours’ duration. Two patients were already ‘isolated’ when tested. Of the remaining six, one was discharged after the test and none of the remainder was isolated subsequently. Three of the eight patients were discharged within 24 hours of the test without antibiotics – two had a provisional diagnosis of asthma, one had influenza/ILI; they were the youngest of all of the eight patients.
| Observation | Patient | |||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
| Temperature on admission, °C | 37.3 | 37.2 | 36.3 | 36.8 | 36.7 | 38.0 | 37.5 | 37.6 |
| Age, years | 22 | 30 | 39 | 71 | 74 | 75 | 83 | 86 |
| Eligible for NIs, based on | ||||||||
| Symptom duration | No | No | No | No | No | No | No | No |
| Comorbidity | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Unknowna |
| NI prescribed | No | No | No | No | No | No | No | No |
| Isolated | ||||||||
| At time of diagnostic test | No | Yes | Yes | No | No | No | No | No |
| Within 24 hours of test | Discharged | Discharged | Discharged | No | No | No | No | No |
| Antibiotics during the admission: | Unknowna | No | Yes | Yes | No | Yes | No | Yes |
| Oral, narrow spectrum | Unknowna | – | Yes | – | – | – | – | – |
| Oral, broad spectrum | Unknowna | – | – | Yes | – | Yes | – | – |
| Intravenous, broad spectrum | Unknowna | – | – | Yes | – | Yes | – | Yes |
| Changed within 24 hours of testing | Unknowna | – | Yes, stopped | Yes, stepped up from oral to i.v. | – | Yes, from i.v. to oral | – | No |
| Discharged within 24 hours of test | Yes | Yes | Yes | No | No | No | No | Nob |
| Provisional diagnosis at admission | Exacerbation of asthma, chest infection | Influenza/ILI | Exacerbation of asthma | Exacerbation of airways disease, heart failure | Heart failure | LRTI | Exacerbation of airways disease | LRTI |
There was no consistent pattern of treatment after diagnostic testing for the four patients who are known to have received antimicrobials. One patient (No. 3), who had an exacerbation of asthma and was apyrexial on admission, had treatment with a narrow-spectrum oral antibiotic stopped and was discharged within 24 hours of testing. A 71-year-old patient (No. 4), who was pyrexial (38.8 °C) and was diagnosed with heart failure and an exacerbation of chronic airways disease on admission, had broad-spectrum oral antibiotics switched to the i.v. route for a presumed bacterial co-infection. Conversely, a 75-year-old patient (No. 6) with LRTI had broad-spectrum oral antibiotics switched from the i.v. to the oral route. Finally, an 86-year-old patient (No. 8) with LRTI was kept on broad-spectrum i.v. antibiotics but died from methicillin-resistant Staphylococcus aureus (MRSA) bacteraemia about 48 hours after admission.
BinaxNOW Streptococcus pneumoniae test
Twenty-one admissions that were randomised to the ‘near-patient’ group had a positive BinaxNOW S. pneumoniae test result. Characteristics of these patients are shown in Box 3 . Details of the spectrum and route of delivery of the antibiotics they received in hospital before and after the urinary antigen test are shown in Table 20 .
Median age (IQR), years: 59 (43–76.5).
Febrile on admission (≥ 37.3 °C), numerator/denominator (%), median (IQR) °C: 8/20a (40%), 37.1 °C (36.5–37.65 °C).
Symptom duration on admission, median (IQR) hours: 120 (48–168).
Received pre-admission antibiotics from GP, numerator/denominator (%, 95% CI): 3/20a (15%, 95% CI 3.2% to 37.9%).
Provisional diagnoses on admission:
-
CAP (n = 7).
-
AE airways disease: COPD (n = 3), asthma (n = 2).
-
ARI (n = 4).
-
Cardiac: heart failure (n = 3), arrhythmia (n = 1).
-
None entered (n = 1).
a Case notes missing for one patient.
| Subject | None | Narrow spectrum | Broad spectrum | Summary of changes after result | ||
|---|---|---|---|---|---|---|
| Oral | i.v. | Oral | i.v. | |||
| 1c | ✓ | None | ||||
| 2 | ✓ | [✓]d | Step-up | |||
| 3 | ✓ | None | ||||
| 4 | ✓ | None | ||||
| 5 | ✓ | [✓] | Step-up | |||
| 6 | ✓ | [✓] | Step-up | |||
| 7e | ✓ | None | ||||
| 8 | ✓ | None | ||||
| 9 | ✓ | None | ||||
| 10 | ✓ | None | ||||
| 11 | ✓ | None | ||||
| 12 | ✓ | None | ||||
| 13 | ✓ | None | ||||
| 14 | ✓ | None | ||||
| 15 | ✓ | None | ||||
| 16 | ✓ | None | ||||
| 17 | ||||||
| 18 | ✓ | None | ||||
| 19f | ✓ | None | ||||
| 20 | ✓ | [✓] | Step-up | |||
| 21 | ✓ | None | ||||
Table 20 shows the spectrum and route of delivery of antibiotics before and after the results of urinary antigen tests for 20 patients in the ‘near-patient’ group. Antibiotics were given to everyone except four patients before the test and one person afterwards. All but five patients received broad-spectrum antibiotics before or after the test. Table 20 shows that four patients had a ‘step-up’ in treatment within 24 hours of the test result. Although it is unclear from the CRFs whether these changes in treatment occurred in response to the test or slow clinical recovery, Table 20 shows that none of the patients had a ‘step-down’ in treatment within 24 hours of the result, i.e. there was no evidence from this small number of patients that the test led to a ‘step-down’ in treatment.
All but two of the 21 patients had comorbidities placing them at risks from the consequences of influenza and could reasonably be expected to be considered for treatment with NIs on admission, assuming that their symptoms were of < 48 hours’ duration. We examined the data of all 21 patients to see whether any were eligible for treatment, and whether it had been administered and stopped within 24 hours of the urinary antigen test result. Only 4 of the 21 patients had symptoms of < 48 hours’ duration when admitted (all four had symptoms of 24 hours’ duration) – two were pyrexial (temperature ≥ 37.3 °C). All four patients had symptoms compatible with influenza, three had underlying respiratory comorbidity, and all four had oxygen saturations of 89–95% on admission, the latter on treatment with 5 l of oxygen. None of the 21 patients received treatment with a NI.
We examined data on ‘isolation’ for all 21 patients before and within 24 hours of the urinary antigen test result. One of the 21 patients was nursed in single-room accommodation between admission and the urinary pneumococcal antigen test result. This patient had a fever of 40.3 °C on admission and remained in single-room accommodation after the test result. One other patient was isolated within 24 hours of the urinary antigen test result – for diarrhoea. Overall, there was no evidence that the BinaxNOW S. pneumoniae test influenced decisions on ‘isolation’.
We examined data regarding the time of discharge in relation to the urinary pneumococcal antigen test result. Four patients were discharged within 24 hours of the urinary antigen test result. It is unknown whether any of these discharges occurred as a consequence of the test.
Discussion
In this chapter we explored the relationship between the diagnostic group and clinical measures, notably antiviral and antibacterial prescribing, the duration of fever and hospitalisation, treatment with oxygen and CPAP, intensive care support and death. We also examined the use of single-room accommodation to see whether the location of care was influenced by different diagnostic tests. Finally, we explored whether the randomisation resulted in any differences in the QoL of survivors across groups. By each measure, we failed to demonstrate any clinical benefit to first admission patients from POCTs for influenza A and B and S. pneumoniae, or molecular diagnostic tests for influenza A and B, or RSV A and B. We also found no evidence that the microbiological diagnosis, whether obtained by POCTs or by RT-PCR, had any effect on containment.
This study was carried out in a busy teaching hospital environment that caters for a local population of approximately one million people. The patients who were recruited to our study had a broad range of acute cardiopulmonary conditions that are typically seen in acute care facilities around the country. Our cohort included substantial numbers of patients who were diagnosed with acute exacerbations of asthma, bronchitis, acute exacerbations of COPD, CAP – conditions that are associated with influenza A and B, RSV and S. pneumoniae. In our study, influenza A or B occurred in 91 of 1172 first admissions (7.8%, 6.3–9.4%), RSV in 29 (2.5%, 1.7–3.5%), and S. pneumoniae in 99 (8.4%, 6.9–10.2), i.e. infection with these pathogens occurred in more than one in six patients.
Two recent studies68,69 suggest that rapid influenza testing may influence the management of adults with influenza. Falsey et al. 68 retrospectively evaluated the clinical management of patients with positive rapid antigen tests for influenza upon admission, during a period when the hospital infection control policy mandated such testing. Viral culture tests and/or RT-PCR, or serological testing were performed if the test was negative using the Directigen POCT for influenza. Antibiotic use was cut modestly from 98.7% (79 out of 80) in patients whose antigen test was negative or unknown to 86.0% (74 out of 86) (relative risk reduction 12.8%: 95% CI 5.7 to 21.9%) in those in whom it was positive. This reduction was not associated with significant differences in either antibiotic days, length of hospital stay among survivors or antibiotic complications; rather, the mean length of stay was 41 hours longer (p = 0.16) in those found positive using rapid influenza testing. Although antiviral use was high in the antigen-positive test group (63 out of 86, 73%) compared with patients whose test was negative, or in whom the test was not performed (6 out of 80, 6%), 39% of patients treated in the study presented outside the 48-hour therapeutic window recommended for prescribing antiviral therapy. Regarding infection control, the antigen test was falsely negative in 41.5% out of 147 patients tested; it is unclear whether any of these patients were isolated. It is also uncertain how many antigen-positive cases had false-positive results and were isolated and given antiviral therapy treatment inappropriately.
D’Heilly et al. 69 evaluated the influence on clinical care of different diagnostic tests (an immunoassay and two cell culture assays) that were used to detect influenza in adults with a mean age of 57.4 years who presented to an urgent care or outpatient medical clinic at a Veterans Affairs medical centre in Minneapolis, USA. The rapid antigen test had a sensitivity of 65% (84 patients tested positive overall); 20 of the 55 cases of influenza that were detected by rapid antigen testing had symptoms of < 48 hours and received antiviral medication. Thus, in this study, rapid testing led to treatment of 23.8% of the 84 patients with influenza. Altogether, 91% of patients with a positive rapid antigen test who described symptoms of ≤ 48 hours’ duration received antiviral therapy compared with only 8% of patients with a positive rapid cell culture test but a negative rapid antigen test. Individuals with a positive rapid antigen test were significantly less likely to be treated with antibiotics.
Both of the above studies focused on older adults tested under clinical rather than controlled study conditions and illustrate the potential for near-patient influenza tests to influence prescribing practices, with certain provisos.
-
First, to meet NICE criteria for treatment of seasonal influenza, it is essential that results are available to clinicians within a period of 48 hours after the onset of symptoms. 70 Our cohort presented with a median duration of symptoms before admission of 120 hours (IQR 72–168 hours) (see Chapter 7 ), and only 12% of all patients who were studied could potentially benefit from treatment with influenza antiviral drugs (on the basis of symptom duration, the turnaround time of the diagnostic tests and NICE guidance), even if they all had influenza. However, as shown in Chapter 4 , influenza occurred in 91 out of 1172 (7.7%) first admissions. Thus, with current referral and admission practice, it seems unlikely that detection and treatment of influenza in the small numbers of patients who are eligible (because of underlying ill health and illness duration of < 48 hours) would impact on the outcomes in our study. However, we did not stratify illness severity as part of the study or take into consideration the possibility that late treatment may benefit severely ill patients who are hospitalised with influenza, as such treatment falls outside NICE guidance. Evidence indicates that oseltamivir (Tamiflu®, Roche) treatment of patients hospitalised with seasonal71–73 and pandemic 2009 H1N1 influenza74–80 is associated with reductions in radiological pneumonia, illness severity and death. Even when administered > 48 hours after symptom onset, oseltamivir showed considerable potential for reducing pneumonia due to 2009 pandemic H1N1 virus. 79 Survival benefits for H5N1 influenza have also been observed when treatment with oseltamivir was delayed up to 6 days after symptom onset. 81
-
Second, the ability to diagnose influenza is dependent on the sensitivity of the test. Many of the studies that have compared rapid tests with RT-PCR and/or cell culture have been carried out in young healthy adults or in children. Children generally present earlier to medical practitioners with ILI and they shed higher titres of virus than older people, who also present later. 82 In our study, the sensitivity of the POCT in comparison with RT-PCR was low at 24.4% (see Chapter 5 ), indicating that opportunities to identify and treat patients like ours could be reduced considerably. The low sensitivity in our study may reflect the length of illness and age of the patients prior to sample collection. Nilsson et al. 83 found the sensitivity of the BinaxNOW influenza A/B test against PCR to drop from 71% to 63% and 14% when specimens were collected from adults presenting to the emergency department of Malmö University Hospital on days 1–3, 1–5 and > 5 days, respectively, following onset of symptoms. Similarly, Stripeli et al. 84 observed a fall in sensitivity from 76% to 65% for The QuickVue Influenza Test when specimens were collected from hospitalised children with disease duration of < 48 hours compared with children with disease duration of longer than 48 hours.
-
Third, the sensitivity of near-patient immunoassay tests can also be influenced by virus type, and pandemic 2009 H1N1 virus compared with seasonal influenza. 85 Heinonen et al. 86 observed a sensitivity of 90% for influenza A viruses but only 25% for influenza B viruses when specimens were collected from young children aged 1–3 years who presented within 24 hours of onset of fever, and tested using the Actim Influenza A&B POCT. Reduced sensitivity for influenza B virus has also been noted by other investigators using other rapid tests. 87–89 In our study more than one-third of the specimens positive for influenza by RT-PCR were identified as influenza B virus, which may, in part, account for the low sensitivity of the near-patient QuickVue Influenza A + B test in our study (see Chapter 5 ), and the lack of clinical benefit from the POCT in comparison with molecular or conventional diagnostic tests in our study. Our study was carried out before the 2009 H1N1 pandemic.
-
Fourth, the quality of the specimen and method of collection can also impact upon test sensitivity. Scansen et al. 90 compared the performance of the Quidel QuickVue Influenza A + B test on secretions from the anterior nares when a polyurethane foam swab was used for collection to that of a nylon flocked swab. The QuickVue test was positive for 40 foam and 30 flocked swabs, for sensitivities of 71% and 54%, respectively, a difference that achieved statistical significance. Agoritsas et al. 91 showed that the sensitivity of the QuickVue test was 85% with nasopharyngeal swabs, 78% with nasal swabs and 69% with nasopharyngeal washes, when specimens were collected from children with a mean age of 5 years. Anterior nasal swab collection was performed first with an absorbent foam swab, posterior nasopharyngeal swab collection was performed second with a Dacron swab, and nasopharyngeal washing was performed last with sterile saline. The difference in sensitivity between nasopharyngeal swabs and nasopharyngeal washes was significant. There was no difference in sensitivity between anterior nasal swab collection and the two nasopharyngeal collection methods. Walsh et al. 92 compared nylon flocked swab/universal transport medium collection method (sampling at the mid-turbinate level) with nasal aspirates collected from infants and toddlers in a PCR test – the swab collection method significantly outperformed the use of nasal aspirates. Schmid et al. 93 evaluated traditional and rapid culture methods for influenza using nasopharyngeal aspirates and throat swabs from adults with symptoms and signs consistent with influenza. Nasopharyngeal aspirates were twice as sensitive as throat swabs. Similarly, Smit et al. 94 tested upper respiratory tract samples in parallel with the BinaxNOW Influenza A&B combination assay, BinaxNOW Flu A and BinaxNOW Flu B assays, the Becton Dickinson Directigen Flu A + B assay and IF, and the results were compared with viral culture. Altogether 521 samples – including 338 nasopharyngeal swabs, 162 throat swabs, 19 nasal washes and two swabs from unspecified sites – were collected from 448 adults and children. There were no significant differences in the performances of all rapid antigen tests, with sensitivities of 53–59% compared with culture and IF but the sensitivities of all the rapid antigen tests were significantly higher for nasopharyngeal samples than for throat swabs. In our study, nasopharyngeal specimen collection was undertaken by trained research staff, and specimens were handled as per the manufacturer’s instructions. It is therefore unlikely that specimen quality was responsible for the observed outcomes.
Most antibiotics prescribed in general practice are for respiratory tract infections that represent about 70% of all infections treated in primary care. 95,96 In the USA, antibiotic prescribing for acute respiratory illness accounts for over 50% of all antibiotics used in primary care. 97 Antibiotic prescribing has decreased in UK general practices in recent years mainly because there are fewer consultations for common respiratory infections, and partly because GPs are prescribing antibiotics less frequently for conditions that are primarily viral in aetiology. 98 There is a paucity of evidence showing benefit from antibiotic use in many acute respiratory conditions including acute bronchitis,99 acute asthma exacerbations,100–102 and in many cases of acute exacerbation of chronic obstructive airways disease. 103–105 Although overprescription of antibiotics has led to the emergence and proliferation of antibiotic-resistant bacteria,106–109 a meta-analysis of 122 reports of CAP showed that approximately 6000 (18%) of 33,148 cases had a bacterial pathogen, with S. pneumoniae accounting for 73% of all cases and 66% of fatal cases. 16,110 Thus, clinicians face the problem of unnecessary use of antibiotics for some respiratory conditions – and the requirement for prompt (empiric) antimicrobial treatment for others.
National and international bodies including the British Thoracic Society (BTS) provide guidance for empiric treatment of CAP due to the risks from delayed treatment and limitations of diagnostic tests. 17 Isolation of S. pneumoniae from the blood is specific but lacks sensitivity. 111–113 Culturing blood before antibiotic therapy is recommended by the BTS (and similar bodies) for all hospitalised patients with moderate and high-severity CAP, but studies have suggested that blood cultures rarely alter antibiotic therapy for patients presenting with pneumonia because of the low overall positive rate of blood cultures. 112,114–118 Moreover, clinicians did not follow protocols for narrowing an antibiotic spectrum even when appropriate. Sputum culture is often used to help identify aetiological agents and is recommended by BTS to investigate moderate severity CAP and severe CAP that fails to improve. 17 The test lacks specificity, and isolation of S. pneumoniae from sputum may represent colonisation. 119 In our study, sputum culture had a sensitivity of 100% (2.5–100%) (see Chapter 5 ) when compared with blood culture as the reference standard but the CIs were extremely wide due to the infrequency of a productive cough in our cohort. Musher et al. 120 analysed 105 patients with pneumococcal pneumonia proven by blood culture – sputum culture was positive in only 44% of all cases. These investigators reviewed earlier articles and noted that sensitivity of Gram staining in proven cases of pneumococcal pneumonia ranged from 20% to 69% and that the sensitivity of culture ranged from 29% to 94% (see Chapter 6 ).
Given the limitations of conventional diagnostic tests for S. pneumoniae infection, the development of a S. pneumoniae urinary antigen test offered several advantages – ease in getting a diagnostic specimen, ability to detect pneumococcal infection after antibiotic treatment has been commenced, its diagnostic yield, and a rapid turnaround time. Among adult patients with CAP, this test has shown sensitivities of 64–87% compared with blood culture,111,121 but dropped to 54% when cultures of respiratory tract secretions were included in the reference standard. 122 In our study, the sensitivity of the urinary antigen test in comparison with blood culture was 57.1% (18.4% to 90.1%) (see Chapter 5 ), which is comparable with previous reports. In our study, the POCT for pneumococcal infection was positive in 92 (7.8%; 6.4 to 9.5) of all 1172 admissions and, together with POCT for influenza, it had no evident effect on clinical outcome.
Only one patient in this study was prescribed a NI. Reasons for the low use these drugs might include the length of illness pre-admission together with unfamiliarity with NIs and the diagnostic test. The number of patients in the ‘near-patient’ group with a positive QuickVue Influenza A+B test was too small to draw definitive conclusions – none of eight patients with a positive result was eligible for treatment based on their duration of symptoms pre-admission and NICE guidance.
Regarding ‘containment’, two of the eight patients in the ‘near-patient’ group with a positive QuickVue Influenza A+B test were already in single-room accommodation when the test was carried out. One patient was discharged shortly afterwards. None of the remaining five patients was isolated, but four had fever and respiratory symptoms and were probably still shedding influenza virus. 123–125 We may reason that single-room accommodation was either unavailable, or that the nursing and medical carers were unaware of the possible risk of nosocomial transmission. Altogether, 120 of the 1172 patients with a first admission had positive tests for influenza or RSV, of whom only 14 were given single-room accommodation during the admission.
The US Healthcare Infection Control Practices Advisory Committee recommends that patients with influenza are given single-room accommodation, when available, for a period of 5 days, and masked when transported out of the room. 126 The problem for clinicians is recognising patients with influenza, as exacerbations of asthma,127 cystic fibrosis,128 COPD,129 heart failure,130 bouts of pneumonia,17 acute bronchitis131 and other acute respiratory conditions are all associated with influenza, and such complications may dominate the features of influenza. Nosocomial influenza is a well-recognised problem in acute care hospital settings. 132–135 Evidence indicates that nosocomial infection of health-care workers with influenza in acute hospitals is not uncommon136 and that influenza acquired within hospitals can be fatal. 137 In our study, the rate of isolation for patients with influenza or RSV (14/120; 11%, 95% CI 7.1% to 18.6%) was no different to the overall rate of isolation (112/1172; 9.5%, 95% CI 7.9% to 11.4%), suggesting that hospitals like ours have inadequate resources to contain infection.
The extremely low prescription rate for NIs in this study indicates that near-patient testing for S. pneumoniae could not reduce NI use, even if considered appropriate. Bacterial pneumonia is a well-recognised complication of influenza and occurred in 29% of ≈1200 patients hospitalised during the 2009 H1N1 pandemic. The pneumococcus was by far the most common pathogen,138–145 suggesting that withdrawal of NIs from some patients with a positive urinary pneumococcal antigen test may be inappropriate.
Chapter 5 Diagnostic outcomes
Statistical methods
Data on diagnostic performance of the various tests (blood culture, viral culture, sputum culture, PCR, Quidel and Binax POCTs) for the diagnosis of (1) influenza and (2) S. pneumoniae infection, are summarised as:
-
percentage diagnostic agreement (the percentage of test results, either positive or negative, that are in agreement) (and associated 95% CI), or
-
sensitivity (percentage of true-positives correctly identified)
-
specificity (percentage of true-negatives correctly identified) (and associated 95% CI)
-
positive predictive values (PPVs) (percentage of positive test results that are truly positive) and negative predictive values (NPVs) (percentage of negative test results that are truly negative) (and associated 95% CI), and
-
area under receiver operating characteristic (ROC) curve (area under the curve, AUC) (the probability that two patients – one diseased and one not diseased – would be both correctly classified by the test) (and associated 95% CI).
In the case of influenza, uncertainty remains as to what is currently considered to be the reference standard to which all tests should be compared (i.e. ‘gold standard’). When the 3WS was designed, standard laboratory testing procedures were anticipated to be the then current ‘gold standard’. However, during the course of the 3WS, PCR techniques have been developed and now may be considered to be the ‘gold standard’ – for example in the meta-analysis reported in Chapter 6 of influenza diagnoses, 13 out of 64 (20%) studies used PCR as the ‘gold standard’ and only 24 of 64 (37%) studies considered laboratory culture alone to be the ‘gold standard’. Hence a number of analyses using different ‘gold standards’ have been undertaken to enable comparison between the results from 3WS and those found in the literature.
In the case of influenza, test performance could also be assessed by serology. The primary analysis uses only definite positive serology results and does not allow for month and time between blood samples. However, due to the nature of the 3WS, patients could be discharged from hospital and subsequently have had an influenza vaccination prior to a second (convalescent) blood sample being taken. In order to allow for this, and the fact that positive serology results were graded as definite, probable and possible, two sensitivity analyses were undertaken: (1) excluding those who had their first blood taken between September and December and had > 30 days between samples and (2) including both definite and probable positive serology results.
All analyses were undertaken in Stata version 11.
Results
Influenza
Comparison between tests themselves
Table 21 summarises the diagnostic performance of the tests in terms of influenza using either PCR or viral culture as the ‘gold standard’. Although there is a relatively high level of overall agreement between the tests, the Quidel POCT has relatively low sensitivity compared with either PCR (24.4%, 95% CI 16% to 34.6%) or viral culture (33.3%, 95% CI 14.6% to 57.0%), although the corresponding specificity is always over 90%. Thus, although the Quidel POCT appears to perform acceptably at classifying truly negative patients, in terms of influenza, less than one-third of those with influenza appear to be detected with the test. In terms of sensitivity, this is considerably lower than values reported in the literature. Overall, in the meta-analysis reported in Chapter 6 , the 64 studies included yielded a pooled sensitivity of 74%, although those using PCR and viral culture as ‘gold standards’ produced sensitivities of 51% and 86%, respectively. However, subgroup and sensitivity analyses suggest that comparable estimates to 3WS based on the meta-analysis are lower than these overall headline figures (see Chapter 6 for further details).
| ‘Gold standard’ | Comparator | Measure | Estimate | 95% CI |
|---|---|---|---|---|
| PCR | Quidel POCT | Agreement | 94.3% | 92.8% to 95.5% |
| Sensitivity | 24.4% | 16% to 34.6% | ||
| Specificity | 99.7% | 99.2% to 99.9% | ||
| PPV | 88.0% | 68.8% to 97.5% | ||
| NPV | 94.4% | 93.0% to 95.7% | ||
| ROC AUC | 0.62 | 0.58 to 0.67 | ||
| Viral culture | Agreement | 94.1% | 92.6% to 95.4% | |
| Sensitivity | 21.6% | 13.5% to 31.6% | ||
| Specificity | 99.8% | 99.4% to 100% | ||
| PPV | 90.5% | 69.6% to 98.8% | ||
| NPV | 94.2% | 92.7% to 95.4% | ||
| ROC AUC | 0.61 | 0.56 to 0.65 | ||
| Viral culture | Quidel POCT | Agreement | 97.4% | 96.4% to 98.2% |
| Sensitivity | 33.3% | 14.6% to 57.0% | ||
| Specificity | 98.6% | 97.7% to 99.2% | ||
| PPV | 29.2% | 12.6% to 51.1% | ||
| NPV | 98.8% | 98.0% to 99.3% | ||
| ROC AUC | 0.66 | 0.56 to 0.76 | ||
| PCR | Agreement | 94.1% | 92.6% to 95.4% | |
| Sensitivity | 90.5% | 69.6% to 98.8% | ||
| Specificity | 94.2% | 92.7% to 95.4% | ||
| PPV | 21.6% | 13.5% to 31.6% | ||
| NPV | 99.8% | 99.4% to 100.0% | ||
| ROC AUC | 0.92 | 0.86 to 0.99 |
Comparison with serology
Table 22 summarises the diagnostic performance of the tests in terms of influenza using serology as the ‘gold standard’. Of the three approaches to testing for influenza, all have relatively high specificities, although PCR has the highest sensitivity (42.6%, 95% CI 28.3% to 57.8%), those of viral culture and Quidel POCT are relatively low, at 13.3% and 14.9%, respectively. Both sensitivity analyses indicate that these estimates of sensitivity could increase, although only very slightly.
| Comparator | Measure | Estimate | 95% CI |
|---|---|---|---|
| PCR | |||
| Primary analysis | |||
| Agreement | 92.3% | 90.6% to 93.7% | |
| Sensitivity | 42.6% | 28.3% to 57.8% | |
| Specificity | 94.2% | 92.7% to 95.4% | |
| PPV | 22.2% | 14.1% to 32.2% | |
| NPV | 97.7% | 96.6% to 98.5% | |
| ROC AUC | 0.68 | 0.61 to 0.76 | |
| Sensitivity analysesa | |||
| (i) | Agreement | 91.8% | 90.0% to 93.3% |
| Sensitivity | 48.8% | 32.9% to 64.9% | |
| Specificity | 93.3% | 91.5% to 94.7% | |
| PPV | 22.7% | 14.5% to 32.9% | |
| NPV | 97.7% | 96.6% to 98.5% | |
| ROC AUC | 0.71 | 0.63 to 0.79 | |
| (ii) | Agreement | 92.3% | 90.7% to 93.7% |
| Sensitivity | 43.8% | 29.5% to 58.8% | |
| Specificity | 94.3% | 92.8% to 95.5% | |
| PPV | 23.3% | 15.1% to 33.4% | |
| NPV | 97.7% | 96.6% to 98.5% | |
| ROC AUC | 0.69 | 0.62 to 0.76 | |
| Viral culture | |||
| Primary analysis | |||
| Agreement | 95.4% | 94.1% to 96.5% | |
| Sensitivity | 13.3% | 5.05% to 26.8% | |
| Specificity | 98.7% | 97.9% to 99.3% | |
| PPV | 28.6% | 11.3% to 52.2% | |
| NPV | 96.7% | 95.5% to 97.7% | |
| ROC AUC | 0.56 | 0.51 to 0.61 | |
| Sensitivity analysesa | |||
| (i) | Agreement | 95.1% | 93.7% to 96.4% |
| Sensitivity | 15.4% | 5.8% to 30.5% | |
| Specificity | 98.5% | 97.5% to 99.1% | |
| PPV | 28.6% | 11.3% to 52.2% | |
| NPV | 96.6% | 95.3% to 97.6% | |
| ROC AUC | 0.57 | 0.51 to 0.63 | |
| (ii) | Agreement | 95.1% | 94.1% to 96.5% |
| Sensitivity | 13.0% | 4.9% to 26.3% | |
| Specificity | 98.7% | 97.9% to 99.3% | |
| PPV | 28.6% | 11.3% to 52.2% | |
| NPV | 96.6% | 95.4% to 97.6% | |
| ROC AUC | 0.56 | 0.51 to 0.61 | |
| Quidel POCT | |||
| Primary analysis | |||
| Agreement | 95.3% | 93.9% to 96.4% | |
| Sensitivity | 14.9% | 6.2% to 28.3% | |
| Specificity | 98.5% | 97.6% to 99.1% | |
| PPV | 28.0% | 12.1% to 49.4% | |
| NPV | 96.7% | 95.6% to 97.7% | |
| ROC AUC | 0.57 | 0.52 to 0.62 | |
| Sensitivity analysesa | |||
| (i) | Agreement | 95.1% | 93.6% to 96.3% |
| Sensitivity | 17.1% | 7.15% to 32.1% | |
| Specificity | 98.3% | 97.3% to 99.0% | |
| PPV | 29.2% | 12.6% to 51.1% | |
| NPV | 96.6% | 95.4% to 97.6% | |
| ROC AUC | 0.58 | 0.52 to 0.64 | |
| (ii) | Agreement | 95.3% | 93.9% to 96.4% |
| Sensitivity | 14.6% | 6.1% to 27.8% | |
| Specificity | 98.5% | 97.6% to 99.1% | |
| PPV | 28.0% | 12.1% to 49.4% | |
| NPV | 96.7% | 95.5% to 97.6% | |
| ROC AUC | 0.57 | 0.52 to 0.62 | |
Streptococcus pneumoniae infection
Table 23 summarises the diagnostic performance of the tests in terms of identifying S. pneumoniae infection using either blood culture or sputum culture as the ‘Gold standard’. In all cases specificity is > 90%, whereas the Binax POCT produces a sensitivity of 57.1% (95% CI 18.4% to 90.1%) compared with blood culture and 30.0% (95% CI 6.7% to 65.2%) compared with sputum culture.
| ‘Gold Standard’ | Comparator | Measure | Estimate | 95% CI |
|---|---|---|---|---|
| Blood culture | Binax | Agreement | 92.3% | 90.3% to 93.9% |
| Sensitivity | 57.1% | 18.4% to 90.1% | ||
| Specificity | 92.5% | 90.6% to 94.1% | ||
| PPV | 5.5% | 1.5% to 13.4% | ||
| NPV | 99.6% | 99.0% to 99.9% | ||
| ROC AUC | 0.75 | 0.55 to 0.95 | ||
| Sputum culture | Agreement | 97.2% | 94.4% to 98.9% | |
| Sensitivity | 100% | 2.5% to 100.0% | ||
| Specificity | 97.2% | 94.3% to 98.9% | ||
| PPV | 12.5% | 0.3% to 52.7% | ||
| NPV | 100% | 98.5% to 100% | ||
| ROC AUC | 0.99 | 0.48 to 1.00 | ||
| Sputum culture | Binax | Agreement | 90.0% | 85.6% to 93.1% |
| Sensitivity | 30.0% | 6.7% to 65.2% | ||
| Specificity | 92.0% | 88.1% to 95.0% | ||
| PPV | 12.5% | 2.7% to 32.4% | ||
| NPV | 97.2% | 94.3% to 98.9% | ||
| ROC AUC | 0.61 | 0.46 to 0.76 |
Discussion
We compared the diagnostic performances of viral culture, Quidel QuickVue Influenza A + B test and RT-PCR, using either PCR or viral culture as the ‘gold standard’. With PCR as the gold standard, viral culture carried out at the UHL NHS Trust laboratory had a sensitivity of just 21.6% (95% CI 13.5% to 31.6%), which, although similar to the sensitivity reported by the Portuguese national surveillance network, during the 7-year period 1992–93 to 1998–99,146 is suboptimal in comparison with sensitivities of 61–96% reported in nine studies90,91,147–153 ( Table 24 ) that were identified for the systematic review and meta-analysis of POCTs for influenza (see Chapter 6 ). However, the sensitivity of viral culture was similar to that of the Quidel POCT (24.4%, 95% CI 16% to 34.6%). Although the performance of both the viral culture and the Quidel POCT appears relatively low in terms of identifying truly positive cases, especially in comparison with the headline estimates of the meta-analysis of POCTs for influenza (see Chapter 6 ), any comparisons need to be approached with caution owing to the extremely heterogeneous nature of the studies reported, and because subgroup and sensitivity analyses indicate that 3WS provides comparable estimates of diagnostic performance to appropriate comparator studies (see Chapter 6 ).
| Author | Location | Children | Adults | No. in study | Culture sensitivity, % (no. positive by culture/no. positive by PCR) | Culture specificity |
|---|---|---|---|---|---|---|
| Agoritsas, 200691 | USA | From 2 weeks to 18 years | None | 122 | 91.5 (54/59) | |
| Chan, 2002147 | Hong Kong | From < 2 years | To > 55 years | 250 | 91.7 (54/57) | |
| Cheng, 2010148 | China | From 5 months | To 70 years | 5740 | 95.7 (551/576) | |
| Mehlmann, 2007149 | USA | From 3 months | To 86 years | 102 | 93.4 (57/61) | |
| Poehling, 2002150 | USA | From 6 months to 18 years | None | 233 | 61.1 (11/18) | 99.5 (214/215) |
| Rahman, 2008151 | USA | From 6 months | To ≥ 65 years | 932 | 94.9 (93/98) | 100 |
| Ruest, 2003152 | Canada | From 1 day | To 98 years | 200 | 83.5 (71/85) | |
| Scansen, 201090 | USA | ≤ 17 | None | 100 | 87.5 (49/56) | |
| Simmerman, 2007153 | Thailand | From 1 month | To 86 years | 1092 | 81.3 (205/252) |
Factors that may contribute to lower rates of influenza virus isolation, include the type of swab collection device, the cell culture system, the passage history of the mammalian cells used, the stability of protease supplement to the media and, possibly, alterations in receptor binding properties of isolates of influenza A H3N2 and H1N1 since the late 1980s. 146,154 Specimens were collected according to the methods recommended by the HPA, and by the manufacturer of the Quidel POCT (see www.cliawaived.com/web/items/pdf/QDL-20183-Quidel_Influenza_Tests_Insert∼619file1.pdf) by research nurses dedicated to the project. Thus, it seems unlikely that the expertise of personnel who collected the specimens or the method of specimen collection were important contributory factors to the low sensitivities of culture and the Quidel POCT observed in our study. In our study specimens were received by the laboratory at a median of 3.6 hours after specimen collection, making delayed transportation and inappropriate specimen handling unlikely as contributory factors. A more plausible explanation is the length of illness prior to sampling, which in our study was a median of 120 hours. Lee et al. 125 evaluated secular shedding of seasonal influenza virus from 147 inpatients aged > 16 years (mean age 72 years, ± 16 years) in Hong Kong. The results of virus isolation showed that among untreated (i.e. by NIs) patients, 38.5% and 21.2% remained culture positive by symptom day ≥ 4 and ≥ 5, respectively, and in patients with comorbidities the proportions were somewhat higher, at 41.7% and 33.3%, respectively. Leekha et al. 155 examined secular shedding of seasonal influenza virus by 50 hospitalised patients by culture. Results showed that by symptom days 3, 4, 5 and ≥ 7, 91%, 75%, 52% and 20% of patients, respectively, remained culture positive. Although these observations have implications for infection control, they show that by day 5 of symptoms (i.e. the median duration of symptoms upon sampling in our study), approximately 50–80% of patients with influenza were unlikely to be shedding infectious virus. Presence of viral nucleoproteins that are detectable by the Quidel POCT is likely to fall at a similar rate.
Despite being the most common pathogen in CAP, S. pneumoniae is underdiagnosed because of the limitations of conventional diagnostic tests. Although blood culture is used as the ‘gold standard’ for diagnosis of pneumococcal CAP in many studies, it has low sensitivity and premedication with antibiotics reduces it further. The observed sensitivity (57.1%, 95% CI 18.4% to 90.1%) and specificity (92.5%, 95% CI 90.6% to 94.1%) for the BinaxNOW urinary antigen detection test in our study are comparable to sensitivities of 75% to 88%49,156–160 compared with blood culture (or culture of blood and pleural aspirate) in previous studies. The BinaxNOW POCT has a high specificity, in excess of 90%, and contributed to a reduction in the spectrum of antibiotic cover of 41 of 474 episodes of CAP in one recent study. 161 In comparison with blood and sputum cultures, the pneumococcal urinary antigen test has advantages of being a simple and rapid method with visually detectable results (see Chapter 7 ), providing speedy diagnosis without any additional equipment or reagents in a clinic or triage setting; is non-invasive; positive results persist over a period of days and occur despite treatment with antibiotics; and the test has high sensitivity and specificity. However, a negative result cannot rule out pneumococcal infection.
In our study, sputum culture had a sensitivity of 100% (2.5–100%) when blood culture was used as the gold standard. The CIs are wide due to the small number of patients with a positive blood culture and productive cough. Musher et al. 120 analysed 105 patients with pneumococcal pneumonia proven by blood culture. Sputum culture yielded a pneumococcus in 44% of cases. Sensitivity data for sputum culture from that review and several recent studies are shown in Table 25 . 119,120,162–167 Random-effects meta-analysis of the data in Table 25 reveals sensitivities of 43% (33–53.0%) when cases who received antibiotics are included, and 59% (21–92%) when they are excluded. Specificity data are not available, but the specificity of sputum culture is generally poor due to colonisation of the respiratory tract. The poor sensitivity and specificity of sputum culture therefore question its value in the management of lower respiratory infections. Sputum culture also has downsides of cost and ease and speed of use, but empiric antibiotic recommendations are ultimately dependent upon the accumulated information available from such tests.
| Author | Location | No. in study | Sensitivity: % (95% CI) (no. positive by sputum culture/no. of positive by blood culture) | Patients with prior antibiotics excluded |
|---|---|---|---|---|
| Davidson, 1976119 | USA | 25 | 100 (15.8 to 100) (2/2) | No |
| Kalin, 1982162 | Sweden | 89 | 44.1 (31.1 to 57.6) (26/59) | No |
| Musher, 2004120 | USA | 105 | 43.8 (34.1 to 53.8) (46/105) | No |
| Torres, 1998163 | USA | 71 | 48.8 (33.3 to 64.5) (21/43) | No |
| Watanakunakorn, 2002164 | USA | 59 | 28.8 (33.3 to 64.5) (21/43) | No |
| Drew, 1977165 | USA | 31 | 93.5 (78.6 to 99.2) (29/31) | Yes |
| Fiala, 1969166 | USA | 25 | 56.0 (34.9 to 75.6) (14/25) | Yes |
| Garcia-Vazquez, 2004167 | Spain | 133 | 17.3 (11.3 to 24.8) (23/133) | Yes |
| Kalin, 1982162 | Sweden | 89 | 66.7 (49.8 to 80.9) (26/39) | Yes |
Chapter 6 Systematic review and meta-analysis of near-patient tests for influenza A and B
Introduction
Influenza resembles other acute viral respiratory infections with respect to its seasonality, clinical presentation and complications, but it differs from other respiratory viral conditions by being preventable by annual vaccination and ameliorated by antiviral drugs if given within 48 hours of symptom onset. The gold standard for influenza diagnosis is viral culture, which, although specific, had low sensitivity compared with real-time RT-PCR in our study and gave results long after hospital discharge or death (see Chapters 5 and 7 ). In contrast, we were able to correlate the results of PCR and serology (see Chapter 5 ), confirming the accuracy of RT-PCR, and we showed that it gave a diagnosis in a busy clinical setting within a median of 29 hours (IQR 13.5–31.6 hours) (see Chapter 7 ). This turnaround time might facilitate timely antiviral therapy but concerns have been raised that the demands of the test, requiring transportation of specimens to a laboratory with specialised expertise and equipment, and its turnaround time make it too slow for therapeutic or infection control purposes. 168–170
Commercial POCTs were used to manage patients with ILI during the 2009 H1N1 pandemic. 168,169,171,172 However, there have been mixed reports of the diagnostic accuracy of such tests, perhaps reflecting the test used, patient age, the method of sample collection and transport, the ‘gold standard’ used, and the type and subtype of influenza (i.e. whether seasonal or pandemic influenza, or influenza B). According to the manufacturer, the QuickVue Influenza A + B test detected all 24 influenza A viruses, subtypes H1–H15, which were isolated from birds and mammals, although performance characteristics were not established (see www.cliawaived.com/web/items/pdf/QDL-20183-Quidel_Influenza_Tests_Insert∼619file1.pdf).
We used the QuickVue (Quidel, USA) POCT in our study and found that it had a sensitivity of 33.3% and a specificity of 98.6% when compared with culture and 24.4% and 99.7%, respectively, when compared with RT-PCR (see Chapter 5 ). To compare our findings with other studies, we systematically reviewed published articles on the diagnostic accuracies of commercially available influenza POCTs. To assess the quality of the methodology and the completeness of the reporting of each manuscript, we ‘scored’ each publication using the QUADAS (quality assessment tool for diagnostic accuracy studies) tool and the Standards for Reporting of Diagnostic Accuracy (STARD) initiative. 173–176
Methods
Literature searches
On-line searches were made on MEDLINE/PubMed on 28 April 2011 and on the Bioscience Information Service (BIOSIS) and The Cochrane library on 27 May 2011 for publications on influenza POCT diagnostic accuracy studies between 1991 and 2011 (inclusive) that met the following five criteria:
-
Articles written in English.
-
Commercially available test kits.
-
Testing done in human seasonal and pandemic influenza.
-
Sensitivity results with specific numerators and denominators.
-
We had authorised journal access.
Medical subject heading (MeSH) search phrases included:
-
“QuickVue test influenza”.
-
“Rapid influenza test”.
-
“Rapid antigen test influenza”.
-
“POCT influenza”.
-
“Immunochromatographic test influenza”.
-
“Bedside test influenza”.
Figure 7 shows a flow diagram of the manuscript screening process, taken from the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guideline on systematic reviews. 177
FIGURE 7.
The manuscript screening process. a. Partial data from three publications.
Manuscript scoring
The QUADAS tool is an evidence-based scheme for the determination of the quality of both the methodology and the reporting of findings when doing systematic reviews. It consists of 14 questions called ‘items’ about the study patients, selection criteria, testing standards, results and clinical data that were answered with a ‘yes,’ ‘no’ or ‘unclear.’
The STARD initiative was another evidence-based tool used to assess the accuracy, completeness, and risk of bias in the systematic review of diagnostic accuracy studies. The STARD checklist consists of 25 questions pertaining to a study’s title, introduction, methodology, results and discussion sections; the more of the 25 items that are identified or described in the report, the more favourable the outcome.
Data analysis
For those studies that reported a full 2 × 2 table, i.e. numbers of true-positives, false-positives, false-negatives and true-negatives, or for which these could be calculated, pooled sensitivity and specificities were estimated using a bivariate mixed-effects meta-analysis model. 178 As well as summarising the overall diagnostic measures a hierarchical summary ROC curve was also estimated using the derived logit estimates of sensitivity, specificity and their respective variances. 179 Heterogeneity was assessed using the I 2 measure and explored using subgroup analyses using study-level covariates. 180 A number of studies reported only estimates of sensitivity, and in order to explore whether there was a selection effect between those studies that reported both sensitivity and specificity, and those that reported only sensitivity, a sensitivity analysis was undertaken in which the sensitivities were pooled separately for the two groups of studies using a standard random-effects meta-analysis model on the logit scale, and the pooled estimates compared.
Results
More than 2000 publications were found using the MeSH terms and 490 of these were relevant. In total, 70 out of the 490 publications met all five criteria and were selected for the systematic review. Some of the 70 had more than one finding, which we called ‘studies’. There were 143 studies altogether. Twenty-eight of the 70 publications reported full 2 × 2 data and were used for meta-analysis. There were a total of 68 studies in the 28 publications but four studies from three publications were excluded. 88,172,181 Thus 64 studies from 28 publications were included in the meta-analysis. Figure 7 shows a flow diagram of the manuscript screening process, taken from the PRISMA guidance on systematic reviews. 178
Appendix 6 summarises the publications that were screened and Table 26 summarises the sensitivities and specificities of the 64 POC studies that were included in the qualitative synthesis. Table 27 presents the QUADAS results and Table 28 the STARD results. Figure 8 shows the percentage of studies satisfying each of the QUADAS items and Figure 9 displays the distribution of the overall QUADAS score. As can be seen from Figure 9 , 40 of the 64 (62.5%) studies that were included in the meta-analysis scored > 10 indicating that the studies were of a reasonably high quality overall.
| Author | Location | Children, age | Adults, age | Influenza | Gold standard | POCT | Sensitivity (%) | Specificity (%) | PPV% | NPV% |
|---|---|---|---|---|---|---|---|---|---|---|
| Agoritsas, 200691 | USA | 2 weeks to 18 years | None | Seasonal | PCR and culture (R-mix, Diagnostics Hybrids, Athens, OH, USA) | QuickVue Influenza A + B | NPS: 85 (50/59) | NPS: 98 (62/63) | NPS: 98 (50/51) | NPS: 87 (62/71) |
| NPW: 69 (41/59) | NPW: 98 (62/63) | NPW: 98 (41/42) | NPW: 78 (62/80) | |||||||
| NS: 78 (46/59) | NS: 97 (61/63) | NS: 96 (46/48) | NS: 82 (61/74) | |||||||
| Bellei, 2003189 | Brazil | None | 18–56 years | Seasonal | Culture on MDCK cells | QuickVue Influenza A + B | 74 (29/39) | 83 (48/58) | – | – |
| Booth, 2006190 | Australia | 1–16 years | Included, age not specified | Seasonal, influenza A | Culture on MDCK cells and IFA | ImmunoCard Stat! Flu A&B (Meridian Bioscience, Cincinnati, OH, USA) | 80 (28/35) | 98 (185/189) | 88 (28/32) | 96 (185/192) |
| 1–16 years | Included, age not specified | Seasonal, influenza B | Culture on MDCK cells and IFA | ImmunoCard Stat! Flu A&B | 47 (7/15) | 100 (209/209) | 100 (7/7) | 96 (209/217) | ||
| 1–16 years | Included, age not specified | Seasonal, influenza A | Culture on MDCK cells and IFA | NOW Flu A + B (Binax, Portland, ME, USA) | 80 (28/35) | 99 (188/189) | 97 (28/29) | 96 (188/195) | ||
| 1–16 years | Included, age not specified | Seasonal, influenza B | Culture on MDCK cells and IFA | NOW Flu A + B | 47 (7/15) | 100 (208/209) | 88 (7/8) | 96 (208/216) | ||
| Cazacu 2004200 | USA | From 3 days | To 55.8 years | Seasonal | Culture on RMK cells | Directigen Flu A + B | 43.8 (96/219) | 99.7 (3863/3873) | 90.5 (96/106) | 96.9 (3863/3986) |
| Cazacu 2004201 | USA | Included, age not specified | Included, age not specified | Seasonal | Culture on RMK cells | XPECT® Flu A + B (Remel, Lenexa, KS, USA) | 94.4 (118/125) | 100 (275/275) | 100 (118/118) | 97.5 (275/282) |
| Chan, 2002147 | Hong Kong | From < 2 years | To > 55 years | Seasonal, influenza A | Culture on MDCK cells | Directigen EZ Flu A + B | 100 (22/22) | 99 (225/228) | 88 (22/25) | 100 (225/225) |
| From < 2 years | To > 55 years | Seasonal, influenza B | Culture on MDCK cells | Directigen EZ Flu A + B | 88 (28/32) | 97 (211/218) | 80 (28/35) | 98 (211/215) | ||
| Chen, 2002191 | China | Included, age not specified | Included, age not specified | Influenza A | Culture on MDCK cells | FluA Dot (Creative Diagnostics, Shirley, NY, USA) | 88.5(69/78) | 99 (146/147) | 99 (69/70) | 94 (146/155) |
| Chomel, 1992181 | France | Included, age not specified | Included, age not specified | Seasonal, influenza A | ELISA | Directigen Flu A | 87.9 (87/99) | 97 (59/61) | 83 (59/71) | 97 (87/89) |
| Covalciuc, 1999193 | USA | From 2 months | To 76 years | Seasonal | Culture on RMK cells | BioStar FLU OIA (Biostar, Boulder CO, USA) | 80.1 (121/151) | 73 (185/253) | ||
| Diederen, 2010195 | Netherlands | From birth | To 81 years | Pandemic | PCR | NOW Flu A + B | 47 (18/38) | 95 (92/97) | ||
| Hamilton, 2002192 | USA | 12 days to 19 years | None | Seasonal | Culture on RMK cells | Directigen Flu A + B | 75 (49/65) | 93 (218/235) | 74 (49/66) | 93 (218/234) |
| 12 days to 19 years | None | Seasonal | Culture on RMK cells | Zstat Flu-II (Zyme Tx, Oklahoma City, OK, USA) | 88 (57/65) | 92 (216/235) | 75 (57/76) | 96 (216/224) | ||
| Hara, 2008187 | Japan | 4 months to 19 years | None | Seasonal, influenza A | Culture on MDCK cells | Directigen EZ Flu A + B | 100 (53/53) | 99 (438/441) | 100 (53/53) | 100% (441/441) |
| 4 months to 19 years | None | Seasonal, influenza B | Culture on MDCK cells | Directigen EZ Flu A + B | 76 (206/270) | 99 (221/224) | 99 (206/209) | 78 (221/285) | ||
| 4 months to 19 years | None | Seasonal, influenza A | Culture on MDCK cells | Fujirebio Espline® Influenza A&B-N (Fujirebio Diagnostics, Malvern, PA, USA) | 100 (53/53) | 100 (441/441) | 100% (53/53) | 100 (441/441) | ||
| 4 months to 19 years | None | Seasonal, influenza B | Culture on MDCK cells | Fujirebio Espline Influenza A&B-N | 86 (232/270) | 100 (224/224) | 100 (232/232) | 85% (224/262) | ||
| 4 months to 19 years | None | Seasonal, influenza A | Culture on MDCK cells | NOW Flu A + B | 100 (53/53) | 98 (430/441) | 100 (531/531) | 100 (441/441) | ||
| 4 months to 19 years | None | Seasonal, influenza A | Culture on MDCK cells | NOW Flu A + B | 78 (211/270) | 99 (221/224) | 99 (211/214) | 79 (221/280) | ||
| Hawkes, 2010184 | Canada | 1 month to 17 years | None | Pandemic | PCR | NOW Flu A + B | 62 (66/107) | 99 (70/71) | 99 (66/67) | 63% (70/111) |
| 1 month to 17 years | None | Influenza A | DFA | NOW Flu A + B | 69 (72/104) | 99 (567/575) | 90 (72/80) | 95 (567/599) | ||
| 1 month to 17 years | None | Influenza A | Culture and DFA | NOW Flu A + B | 68 (75/111) | 99 (590/596) | 93 (75/81) | 94 (590/626) | ||
| 1 month to 17 years | None | Influenza B | DFA | NOW Flu A + B | 72 (51/71) | 100 (605/608) | 93 (75/81) | 97 (605/625) | ||
| 1 month to 17 years | None | Influenza B | Culture and DFA | NOW Flu A + B | 68 (52/77) | 100 (628/630) | 96 (52/54) | 96 (628/653) | ||
| Hurt 200788 | Australia | From 4 days | To 64 years | Influenza A | RETCIF | Directigen EZ Flu A + B | 69 (34/49) | 100 (128/128) | 100 (34/34) | 90 (128/143) |
| From 4 days | To 64 years | Influenza B | RETCIF | Directigen EZ Flu A + B | 30 (3/10) | 100 (167/167) | 100 (3/3) | 96 (167/174) | ||
| From 4 days | To 64 years | Influenza A | RETCIF | Denka Seiken Quick Ex-Flu (Denka Seiken, Tokyo, Japan) | 71 (35/49) | 100 (128/128) | 100 (35/35) | 90 (128/142) | ||
| From 4 days | To 64 years | Influenza B | RETCIF | Denka Seiken Quick Ex-Flu | 30 (3/10) | 100 (167/167) | 100 (3/3) | 96 (167/174) | ||
| From 4 days | To 64 years | Influenza A | RETCIF | Fujirebio Espline Influenza A&B-N | 67 (33/49) | 100 (128/128) | 100 (5/5) | 89 (128/144) | ||
| From 4 days | To 64 years | Influenza B | RETCIF | Fujirebio Espline Influenza A&B-N | 30 (3/10) | 100 (167/167) | 100 (3/3) | 96 (167/174) | ||
| From 4 days | To 64 years | Influenza B | RETCIF | Influenza Antigen test (Rockeby BioMed, Singapore) | 10 (5/49) | 100 (128/128) | 100 (5/5) | 74 (128/172) | ||
| From 4 days | To 64 years | Influenza A | RETCIF | NOW Flu A + B | 73 (36/49) | 99 (127/128) | 97 (36/37) | 91 (127/140) | ||
| From 4 days | To 64 years | Influenza B | RETCIF | NOW Flu A + B | 30 (3/10) | 100 (167/167) | 100 (3/3) | 96 (167/174) | ||
| Kim, 2010202 | Korea | From 2 months | To 78 years | Pandemic | PCR | SD Bioline influenza A/B/A H1N1 pandemic rapid test | 70 (182/260) | 98 (677/688) | 94 (182/193) | 90 (677/755) |
| Lucas, 2011198 | USA | From < 5 years | To < 60 years | Influenza A | PCR | QuickVue Influenza A + B | 15 (2/13) | 99 (1464/1479) | 12 (2/17) | 99 (1464/1475) |
| From < 5 years | To < 60 years | Influenza B | PCR | QuickVue Influenza A + B | 31 (4/13) | 99 (610/615) | 44 (4/9) | 98 (610/619) | ||
| From < 5 years | To < 60 years | Pandemic | PCR | QuickVue Influenza A + B | 20 (6/30) | 99 (1464/1479) | 29 (6/21) | 98 (1076/1096) | ||
| Nougairede, 2010183 | France | Included, age not specified | Included, age not specified | Pandemic | PCR | Directigen EZ Flu A + B | 57.7 (64/111) | 100 (1910/1910) | 100 (64/64) | 97.5 (1910/1957) |
| Quach, 2002199 | Canada | Included, age not specified | None | Seasonal | Culture on RMK and MDCK cells | QuickVue Influenza A + B | 79.2 (42/53) | 82.6 (204/247) | 49.4 (42/85) | 94.9 (204/215) |
| Rahman, 2007196 | USA | From 6 months | To ≥ 65 years | Seasonal | Culture on RMK cells | Directigen EZ Flu A + B | 42 (18/43) | 96 (72/75) | 86 (18/21) | 74 (72/97) |
| Rahman, 2008151 | USA | From 6 months | To ≥ 65 years | Seasonal | PCR and culture on RMK cells | NOW Flu A + B | 61 (11/18) | 100 (55/55) | 100 (11/11) | 89 (55/62) |
| Rashid, 2007182 | UK and Saudi Arabia | From 1 year | To 85 years | Seasonal | PCR | QuickVue Influenza A + B | 22 (13/58) | 99 (492/497) | 72 (13/18) | 92 (492/537) |
| Rodriguez, 2002185 | USA | From < 3 years | To > 21 years | Seasonal | Culture and DFA | BioStar OIAFlu A/B | 93 (54/58) | 82 (47/57) | 84% (54/64) | 92% (47/51) |
| From < 3 years | To > 21 years | Seasonal | Culture and DFA | Directigen EZ Flu A + B | 95 (55/58) | 84 (49/58) | 86 (55/64) | 94% (49/52) | ||
| From < 3 years | To > 21 years | Seasonal | Culture and DFA | QuickVue Influenza A + B | 95 (54/57) | 76 (42/55) | 81 (54/67) | 93 (42/45) | ||
| From < 3 years | To > 21 years | Seasonal | Culture and DFA | Zstat Flu AB | 72 (41/57) | 83 (48/58) | 80 (41/51) | 75 (48/64) | ||
| Ruest, 2003152 | Canada | From 1 day | To 98 years | Seasonal | Culture on MDCK cells | Directigen Flu A + B | 86 (57/66) | 94 (115/122) | 89% (57/64) | 92 (115/124) |
| From 1 day | To 98 years | Seasonal | PCR | Directigen Flu A + B | 80 (63/79) | 98 (121/123) | 97 (63/65) | 88 (121/137) | ||
| From 1 day | To 98 years | Seasonal | Culture on MDCK cells | QuickVue Influenza A + B | 91 (64/70) | 86 (105/122) | 79 (64/81) | 95 (105/111) | ||
| From 1 day | To 98 years | Seasonal | PCR | QuickVue Influenza A + B | 86 (72/84) | 92 (127/138) | 87 (72/83) | 90 (114/127) | ||
| Sabetta, 2009186 | USA | Included, age not specified | Included, age not specified | Pandemic | PCR | XPECT Flu A + B | 47 (23/49) | 86 (12/14) | 92 (23/25) | 32 (12/38) |
| Scansen, 201090 | USA | ≤ 17 years | None | Influenza A | PCR | QuickVue Influenza A + B | Swabs: | Swabs: | Swabs: | Swabs: |
| Foam 81 (30/37) | Foam 98 (62/63) | Foam 97 (30/31) | Foam 90 (81/90) | |||||||
| Flocked 59 (22/37) | Flocked 98 (62/63) | Flocked 96 (22/23) | Flocked 81 (62/77) | |||||||
| ≤ 17 years | None | Influenza B | PCR | QuickVue Influenza A + B | Swabs: | Swabs: | Swabs: | Swabs: | ||
| Foam 53 (10/19) | Foam 100 (81/81) | Foam 100 (10/10) | Foam 90 (81/90) | |||||||
| Flocked 42 (8/19) | Flocked 100 (81/81) | Flocked 100 (8/8) | Flocked 88 (81/92) | |||||||
| ≤ 17 years | None | Influenza A | Culture: R-Mix and DFA | QuickVue Influenza A+B | Swabs: | Swabs: | Swabs: | Swabs: | ||
| Foam 85 (29/34) | Foam 97 (64/66) | Foam 94 (29/31) | Foam 93 (64/69) | |||||||
| Flocked 65% (22/34) | Flocked 97 (64/66) | Flocked 92 (22/24) | Flocked 84 (64/76) | |||||||
| ≤ 17 years | None | Influenza B | Culture: R-Mix and DFA | QuickVue Influenza A + B | Swabs: | Swabs: | Swabs: | Swabs: | ||
| Foam 60 (9/15) | Foam 99 (84/85) | Foam 90 (9/10) | Foam 93 (84/90) | |||||||
| Flocked 53 (8/15) | Flocked 100 (85/85) | Flocked 100 (8/8) | Flocked 92 (85/9) | |||||||
| Shoji, 2009188 | Japan | From 1 year | To 88 years | Seasonal | Culture | QuickVue Influenza A + B | 87.8 (72/82) | Flu A: 90.2 (322/357) | ||
| Waner, 1991194 | USA | From < 2 months | None | Seasonal | Culture on RMK cells | Directigen Flu A | 100 (23/23) | 91.6 (153/167) | 62 (23/37) | 100 (153/153) |
| From < 2 months | None | Seasonal | IFA | Directigen Flu A | 100 (24/24) | 95 (152/160) | 75 (24/32) | 100 (152/152) | ||
| Yoo, 2007197 | Korea | 3 months to 15 years | 18.5–81.9 years | Seasonal | Culture (R-mix) | QuickVue Influenza A + B | 46.7% (35/75) | 94.5% (208/220) | 74.5% (35/47) | 83.9% (208/248) |
| 3 months to 15 years | 18.5–81.9 years | Influenza A | Culture (R-mix) | SD Bioline Influenza A/B/A(H1N1) Pandemic rapid test | 61.9 (26/42) | 96.8 (245/253) | 76.5 (26/34) | 93.9 (245/261) | ||
| 3 months to 15 years | 18.5–81.9 years | Influenza B | Culture (R-mix) | SD Bioline Influenza A/B/A(H1N1) Pandemic rapid test | 54.5 (18/33) | 100 (262/262) | 100 (18/18) | 94.6 (262/277) |
| Methodology checklist | Agoritsas, 200691 | Bellei, 2003189 | Booth, 2006190 | Cazacu, 2004201 | Cazacu, 2004200 | Chan, 2002147 | Chen, 2010191 | Chomel, 1992181 | Covalciuc, 1993193 | Diederen, 2010195 | Hamilton, 2002192 | Hara, 2008187 | Hawkes, 2010184 | Hurt, 200788 | Kim, 2010202 | Lucas, 2011198 | Nougairede, 2010183 | Quach, 2002199 | Rahman, 2007196 | Rahman, 2008151 | Rashid, 2007182 | Rodriguez, 2002185 | Ruest, 2003152 | Sabetta, 2009186 | Scansen, 201090 | Shoji, 2009188 | Waner, 1991194 | Yoo, 2007197 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patients representative of who will receive test in practice? | N | N | Y | U | Y | Y | U | U | Y | Y | N | N | N | Y | Y | Y | Y | N | Y | Y | Y | N | Y | N | N | U | N | Y |
| Selection criteria clearly described? | Y | Y | Y | Y | Y | Y | N | Y | Y | Y | Y | N | Y | Y | N | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Reference standard likely to correctly classify target condition? | Y | N | N | N | N | Y | N | N | N | Y | Y | Y | Y | N | Y | Y | Y | N | N | Y | Y | N | Y | Y | Y | Y | N | N |
| Short time period between index and standard tests? | U | N | N | N | N | N | N | N | N | U | N | U | U | U | Y | U | Y | U | N | U | N | Y | N | U | Y | U | N | U |
| Verification with reference standard? | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | N | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Samples received same reference standard? | Y | Y | Y | Y | Y | Y | Y | N | Y | Y | Y | Y | Y | Y | Y | N | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Reference standard independent of index test? | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Execution of index test described in detail? | Y | Y | Y | Y | Y | Y | N | Y | Y | N | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | N | Y | Y | Y | U | Y | Y |
| Execution of reference standard described in detail? | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | N | Y | N | Y | U | Y | Y |
| Index test results interpreted without knowledge of reference test result? | Y | N | N | Y | Y | Y | U | N | U | U | Y | Y | Y | Y | Y | U | U | Y | Y | Y | Y | U | U | N | U | U | U | Y |
| Reference test results interpreted without knowledge of index test result? | U | Y | Y | U | U | U | U | Y | U | U | U | U | U | U | U | U | U | Y | U | U | U | U | U | N | U | U | U | U |
| Routine clinical data available? | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | N | Y | Y | Y | Y | Y |
| Uninterpretable/intermediate results reported? | N | Y | Y | N | N | Y | N | N | N | N | Y | N | N | N | N | N | N | Y | N | N | N | N | Y | N | N | U | Y | N |
| Study withdrawals explained? | Y | Y | Y | Y | Y | Y | U | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | N |
| STARD checklist | Agoritsas, 200691 | Bellei, 2003189 | Booth, 2006190 | Cazacu, 2004201 | Cazacu, 2004200 | Chan, 2002147 | Chen, 2010191 | Chomel, 1992181 | Covalciuc, 1993193 | Diederen, 2010195 | Hamilton, 2002192 | Hara, 2008187 | Hawkes, 2010184 | Hurt, 200788 | Kim, 2010202 | Lucas, 2011198 | Nougairede, 2010183 | Quach, 2002199 | Rahman, 2007196 | Rahman, 2008151 | Rashid, 2007182 | Rodriguez, 2002185 | Ruest, 2003152 | Sabetta, 2009186 | Scansen, 201090 | Shoji, 2009188 | Waner, 1991194 | Yoo, 2007197 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Identify the article as diagnostic accuracy study | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Stated study aims | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Specified inclusion and exclusion criteria? | Y | Y | Y | Y | Y | Y | N | Y | Y | Y | Y | N | Y | Y | N | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| What was recruitment based upon? | PS | a | PS | PS | PS | PS | PS | PS | PS | PS | PS | PS | PS | PS | PS | PS | PS | PS | PS | PS | PS | PS | PS | PS | PS | PS | PS | PS |
| Consecutive series of participants? | Y | Y | Y | Y | Y | Y | N | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| If not, specify further selection process? | U | |||||||||||||||||||||||||||
| Prospective or Retrospective study? | P | R | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P |
| Rationale for reference standard? | Y | Y | Y | N | Y | Y | N | Y | Y | Y | Y | Y | Y | Y | Y | Y | N | Y | Y | Y | N | N | Y | Y | Y | NA | Y | Y |
| Technical specification of materials and methods? | N | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | NA | Y | Y |
| Definition of, and rationale for, units, cut-offs and/or categories of results? | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | NA | Y | Y |
| Number, training and expertise of the technicians? | Y | N | N | Y | N | Y | N | N | N | N | N | N | N | N | N | N | Y | N | Y | Y | N | Y | N | Y | N | NA | N | Y |
| Blinding of results? | N | N | Y | N | N | Y | N | N | N | N | N | N | N | N | N | N | N | Y | N | N | N | Y | Y | N | N | NA | N | N |
| Specify data analysis methods? | Y | N | Y | Y | Y | Y | N | N | Y | N | Y | Y | Y | N | Y | Y | N | N | Y | Y | N | N | Y | N | Y | NA | N | Y |
| Specify methods for calculating reproducibility, if done? | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Include beginning and end dates of recruitment? | Y | Y | Y | Y | Y | Y | N | Y | Y | N | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | N | Y |
| Include participant clinical and demographic data? | Y | Y | Y | N | Y | Y | N | N | Y | Y | Y | Y | Y | Y | Y | Y | Y | N | Y | Y | Y | Y | Y | Y | N | NA | Y | Y |
| Include no. of participants satisfying the inclusion criteria? | Y | Y | Y | Y | Y | Y | N | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | N | Y | Y | Y |
| Stated time interval between index and reference test? | N | N | Y | Y | Y | Y | N | Y | N | N | Y | N | N | N | Y | N | Y | N | Y | N | N | Y | Y | N | N | NA | Y | N |
| Included distribution of disease severity? | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | N | N | NA | NA | NA |
| Cross-tabulation of index test results with reference standard? | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Indicated any adverse events from the test procedure? | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Provided estimates of diagnostic accuracy and statistical uncertainty (e.g. 95% CI)? | Y | N | N | Y | N | N | N | N | N | Y | N | N | Y | N | Y | Y | N | Y | Y | Y | N | N | Y | N | Y | NA | N | N |
| Specify how indeterminate results and outliers were handled? | N | Y | Y | N | N | Y | N | Y | N | N | Y | N | N | N | N | N | N | Y | N | N | N | N | Y | N | N | NA | Y | N |
| Provided estimates of variability of diagnostic accuracy among subgroups, readers or centres? | Y | NA | Y | N | Y | NA | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | NA | Y | Y |
| Provided estimates of test reproducibility, if done? | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Discuss clinical applicability of findings? | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | N | Y | Y | Y | Y | Y | Y | Y | Y |
FIGURE 8.
Percentage of studies satisfying QUADAS items.
FIGURE 9.
Distribution of QUADAS Score.
Of the 143 studies in Appendix 6 for which data could be extracted, 64 (45%) reported the full 2 × 2 table, and using a bivariate mixed-effects meta-analysis model produced an overall estimate of sensitivity of 0.73 (95% CI 0.66 to 0.80) and of specificity of 0.99 (95% CI 0.98 to 0.99). However, there was a high level of heterogeneity between the studies for both outcomes (sensitivity: Q = 777.5, p < 0.01, I 2 = 91.9%, 95% CI 90.5% to 93.3%; specificity: Q = 2128.9, p < 0.01, I 2 = 97.0%, 95% CI 96.7% to 97.4%), as can be seen from the Forest plots in Figure 10 . Figure 11 displays the summary receiver operating characteristic (SROC) curve derived using the estimated overall pooled sensitivity and specificity. Superimposed on the SROC curve are the results from 3WS using (1) PCR (sensitivity 24.4%, specificity 99.7%) and (2) viral culture (sensitivity 33.3%, specificity 98.6%) as the ‘Gold standard’ tests. As can be seen from Figure 11 , the 3WS results, although being within the associated prediction region, are nevertheless considerably lower, in terms of sensitivity, than those estimates from other studies in the meta-analysis.
FIGURE 10.
Forest plot of sensitivity and specificity for 64 studies reporting full 2 × 2 data table.
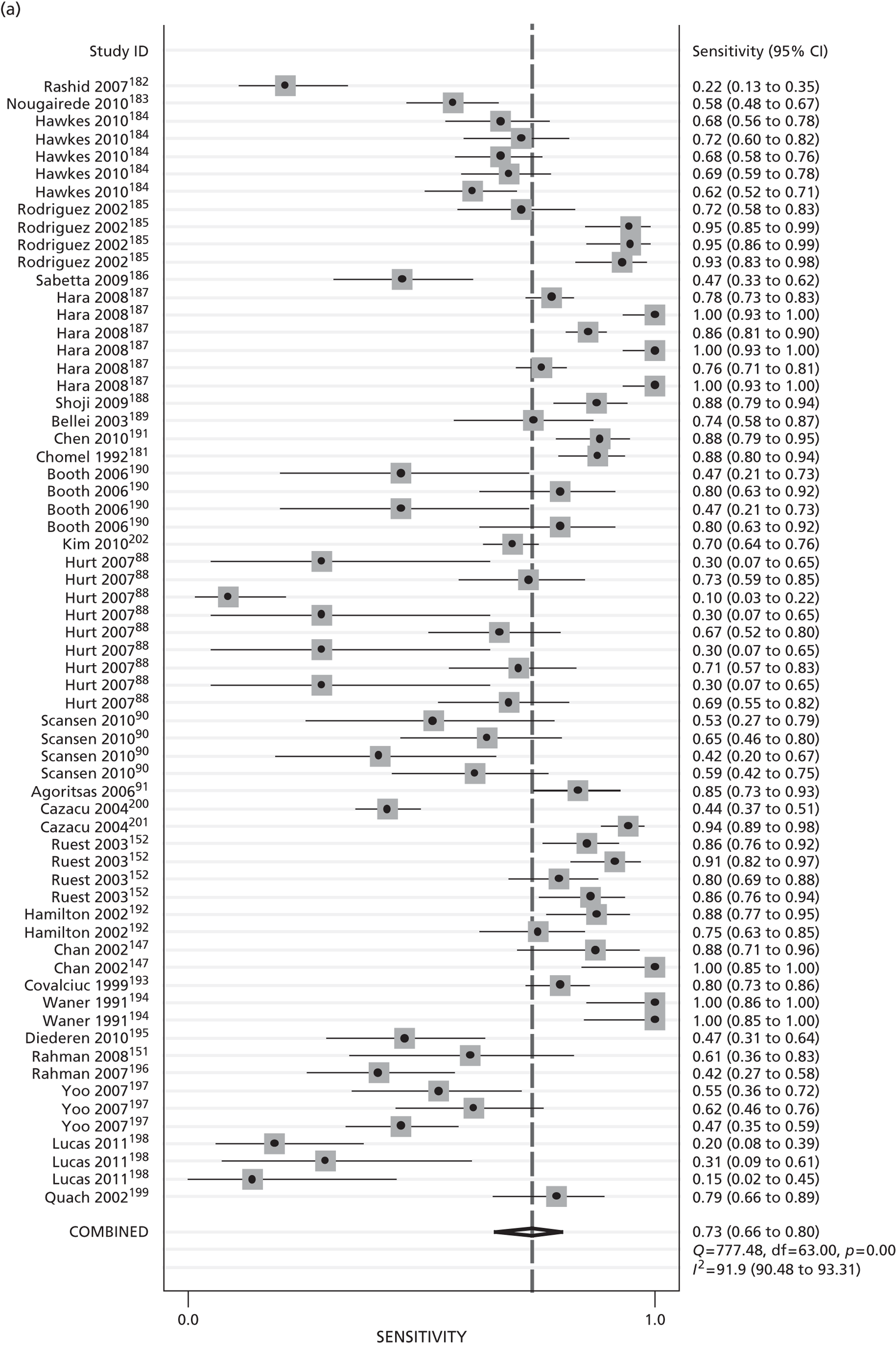
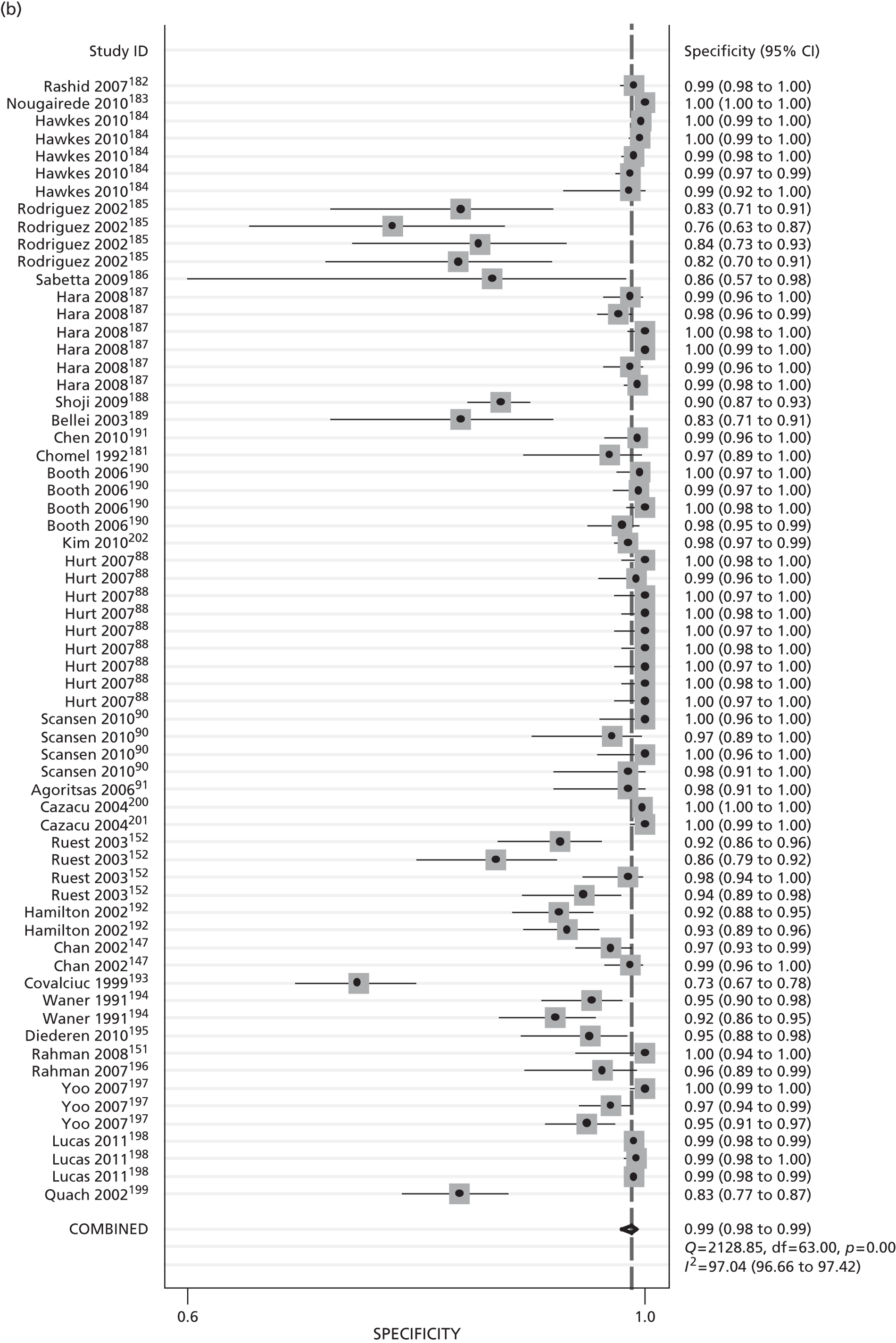
FIGURE 11.
Summary receiver operating characteristic estimated using bivariate mixed-effects meta-analysis model with 3WS results using PCR (a) and viral culture (b) as ‘gold standard’ superimposed.
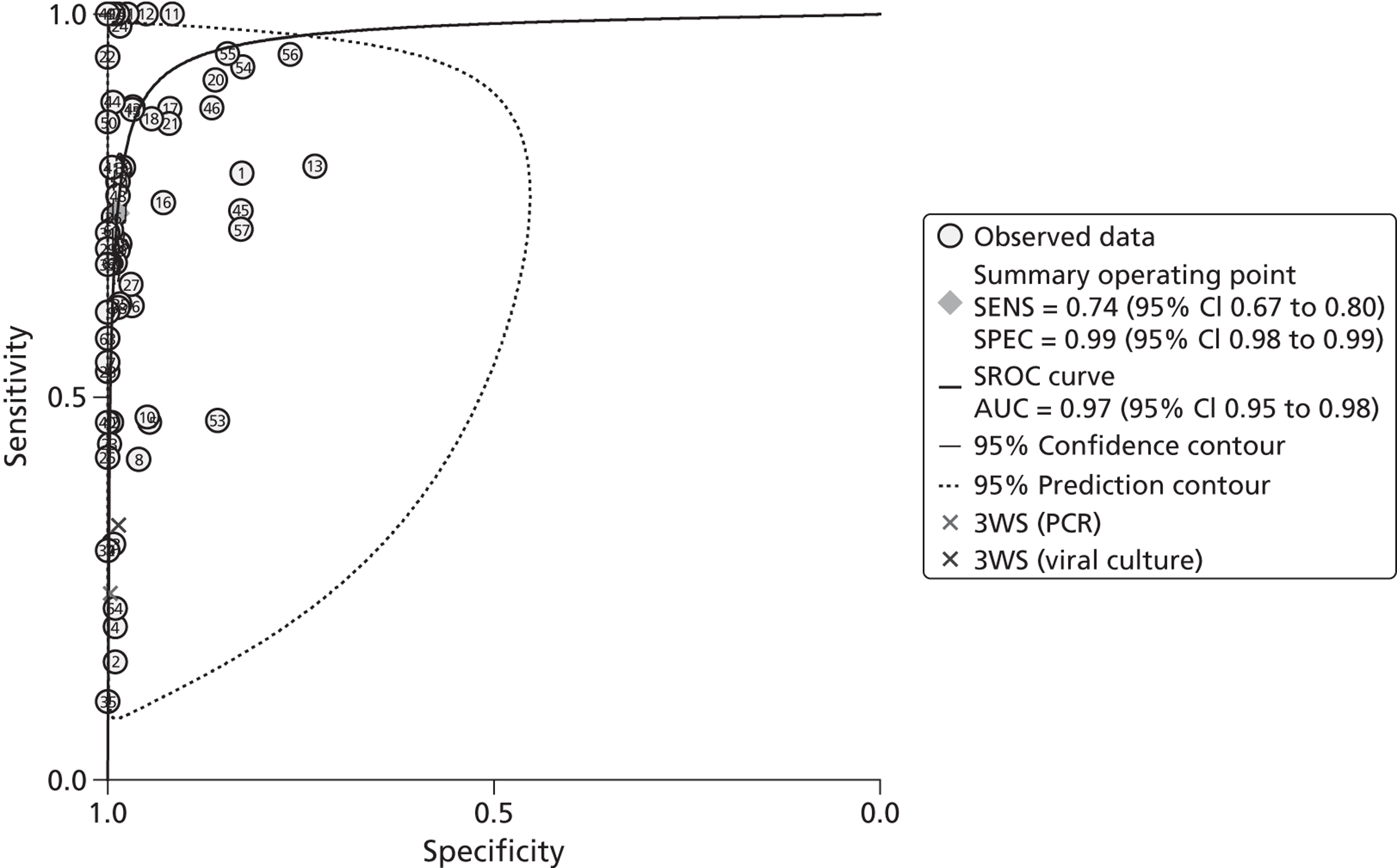
To explore the between-study heterogeneity observed, a number of subgroup-specific models were estimated using study-level covariates (age of participants, ‘gold standard’ used, geographical region in which study was conducted, type of influenza tested for, type of POCT, and study quality assessed using the QUADAS tool). Owing to the relatively small numbers of studies in some specific subgroups (i.e. < 4) it was not always possible to estimate the associated effects. It can seen from Table 29 that the pooled estimates of specificity across subgroups were consistently high with relatively little subgroup-to-subgroup variation; however, for sensitivity there was variation in the pooled estimates across subgroups with a mixed age distribution, use of PCR as a ‘gold standard’, and testing for influenza of swine origin all yielding lower estimates of sensitivity. There was also some variation in sensitivity depending on the geographical region in which the study was conducted, with Europe and Australasia yielding lower estimates; this was also seen for study quality, with ‘higher’-quality studies producing a lower pooled sensitivity than those of ‘lower’ quality.
| Covariate/level | No. of studies | Measure | Estimate | 95% CI |
|---|---|---|---|---|
| Age | ||||
| Children and adolescents | 21 | Sensitivity | 0.86 | 0.75 to 0.93 |
| Specificity | 0.99 | 0.97 to 0.99 | ||
| Mixed | 43 | Sensitivity | 0.67 | 0.58 to 0.75 |
| Specificity | 0.99 | 0.98 to 0.99 | ||
| ‘Gold standard’ | ||||
| PCR | 13 | Sensitivity | 0.51 | 0.38 to 0.64 |
| Specificity | 0.99 | 0.97 to 0.99 | ||
| Culture | 24 | Sensitivity | 0.86 | 0.77 to 0.92 |
| Specificity | 0.98 | 0.95 to 0.99 | ||
| Culture and IF | 13 | Sensitivity | 0.78 | 0.66 to 0.87 |
| Specificity | 0.98 | 0.94 to 0.99 | ||
| IF | 11 | Sensitivity | 0.51 | 0.35 to 0.66 |
| Specificity | 1.00 | 0.97 to 1.00 | ||
| Region | ||||
| North America | 32 | Sensitivity | 0.76 | 0.66 to 0.83 |
| Specificity | 0.97 | 0.95 to 0.98 | ||
| Australasia | 13 | Sensitivity | 0.53 | 0.38 to 0.67 |
| Specificity | 1.00 | 0.99 to 1.00 | ||
| Asia | 14 | Sensitivity | 0.89 | 0.75 to 0.96 |
| Specificity | 0.99 | 0.97 to 0.99 | ||
| Europe | 4 | Sensitivity | 0.56 | 0.29 to 0.80 |
| Specificity | 0.99 | 0.93 to 1.00 | ||
| Type of influenza | ||||
| Influenza A | 19 | Sensitivity | 0.81 | 0.64 to 0.91 |
| Specificity | 0.99 | 0.98 to 0.99 | ||
| Influenza B | 16 | Sensitivity | 0.59 | 0.47 to 0.70 |
| Specificity | 1.00 | 0.99 to 1.00 | ||
| Seasonal | 23 | Sensitivity | 0.84 | 0.74 to 0.90 |
| Specificity | 0.94 | 0.90 to 0.97 | ||
| Pandemic (swine origin) | 6 | Sensitivity | 0.52 | 0.39 to 0.65 |
| Specificity | 0.99 | 0.94 to 1.00 | ||
| Testing kit | ||||
| Directigen EZ | 16 | Sensitivity | 0.85 | 0.71 to 0.93 |
| Specificity | 0.99 | 0.97 to 0.99 | ||
| BinaxNOW | 13 | Sensitivity | 0.69 | 0.58 to 0.79 |
| Specificity | 0.99 | 0.99 to 0.99 | ||
| Quidel QuickVue | 16 | Sensitivity | 0.66 | 0.48 to 0.80 |
| Specificity | 0.96 | 0.93 to 0.98 | ||
| Study qualitya | ||||
| ‘Low’ (QUADAS ≤ 10) | 24 | Sensitivity | 0.85 | 0.72 to 0.93 |
| Specificity | 0.97 | 0.94 to 0.98 | ||
| ‘High’ (QUADAS > 10) | 40 | Sensitivity | 0.67 | 0.59 to 0.75 |
| Specificity | 0.99 | 0.99 to 1.00 | ||
In terms of the subgroup specific estimates of pooled sensitivity, even the lower estimates were still higher than those found in 3WS – using PCR as the ‘gold standard’, a Quidel POCT produced a sensitivity of 24.4%, whereas using viral culture as the ‘gold standard’ it was 33.3%. Although the distribution of studies across the various study-level covariates (and their levels) makes it difficult to obtain an estimate of sensitivity that closely matches the characteristics of 3WS – it is possible to estimate a pooled effect for those studies (n = 5), which (1) included both adults and children (as opposed to only children) and (2) compared Quidel with PCR. These produced an overall pooled sensitivity of 34% (95% CI 14% to 62%) and specificity of 99% (95% CI 97% to 100%), which are much more similar to those obtained in 3WS.
As only 64 out of 143 studies (45%) reported sufficient data to permit a formal bivariate analysis to be undertaken, a sensitivity analysis only pooling sensitivities for the 64 and 79 studies separately was undertaken to assess whether there was in fact a selection effect. The 79 studies produced a pooled sensitivity of 60.2% (59.0% to 61.0%), whereas the 64 studies produced an estimate of 69.1% (67.5% to 70.6%) – the latter was slightly lower than that produced by the bivariate model (73%, 95% CI 66% to 80%). A formal test of heterogeneity between the two sets of studies was highly significant (p < 0.001) indicating that had the 79 studies reported both sensitivity and specificity the corresponding bivariate model would have produced an estimate of sensitivity lower than that observed.
Discussion
This systematic review concurs with other reviews of diagnostic studies in different disease areas in that there was considerable heterogeneity – both in terms of reporting of data and clinical and methodological characteristics, thus making formal synthesis of study results using appropriate bivariate meta-analysis methods challenging. 203
Overall, the bivariate meta-analysis produced estimates of sensitivity that were considerably higher than that observed in 3WS. However, exploration of the considerable between-study heterogeneity using subgroup analyses showed that for some subgroup combinations the pooled estimate of sensitivity was considerably lower than that estimated for others. In fact, the subgroup combination most closely resembling the characteristics of 3WS (comparing Quidel POCT with PCR in a mixed-age population) produced an estimate of sensitivity in close agreement with 3WS.
Further sensitivity analysis comparing those studies that reported fully data for sensitivity and specificity with those that only reported sensitivity indicated the possibility of a selection effect that would further reduce the true estimate of sensitivity of POCT for testing for influenza.
Published evidence on the usefulness of diagnostic tests has been summarised in four systematic reviews including this review. Uyeki204 reviewed published evidence on clinically useful diagnostic tests and antiviral treatment for influenza virus infections in children, which were published in the English language from 1966 to September 2002. The topics covered were wide-ranging, including clinical diagnosis, IF and rapid influenza diagnostic tests, as well as antiviral treatment. Altogether 28 studies of rapid influenza tests were identified. This was a descriptive study with no formal assessment of study quality or meta-analysis of the findings, rather the author presented median sensitivity values for the tests in comparison with cell culture as the gold standard. Overall, the POCTs had sensitivities and specificities of 40.4% to 100% and 65.2% to 100%, respectively. The median sensitivity of the Zstat Flu test (Zyme Tx, Oklahoma City, OK, USA) was 68.8% (range 28.1–96%) and median specificity was 83% (range 62.7–92.4%). The median sensitivity of the Directigen Flu A test was 87.2% (range 39–100%), and the median specificity was 98.1% (range 84–100%).The median sensitivity of the FLU OIA test (Biostar, Boulder, CO, USA) was 71.8% (range 36.7–93%), and the median specificity was 82% (range 65.2–95.7%). In five studies of the QuickVue Influenza Test the median sensitivity was 79.2% (range 74–95%) and the median specificity was 91.9% (range 76–98%). The studies were evidently heterogeneous in terms of age and the author concluded that rapid influenza diagnostic tests were ‘moderately to reasonably’ accurate for detecting influenza virus infections, and that false-negative results appeared more common than false-positive results.
Petrozzino et al. 205 were supported by the Quidel Corporation, the manufacturer of the QuickVue Influenza A + B test, to undertake a systematic review and meta-analysis of studies reporting sensitivity, specificity, and effects of ‘rapid flu tests’ (RFTs) and clinical diagnosis on decision-making for patients with ILI. Search results were limited to literature published in English between 1984 and 2009. Results from included studies were stratified according to age categories with an approximate cut-off of 15 years of age. It was not possible to stratify results for older people aged > 60 years. No RCTs were found directly comparing RFTs against the clinical diagnostic skills of clinicians. All included studies used an independent gold standard test for confirmatory influenza diagnosis. Separately, these investigators evaluated the clinical diagnosis of influenza.
Among older subjects aged ≥ 15 years, data on the QuickVue test from five studies showed that this POCT had a sensitivity in a fixed-effects model of 57% (52–62%) and specificity of 96% (95–97%). In a random-effects model, the POCT had a sensitivity of 61% (36–81%) and specificity of 96% (94–98%). Data from 11 studies showed the sensitivity of clinical diagnosis in a fixed-effects model to be 64% (51–75%) and much lower specificity of 65% (63–66%). In the random-effects model, clinical diagnosis had a sensitivity of 64% (51–75%) and specificity of 68% (57–77%). Thus clinicians were as able to diagnose influenza clinically as POC testing but wrongly identified other individuals as having influenza, which could be problematic when isolation facilities for adults are scarce, although it is conceivable that these patients also posed an infection risk to others.
Among younger subjects aged < 15 years, data on the QuickVue test from 14 studies showed that this POCT had a sensitivity in a fixed-effects model of 63% (60–67%) and specificity of 94% (92–95%). In a random-effects model, the POCT had a sensitivity of 76% (65–85%) and specificity of 95% (92–98%). Data from five studies showed the sensitivity of clinical diagnosis in a fixed-effects model to be 70% (66–74%), and the specificity of 61% (59–63%) was again lower than that of the POCT. In the random-effects model, clinical diagnosis had a sensitivity of 69% (44–87%) and specificity of 63% (31–87%). For all age groups combined, the sensitivities of the POCT and clinical diagnosis in both the fixed and random-effects models were similar, with sensitivities of 61% (59–64%) and 62% (60–63%) respectively in the fixed-effects model, and 72% (62–81%) and 65% (55–74%) in the random-effects model. The respective sensitivities were 94% (93–95%) and 63% (62–64%) in the fixed model, and 96% (93–97%) and 67% (57–76%) in the random-effects model.
These authors examined 10 studies reporting outcomes relating to patient management associated with the use of POCT for influenza. This overview led the authors to conclude that in various clinical settings and across a wide age range, RFT use in patients presenting with ILI leads to reduced diagnostic testing, antibiotic use and emergency department length of stay, although increases antiviral prescribing.
Babin et al. 206 did a review and meta-analysis of published literature on the 2009 novel swine flu outbreak to assess the potential utility of POCTs for initiating infection treatment and control for this pathogen. Although these POCTs were not developed for swine-origin virus, their speed and ease of use (EoU) made them attractive for clinical and public health use. These authors identified 14 reports on sensitivity and/or specificity of seven different POC influenza tests for diagnosis of 2009 pandemic H1N1 virus on clinical specimens. The pooled sensitivity and specificity for all studies were 67.5% (95% CI 66.2% to 68.9%) and 80.7% (95% CI 80.0% to 81.4%). Pooled data were provided for three POCTs from different manufacturers: BinaxNOW Influenza A&B, with a pooled sensitivity of 31.4% (95% CI 26.3% to 36.7%) (no specificity data); Directigen EZ Flu A + B, with a pooled sensitivity of 52.8% (95% CI 45.9% to 59.6%)%) (no specificity data); and QuickVue A + B, with a pooled sensitivity of 73.6% (95% CI 72.1% to 75.0%), and specificity of 76.6% (95% CI 75.5% to 77.5%). In conclusion, the authors considered that the relatively poor performance of the POCTs affirmed recommendations by the US Centers for Disease Control and Prevention (CDCP) that caution should be applied in the interpretation of negative POCTs.
Our systematic review and meta-analysis confirms and extends the observations in the above reports. In our study, specificity across subgroups was consistently high but sensitivity was higher in studies involving children and adolescents than in ‘mixed’ populations (i.e. mixed age groups). We may speculate that this might reflect decreased virus shedding in adults than in young children (although virus shedding may be high in very elderly hospitalised patients)125 and the effects of vaccination and past infection. We also found that the test sensitivity was a function of the nature of the gold standard used, with PCR setting a higher target than virus culture. The caution issued by the CDCP regarding the sensitivity of POCTs for the detection of the 2009 pandemic ‘swine-origin’ H1N1 virus was affirmed in our analyses, which also showed better performance of POCTs in the detection of seasonal influenza type A virus than type B virus.
Chapter 7 Ease and speed of use of point-of-care, molecular and traditional test methods for the diagnosis of respiratory infections
Background
The benefits of rapid diagnosis are increasingly appreciated in diverse health-care settings. Besides direct clinical benefits, political and economic imperatives, including the UK NHS 4-hour patient assessment rule in A&E departments, are likely to accelerate demand for rapid diagnostic tests. 207–209 However, successful adoption of new technology in the health-care sector, and especially in hospitals, depends on its acceptance by end users. 210,211 The latter is a function of the ease of the required associated procedures, extent of demand on staff time, perceived efficiency, quality and added clinical value of speedy results and the actual reduction in time to a result for the new rapid diagnostic test compared with the method in use. Although there have been various studies comparing one or more of these features of diagnostics tests,212–214 to the best of our knowledge only one formal rating system has been described to score the ease and speed of use of diagnostic methods. 215
In the USA, the Secretary of Health and Human Services has delegated to the Food and Drug Administration (FDA) the authority to determine whether particular tests are ‘simple’ and have ‘an insignificant risk of an erroneous result’. The US Clinical Laboratory Improvement Amendments (CLIA) categorisation criteria grade specific laboratory test system, assay and examination for level of complexity by assigning scores of 1, 2 or 3 for each of seven criteria: (1) knowledge; (2) training and experience; (3) reagents and materials preparation; (4) characteristics of operational steps; (5) calibration, quality control and proficiency testing materials; (6) test system troubleshooting and equipment maintenance; and (7) interpretation and judgement. 215 A score of ‘1’ indicates the lowest level of complexity, and the score of ‘3’ indicates the highest level. These scores are totalled to derive a measure of complexity. Here we evaluate the ease and speed of use of selected POCTs, PCR-based rapid molecular assays and traditional culture for the aetiological diagnosis of respiratory infections. As outlined below, we modified the scoring system proposed by CLIA and included additional criteria, specifically: (1) test site requirements; (2) equipment; (3) storage and disposal of waste test materials and reagents; (4) health and safety implications; and (5) time to reporting (TtR) of results.
Specimen handling and diagnostic tests
Specimens were collected and transported as described in Chapter 2 . We evaluated point of care, molecular, and traditional test methods for the diagnosis of respiratory infections as described in Chapter 2 . These included the following.
POCTs:
-
QuickVUE Influenza A&B (Quidel, USA).
-
BinaxNOW S. pneumoniae test.
Molecular diagnostic tests:
-
Semi-nested multiplex conventional PCR for influenza A subtypes H1 and H3, influenza B, and RSV A and B (as used during Year 1).
-
HPA Cambridge, one-step, quadriplex, real-time RT-PCR for the detection of influenza (as used during Year 2).
-
HPA Colindale, one-step, multiplex, real-time RT-PCR for the detection of influenza (as used during Year 3).
-
RSV and hMPV, one-step (Year 2) and two-step (Year 3), multiplex real-time RT-PCR.
Conventional diagnostic tests:
-
Virus isolation and culture on continuous cell lines.
-
Blood culture.
-
Sputum culture.
-
Sputum Gram stain and microscopy.
Ease-of-use scores
Each specific test procedure was graded by assigning EoU scores of ‘1’, ‘2’ or ‘3’ for each of the 11 criteria listed below. Like the CLIA system, a low score reflects the lowest levels of complexity of test procedures that can be carried out quickly by personnel with minimal training. A high score reflects high levels of complexity. A score of ‘2’ was assigned to scoring criteria when the characteristics for a particular test are intermediate between those listed for scores of ‘1’ and ‘3’.
Scoring criteria
-
Test site:
-
Score 1 Is a POCT.
-
Score 3 A facility with purpose-built accommodation, ventilation systems and dedicated space is essential.
-
-
Equipment:
-
Score 1 (A) No special equipment is required and (B) the test is readily transferred between hospital facilities.
-
Score 3 Specialised equipment is essential and non-portable.
-
-
Materials and reagents:
-
Score 1 (A) Materials and reagents are stable and (B) they are pre-packaged, and/or pre-measured, and require no special handling steps, or storage conditions.
-
Score 3 Materials and reagents are labile requiring special storage conditions or require special handling steps to ensure reliability and (B) materials and reagents preparation requires manual steps, for example volumetric measurement.
-
-
Operational steps:
-
Score 1 (A) Operational steps are either automatically operated (e.g. pipetting, temperature control, mixing of reagents or timing of steps) or (B) are easily controlled.
-
Score 3 Operational steps require close monitoring and control, and may require special preparation (e.g. for manual nucleic acid extraction), precise temperature control or timing of procedural steps, accurate pipetting or extensive calculations.
-
-
Training, experience, and knowledge:
-
Score 1 (A) Minimal scientific and technical knowledge and experience are required to perform the test and (B) knowledge to perform the test may be obtained through on-the-job instruction [i.e. in the UK the test could be done readily by someone at Agenda for Change (AFC) Band 4 or less].
-
Score 3 Specialised scientific and technical knowledge is essential to perform the testing, and (B) substantial experience is required (i.e. in the UK the test requires skills and knowledge commensurate with someone at AFC Band 7 or higher).
-
-
Calibration and quality control:
-
Score 1 (A) Calibration is either automatic or not required or (B) quality control materials are stable and are included in the test, or are readily available.
-
Score 3 (A) Calibration materials, if available, may be labile, or (B) quality control materials, if available, may be labile, or not available, or (C) technical expertise is required for calibration.
-
-
Interpretation and judgement:
-
Score 1 (A) Minimal interpretation and judgement are required to perform the test; and (B) problems require limited interpretation, judgement, and decision-making.
-
Score 3 (A) Extensive interpretation and judgement are required to perform the test; and (B) resolution of problems requires extensive interpretation, judgement and decision-making.
-
-
Test system troubleshooting and equipment maintenance:
-
Score 1 (A) Test system troubleshooting is automatic or self-correcting, or clearly described or requires minimal judgement and (B) equipment maintenance is seldom needed, or can be easily performed.
-
Score 3 (A) Test system troubleshooting is not automatic or self-correcting and requires decision-making and intervention to resolve most problems and (B) equipment maintenance requires special knowledge, skills, and abilities.
-
-
TtR of results (included as possible indicator of EoU):
-
Score 1 Results are reported within 4 hours of collection of almost all specimens.
-
Score 3 The median TtR of results exceeds the median duration of hospitalisation/time to death.
-
-
Health and safety:
-
Score 1 The test is completed using low levels of personal protection, i.e. gloves, and can be conducted outside a laboratory setting.
-
Score 3 One or more operational steps in the testing process require Biosafety level (BSL) category 3 or higher.
-
-
Storage and disposal of waste test materials and reagents:
-
Score 1 Waste materials and reagents from the testing process are stored and disposed using medium duty ‘Clinical Waste’ plastic bags and Sharps Containers (to BS 7320/UN 3291), or a sluice.
-
Score 3 Waste materials or reagents from the testing process include hazardous materials (including highly infectious waste, chemical waste, waste with a high content of heavy metals, genotoxic waste or radioactive waste) that require special attention.
-
Results
As outlined in Chapter 3 , 1252 admissions were enrolled into the study, of which 418 were randomised to the ‘near-patient’ group, 415 to the ‘molecular’ diagnostic group, and 419 to the ‘conventional’ diagnostic group. The median duration of symptoms before recruitment was 120 hours (IQR 72–168 hours), and the median time to discharge or death was 72 hours (IQR 21–180 hours).
Speed of use of test methods for the diagnosis of respiratory infections
Quidel QuickVUE Influenza A&B
The interval between specimen collection and the results of the near-patient Quidel QuickVue Influenza A + B test was recorded for 327 of the 418 admissions who were randomised to ‘near-patient’ test group (Group 1). The median interval from specimen collection to the provision of a positive or negative result for the 327 admissions was 15 minutes (IQR 10–23 minutes) ( Figure 12 ). Altogether, 278 results (85%, 95% CI 80.7% to 88.7%) were reported by 30 minutes.
FIGURE 12.
Time interval between specimen collection and report (QuickVUE test).
BinaxNOW S. pneumoniae test
Urine samples from 412 out of 418 admissions in the ‘near-patient’ group (Group 1) were available on recruitment for POC testing using the BinaxNOW S. pneumoniae test. The interval between specimen collection and the results of the near patient BinaxNOW S. pneumoniae test was recorded for 324 admissions. The median interval from specimen collection to the provision of a positive or negative result was 20 minutes (IQR 15–30 minutes) ( Figure 13 ). Altogether, 270 results (83.3%, 95% CI 78.8% to 87.2%) were reported by 30 minutes.
FIGURE 13.
Time interval between specimen collection and report (BinaxNOW S. pneumoniae test).
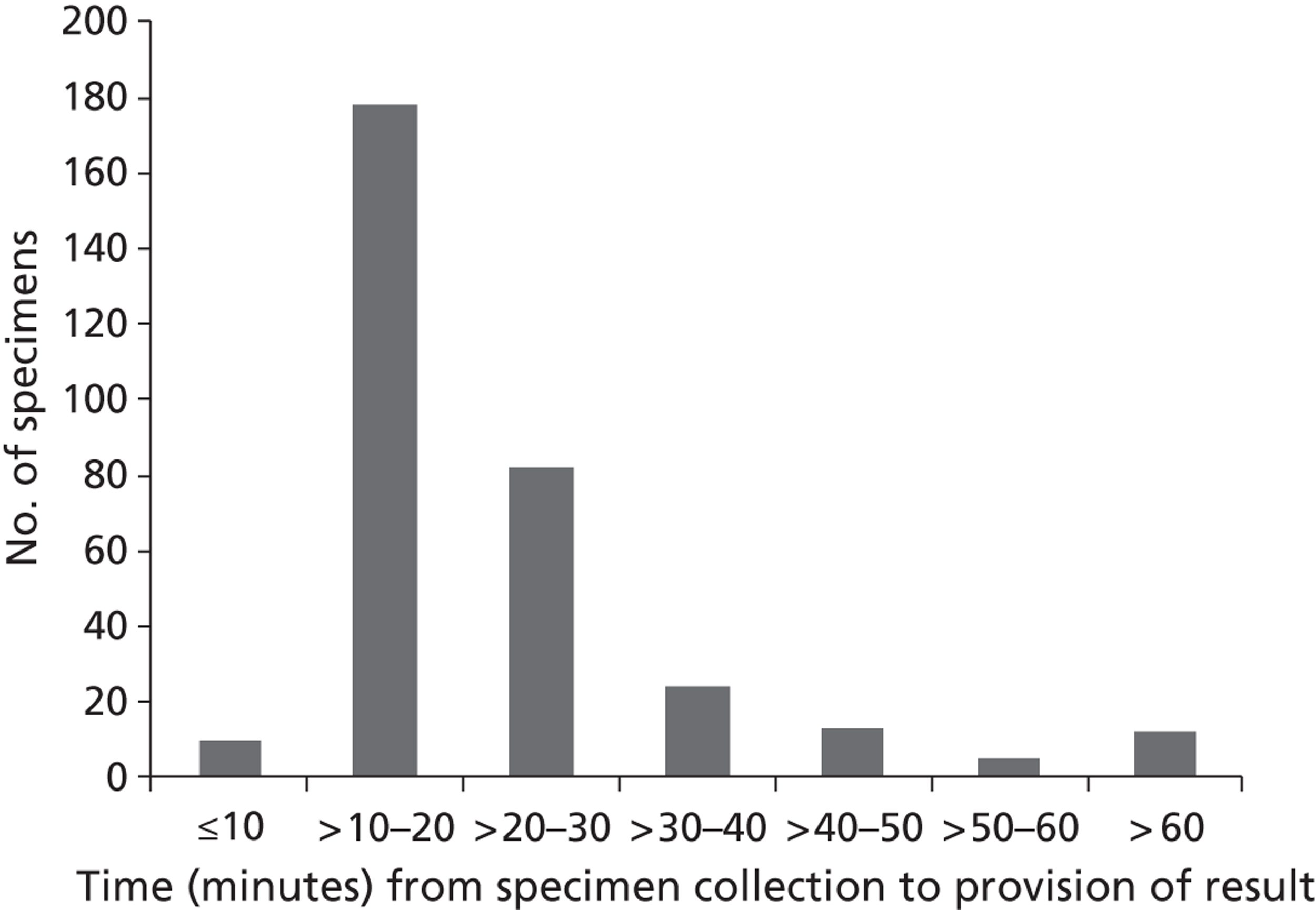
Semi-nested multiplex polymerase chain reaction for influenza A subtypes H1 and H3, influenza B and respiratory syncytial virus A and B
The median TtR of results obtained by ‘conventional’ semi-nested multiplex PCR (n = 57, Group 2) was 50.8 hours (IQR 44.3–92.6 hours). Figure 14 shows a plot of the time interval between specimen collection and the report. Altogether 12 results (21.0%, 95% CI 11.4% to 33.9%) were reported by 36 hours; 18 (31.6%, 95% CI 19.9 to 45.2%) were reported by 48 hours; 36 (63.1%, 95% CI 49.3% to 75.6%) were reported by 72 hours, and 45 (78.9%, 95% CI 66.1% to 88.6%) were reported by 96 hours.
FIGURE 14.
Time interval between specimen collection and report (semi-nested multiplex PCR).
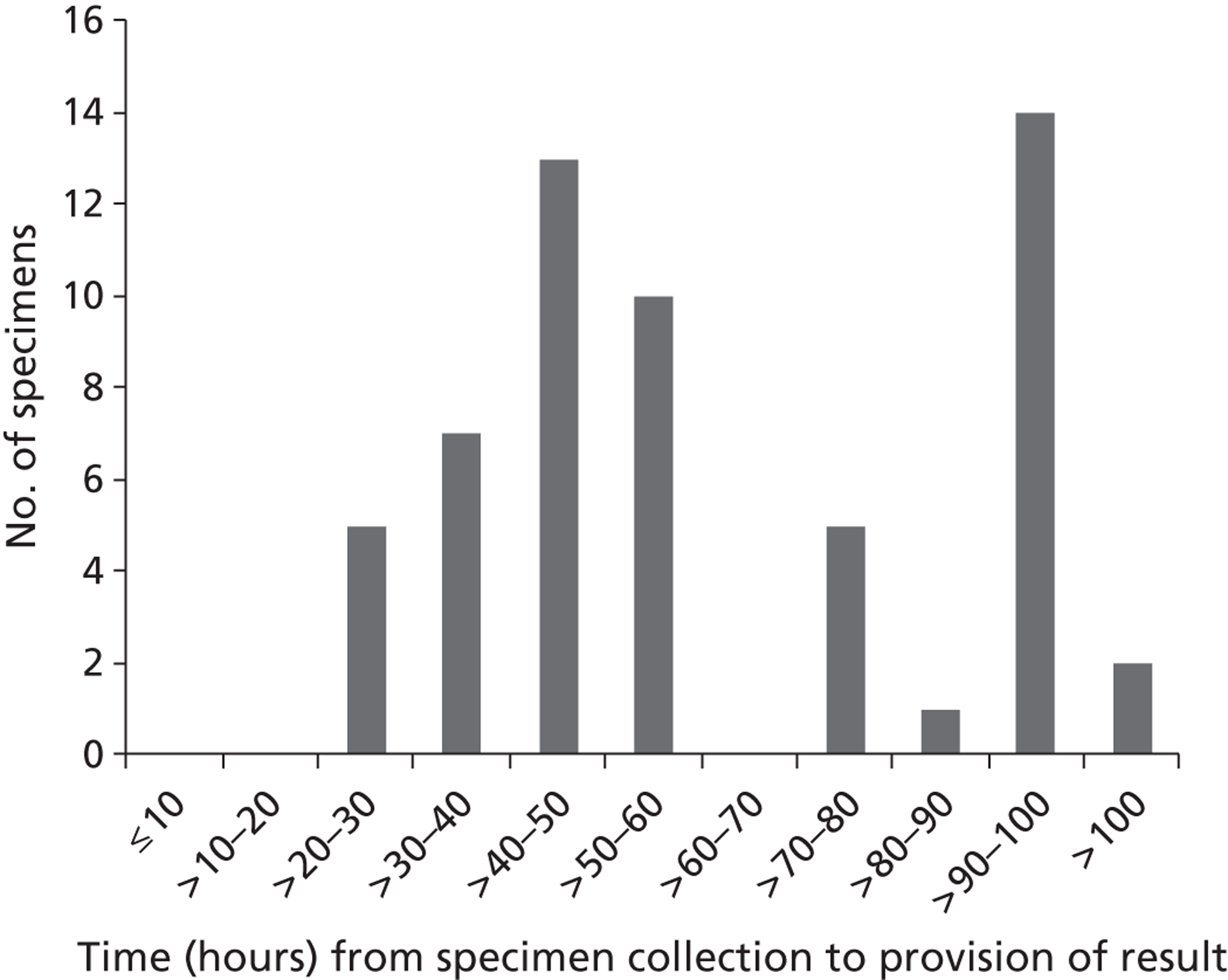
One-step, quadriplex/multiplex, real-time reverse-transcriptase polymerase chain reaction for influenza, and one-step and two-step respiratory syncytial virus and human metapneumovirus multiplex real-time polymerase chain reaction
These real-time PCRs are considered together owing to their similarity. The median TtR of results obtained by ‘real-time’ RT-PCR (n = 358, Group 2) was 29.2 hours (IQR 26–46.9 hours) ( Figure 15 ). The difference between the time taken to analyse specimens by the ‘conventional’ semi-nested multiplex PCR and real-time PCR was 21.9 hours (p < 0.0001, Mann–Whitney U-test). Figure 16 shows the percentage of specimens that are reported as positive or negative by real-time PCR in increments of 12 hours (and at 30 hours) after specimen collection. By 30 hours, results were available for 220 (61.3%, 56.0% to 66.3%) admissions.
FIGURE 15.
Time interval between specimen collection and report (real-time RT-PCR).
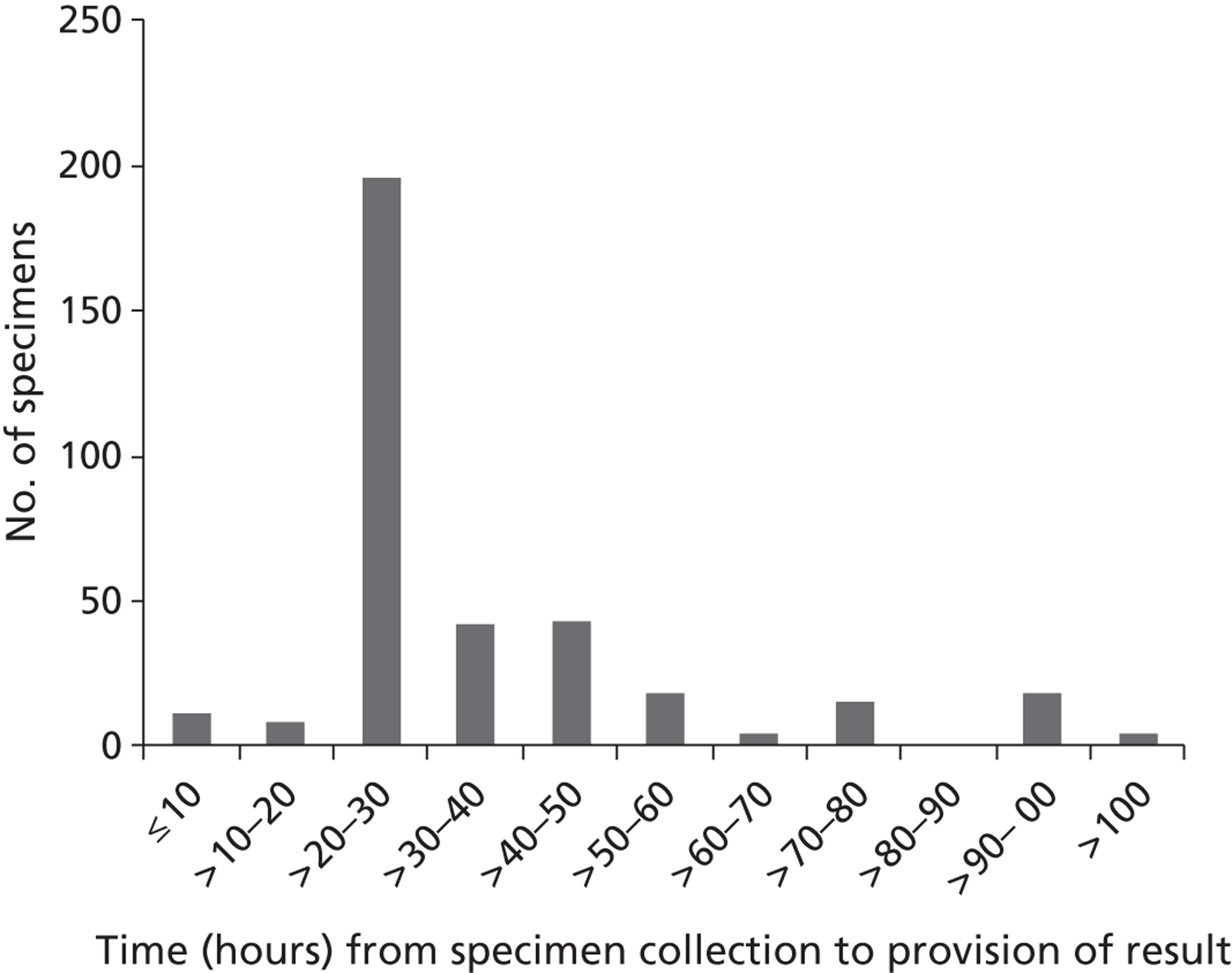
FIGURE 16.
Percentage (± 95% CIs) of specimens reported as positive or negative by real-time RT-PCR at 12-hour intervals after collection.
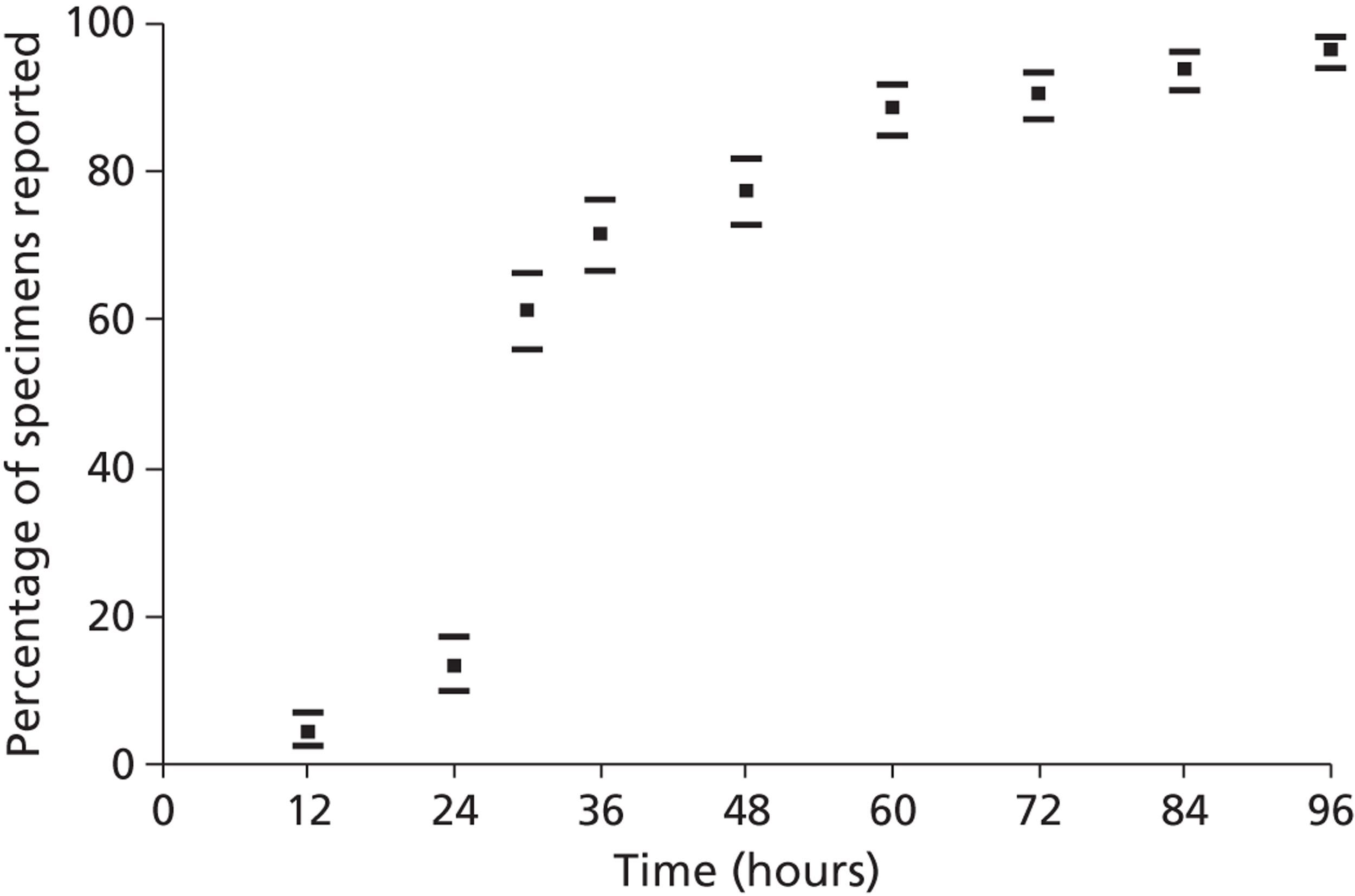
Viral culture
Viral culture results were available for 1245 (99.4%, 95% CI 98.8% to 99.8%) of the 1252 nasopharyngeal specimens that were collected from all admissions in Groups 1, 2 and 3. The cumulative percentage of specimens reported by 24-hour time periods for influenza A and B (n = 21), other viruses (n = 49) and specimens that were culture negative (n = 1175) is shown in Figure 17 . The median time from specimen collection to reporting a positive culture result for admissions with influenza A and B was 629.6 hours (IQR 262.5–846.7), which is approximately nine times greater than the median time to discharge or death (72.08 hours). The median turnaround time of 220.4 hours (IQR 172.1–314.5 hours) for reports of viruses other than influenza was significantly shorter than for the time for influenza (p < 0.0001, Kruskal–Wallis test). The time to report the isolation of influenza A or B did not differ from the time to report culture negative results (454.7 hours, IQR 406.0–621.5 hours). Less than 6% (70 of 1245, 5.6%, 95% CI 4.4% to 7.0%) of all virus culture results were reported within 14 days of specimen collection. The results for 17 out of 21 (80%, 95% CI 56.3% to 94.3%) nasopharyngeal specimens that grew influenza virus became available only a median of 459.1 hours (IQR, 290.1–768.2 hours) after death or discharge. For 33 specimens that grew herpes simplex type 1 virus (another virus for which therapy is available), the results for 26 (78.8%, 95% CI 61.1% to 91.0%) became available only a median of 298.9 hours (IQR 135.95–552.35 hours) after death or discharge – which is outside the therapeutic window for treatment.
FIGURE 17.
Cumulative percentage (± 95% CIs) of nasopharyngeal specimens reported as positive or negative by viral culture at 24-hour intervals after collection.
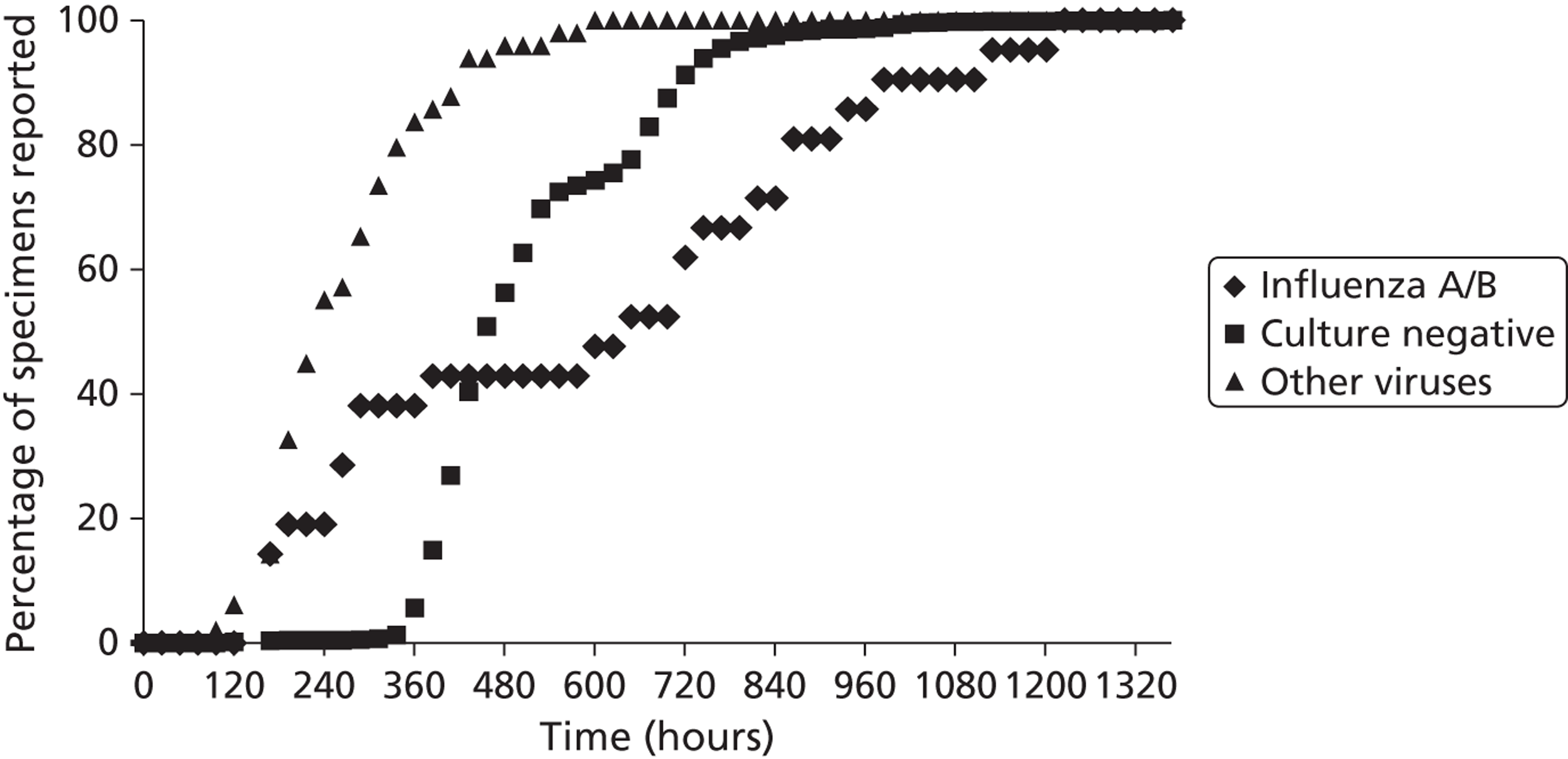
Blood culture
Blood culture results were available for 973 (77.7%, 95% CI 75.3% to 80.0%) patients in Groups 1, 2 and 3. Eighty of the 973 blood cultures grew an organism (including contaminants), and 10 of the 80 grew S. pneumoniae. The median TtR S. pneumoniae cultures was 84.4 hours (IQR 70.7–137.8 hours), but a provisional report was issued a median of 36.8 hours (IQR 22.7–48.75 hours) after specimen collection. The median TtR bacterial growth for the other 70 specimens was 53.8 hours (IQR 47.7–71.8 hours). The median TtR negative culture results for the remaining 893 specimens was 136.2 hours (IQR 124.4–143.25). Blood culture results for three specimens that grew S. pneumoniae were reported a median of 139.2 hours (IQR 27.9–163.4 hours) after death or discharge. The remaining seven pneumococcal culture results were reported a median of 53.2 hours (IQR 30.4–58.1 hours) before discharge. The median interval between specimen collection and reporting all 973 blood culture findings was 130.8 hours (IQR 123.8–142.8 hours). Altogether 619 of the 973 admissions were discharged or died before their blood culture results, i.e. fewer subjects were discharged after the blood culture result was reported than before.
Antimicrobial sensitivity data became available a median of 84.4 hours after specimen collection for the 10 admissions with positive S. pneumoniae blood culture results.
Sputum culture
As in previous studies, we found that a substantial number (941) (75.2%, 95% CI 72.7% to 77.5%) of the 1252 admissions with acute cardiopulmonary conditions were unable to produce sputum for analysis. Sputum was collected from 311 (24.8%) admissions. Test results were only available for 296 (23.6%, 95% CI 21.3% to 26.1%) sputa owing to rejection of 15 samples by the laboratory for reasons of poor quality. Of the 296 specimens, sputum culture and Gram stain results were available for 274 specimens and sputum culture results were available for only an additional 22.
A total of 76 of the 296 sputum cultures grew an organism (excluding Candida species in five additional specimens); of the 76, 10 grew S. pneumoniae after a median interval from specimen collection of 71.4 hours (IQR 69.15–84.0%). The median turnaround time was 60.7 hours (IQR 50.15–71.6 hours) for the remaining 66 culture-positive sputa (p = 0.0065, Kruskal–Wallis test) and 50.5 hours (IQR 48.3–66.9 hours) for the 219 of 220 culture-negative sputa with relevant data (p < 0.0001). The median interval between specimen collection and reporting sputum culture results for 295 out of 296 admissions was 51.4 hours (IQR 48.75–69.2 hours). Overall, 129 of 294 (43.9%) admissions with relevant data were discharged before their sputum cultures were reported.
The median TtR antimicrobial sensitivity was 133 hours (IQR 123.1–148.1 hours) for 8 of the 10 isolates of S. pneumoniae.
Sputum Gram stain
Gram-staining of sputum samples is not routinely performed by the University Hospitals of Leicester NHS Trust, and the median TtR of Gram stain results for 274 sputum samples was 60.7 hours (IQR 50.0–89.4 hours). Figure 18 shows the cumulative percentage of sputa with Gram stain reports at 12-hour intervals after collection.
FIGURE 18.
Cumulative percentage of 274 sputum samples with Gram stain reports at 12-hour intervals after collection.
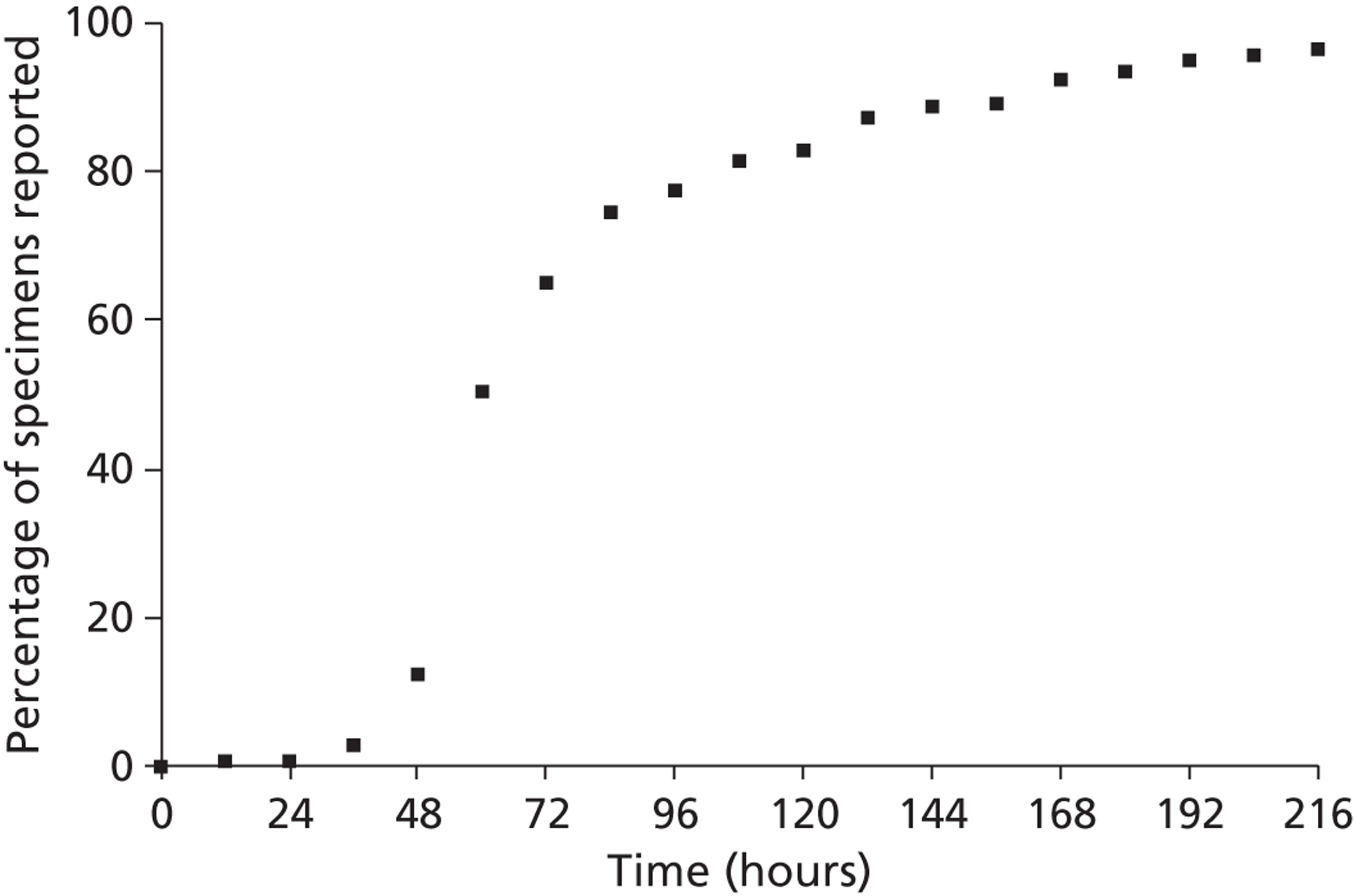
Ease of use
Table 30 shows summary EoU scores for the POCT, molecular and conventional diagnostic tests for the diagnosis of respiratory infections. The component scores for each test are shown in Appendix 7 . EoU scores of 11 (i.e. component scores of ‘1’ for each of the 11 criteria for categorisation) identified test procedures as being straightforward and undemanding to use, scores of 12–22 identified tests as being of moderate complexity and requirements, and scores of ≥ 23 as being ‘complex’, requiring particular skills, training and knowledge, specialised equipment and reagents, and/or accommodation, etc.
| Test | Summary EoU score |
|---|---|
| Point-of-care tests | |
| QuickVUE Influenza A&B | 11 |
| BinaxNOW Streptococcus pneumoniae test | 11 |
| Molecular diagnostic tests | |
| Semi-nested multiplex PCR for influenza A subtypes H1 and H3, influenza B, and RSV A and B | 30 |
| One-step, quadriplex/multiplex, real-time RT-PCR for the detection of influenza, RSV and hMPV | 25 |
| Two-step multiplex real-time RT-PCR for RSV and hMPV | 26 |
| Conventional tests | |
| Virus culture | 26 |
| Blood culture | 20 |
| Sputum culture | 25 |
| Sputum Gram stain and microscopy | 23 |
QuickVUE influenza A&B test
Samples from all 418 patients in the ‘near-patient’ group (Group 1) were tested using the QuickVUE influenza A&B test at the POC. Unequivocal results were reported for all samples tested. The summary EoU score for the QuickVUE influenza A&B test was 11, indicating that the test was straightforward to use, with component scores of ‘1’ for each of the 11 criteria for categorisation (see Appendix 7, Table 38 ).
BinaxNOW Streptococcus pneumoniae test
Urine samples from 412 of the 418 patients in the ‘near-patient’ group (Group 1) were available on recruitment for POC testing using the BinaxNOW Streptococcus pneumoniae test. Unequivocal results were reported for all 412 samples tested. The summary EoU score for the BinaxNOW Streptococcus pneumoniae test was ‘11’, indicating that the test was straightforward to use, having component scores of ‘1’ for each of the 11 criteria for categorisation (see Appendix 7, Table 39 ).
Polymerase chain reaction
Samples from 57 (13.7%) of the 415 patients randomised to the ‘molecular’ diagnostic group (Group 2) were tested using conventional PCR, whereas real-time PCR methods were used to analyse samples from a further 358 patients. All rapid molecular samples were received and tested and unequivocal results were reported for samples from all 415 patients.
Semi-nested multiplex reverse-transcriptase polymerase chain reaction for influenza A subtypes H1 and H3, influenza B, and RSV A and B
The EoU score for the ‘conventional’ semi-nested multiplex RT-PCR was 30, indicating its complexity and requirements. Inspection of the component scores for the 11 categorisation criteria (see Appendix 7, Table 40 ) reveal scores of ‘3’ for eight categories: ‘equipment’; ‘operational steps’; ‘training, experience and knowledge’; ‘calibration and quality control’; ‘interpretation and judgement’; ‘test system troubleshooting and equipment maintenance’; ‘health and safety’ and ‘storage and disposal of waste test materials and reagents’. Scores of ‘2’ were recorded for the remaining three categories: ‘test site’, ‘materials and reagents’ and ‘time to reporting (TtR) of results’. None of the categories scored ‘1’.
One-step, quadriplex/multiplex, real-time reverse-transcriptase polymerase chain reaction for influenza, and one-step and two-step respiratory syncytial virus and human metapneumovirus multiplex real-time polymerase chain reaction
These real-time PCRs are considered together owing to their similarity. The EoU score for real-time PCR was 25 for one-step PCR and 26 for two-step PCR, indicating their complexity and requirements. Component scores for the 11 categorisation criteria are shown in Appendix 7 , Tables 41 and 42 . In contrast with ‘conventional’ semi-nested multiplex PCR, a score of ‘1’ was allocated to one category – ‘materials and reagents’. Besides some minor additional mixing steps, we used real-time PCR kits (SuperscriptTM III Platinum® One-Step qRT-PCR kit and Platinum® Quantitative PCR Supermix-UDG, Invitrogen, Paisley, UK) that contained ready-to-use reagents, which were optimised and quality-controlled by the manufacturer (Invitrogen). In addition, the test calibration was automatic and all reactions were performed in closed tubes, which reduced the level of risk to the operator and environment. With the one-step method, a score of ‘3’ was allocated to four categories: ‘equipment’, ‘calibration and quality control’, ‘health and safety’ and ‘storage and disposal of waste test materials and reagents’. With the two-step method, ‘operational steps’ also scored ‘3’ (a score of ‘2’ was given with the one-step method) because the reverse transcription step involved precise temperature control and timing. All other steps were scored ‘2’.
Viral culture
Although swabs in VTM were received from all 1252 patients, viral culture results were available for 1209 (96.6%); contamination of the PLC/PRF5 and MRC-5 tubes prevented the culture of specimens from 41 patients, and two were rejected by the laboratory due to improper storage of samples at room temperature. A further 29 (2.4%) had only partial results due to contamination of PLC/PRF5 or MRC-5 tubes. The EoU score for virus culture was 26, reflecting the complexity and requirements of the method. The component scores for the 11 categorisation criteria (see Appendix 7 , Table 43 ) included scores of ‘3’ for five categories: ‘equipment’, ‘materials and reagents’, ‘operational steps’, ‘interpretation and judgement’ and ‘time to reporting (TtR)’. Scores of ‘2’ were recorded for all other categories, except ‘Calibration and quality control’, which scored ‘1’.
Blood cultures
Blood cultures from 967 patients were processed using the Bact/ALERT 3D system. The EoU score for blood culture was ‘20’, reflecting moderate complexity and requirements. The component scores for the 11 categorisation criteria (see Appendix 7 , Table 44 ) included scores of ‘3’ for one category – ‘equipment’. Scores of ‘2’ were given to seven categories: ‘test site’; ‘training, experience and knowledge’; ‘calibration and quality control’; ‘interpretation and judgement’; ‘time to reporting (TtR) of results’; ‘health and safety’ and ‘storage and disposal of waste test materials and reagents’. The categories ‘materials and reagents’, ‘operational steps’ and ‘test system troubleshooting and equipment maintenance’ all scored ‘1’.
Sputum culture and Gram stain and microscopy
Sputum culture received an EoU score of 25 (see Appendix 7 , Table 45 ), indicating its complexity or requirements. It scored ‘3’ in five categories: ‘test site’; interpretation and judgement’; test system troubleshooting and equipment maintenance’; ‘health and safety’ and ‘storage and waste disposal.’ It scored ‘2’ in four categories: ‘equipment’; ‘materials and reagents’; ‘training, experience and knowledge’; and ‘time to reporting (TtR) of results’. It scored ‘1’ on ‘operational steps’ and ‘calibration and quality control’.
Sputum Gram staining and microscopy received an EoU score of 23 (see Appendix 7, Table 46 ), primarily reflecting its requirements rather than complexity. It scored ‘3’ in three categories: ‘test site’; ‘training, experience and knowledge’; and ‘health and safety’. It scored ‘2’ in six categories: ‘equipment’; ‘materials and reagents’; ‘operational steps’; ‘interpretation and judgement’; ‘test system troubleshooting and equipment maintenance’; and ‘time to reporting (TtR) of results’. It scored ‘1’ in ‘calibration and quality control’ and ‘storage and disposal of waste test materials and reagents’.
Sensitivity analysis
The EoU score incorporated a score for the time to reporting (TtR) of results as a possible indicator of EoU – the rationale being that tests that provide timely results are easier to use or are less demanding than tests that provide results more slowly. We undertook a sensitivity analysis to examine whether our scoring and ranking of test was unduly influenced by the (TtR) of results or its combination with another measure of EoU and test requirements/complexity. Table 31 shows total EoU scores and their ranking when (1) all 11 criteria are considered; (2) the total EoU score excluding the component score for the ‘time to reporting (TtR) of results’; and (3) the total EoU score excluding the component score for the ‘time to reporting (TtR)of results’ and each remaining component score in addition. The score relating to the (TtR), when considered alone or with other component scores, has no appreciable effect on ranking of tests in terms of their EoU and/or test requirements (see Table 31 ).
| Ranking (score) | Easy to use/least demanding | Gram stain | Sputum culture | Most difficult to use/most demanding | |||||
|---|---|---|---|---|---|---|---|---|---|
| QuickVUE | BinaxNOW | Blood culture | One-step real-time RT-PCR | Virus culture | Two-step real-time RT-PCR | Semi-nested RT-PCR | |||
| TS | 1 = (11) | 1 = (11) | 3 (20) | 4 (23) | 5 = (25) | 5 = (25) | 7 = (26) | 7 = (26) | 9 (30) |
| [TS] – [time to reporting’ (TtR) score] | 1 = (10) | 1 = (10) | 3 (18) | 4 (21) | 5 = (23) | 5 = (23) | 5 = (23) | 8 (24) | 9 (28) |
| [TS] – [‘TtR’ + ‘test site’ scores] | 1 = (9) | 1 = (9) | 3 (16) | 4 (18) | 5 (20) | 6 = (21) | 6 = (21) | 8 (22) | 9 (26) |
| [TS] – [‘TtR’ + ‘equipment’ scores] | 1 = (9) | 1 = (9) | 3 (15) | 4 (19) | 7 = (21) | 5 = (20) | 5 = (20) | 7 = (21) | 9 (25) |
| [TS] – [‘TtR’ and ‘materials and reagents’ scores] | 1 = (9) | 1 = (9) | 3 (17) | 4 (19) | 6 = (22) | 6 = (22) | 5 (20) | 8 = (23) | 9 (26) |
| [TS] – [‘TtR’ + ‘operational steps’ scores] | 1 = (9) | 1 = (9) | 3 (17) | 4 (19) | 6 = (21) | 6 = (21) | 5 (20) | 6 = (21) | 9 (25) |
| [TS] – [‘TtR’ + ‘training, experience and knowledge’ scores] | 1 = (9) | 1 = (9) | 3 (16) | 4 (18) | 5 = (21) | 5 = (21) | 5 = (21) | 8 (22) | 9 (25) |
| [TS] – [‘TtR’ + ‘calibration and quality control’ scores] | 1 = (9) | 1 = (9) | 3 (16) | 4 = (20) | 7 = (22) | 4 = (20) | 7 = (22) | 6 (21) | 9 (25) |
| [TS] – [‘TtR’ + ‘interpretation and judgement’ scores] | 1 = (9) | 1 = (9) | 3 (16) | 4 (19) | 5 = (20) | 7 (21) | 5 = (20) | 8 (22) | 9 (25) |
| [TS] – [‘TtR’ + ‘trouble-shooting/maintenance’ scores] | 1 = (9) | 1 = (9) | 3 (17) | 4 (19) | 5 (20) | 6 = (21) | 6 = (21) | 8 (22) | 9 (25) |
| [TS] – [‘TtR’ + ‘health and safety’ scores] | 1 = (9) | 1 = (9) | 3 (16) | 4 (18) | 5 = (20) | 5 = (20) | 7 = (21) | 7 = (21) | 9 (25) |
| [TS] – [‘TtR’ + ‘storage and disposal of waste’ scores] | 1 = (9) | 1 = (9) | 3 (16) | 4 = (20) | 4 = (20) | 4 = (20) | 7 = (21) | 7 = (21) | 9 (25) |
Discussion
A prime purpose of POCTs and molecular diagnostic tests is to provide information that enables medical and nursing staff to treat patients with optimal medication as soon as possible, and to inform the use of isolation facilities and infection control procedures and equipment. Such measures can potentially cut illness duration, prevent or ameliorate complications, shorten the duration and costs of hospitalisation, improve the cost-effectiveness of health delivery, and also reduce cross-infection. To be of clinical value, such diagnostic tests must have high sensitivity and specificity across all age ranges, be inexpensive and provide test results sufficiently early for a measurable effect. Here we consider the speed and ease of tests rather than their costs, sensitivity and specificity, clinical impact or cost-effectiveness.
The EoU scores used in this study provide a measure of the complexity and requirements of the test procedures that we assessed. We included TtR of results in the scoring system as a possible indicator of EoU. We did a sensitivity analysis and found that exclusion of TtR of results in the scoring system – either alone or with each other component scores – had no appreciable effect on the overall ranking of tests in terms of EoU and/or test requirements.
The patients in our study had a median duration of illness of 120 hours before admission with a lower quartile of 72 hours. Recent systematic reviews and meta-analyses of the therapeutic use of NIs,70,216,217 show that a clinically beneficial effect of oseltamivir and zanamivir (Relenza®, GSK) depends on treatment starting within 48 hours of first symptoms when seasonal (inter-pandemic) influenza is circulating. In our study, only the Quidel QuickVUE Influenza A&B test provided results soon enough to influence treatment decisions that might benefit patients – and then only to a small percentage (150/1240, 12.1%) of admissions. In our study, the Quidel QuickVUE Influenza A&B test had a median turnaround time of < 30 minutes, and 85% of results were reported within half an hour of specimen collection. The PCR tests were much slower in comparison – the median TtR of results using molecular tests was 50.8 hours for conventional PCR, and 29.2 hours for real-time RT-PCR. For patient cohorts as in our study, these turnaround times preclude RT-PCR from guiding decisions to start antiviral treatment but could influence infection control procedures in hospitals with limited single-room accommodation, as influenza virus can still be detected in nasopharyngeal aspirates by real-time RT-PCR up to 1 week or more among patients who do not receive antiviral therapy. 125,155 In contrast, conventional virus culture provided results of no clinical or public health relevance to the patient or hospital. In our study, the median TtR of results obtained by virus culture was 452.6 hours (IQR 403.6–620.8 hours) and fewer than 5% of results were available within 14 days of specimen collection – too late to initiate antiviral therapy for influenza or recurrent orolabial herpes simplex virus (HSV) infection. The virus culture results usually became available when the patient had either been discharged or died; this occurred for 80% of the specimens that grew influenza A or B, and 57% of those that grew HSV.
In the EoU evaluation of the diagnostic tests for influenza, the Quidel QuickVUE Influenza A&B POCT was allocated a score of ‘11’ (i.e. component scores of ‘1’ for each of the 11 criteria for categorisation), indicating that it was straightforward and undemanding to use, and can be used with minimum levels of training, knowledge and technical skills. The tests kits can be stored at room temperature and have a 24-month shelf life from date of manufacture, are readily transferable and convenient to use. In contrast, the ‘conventional’ RT-PCR test for influenza A and B and RSV A and B had the highest of all EoU scores in the analysis (‘30’), reflecting its complexity and requirements. In comparison with ‘real-time’ PCR tests, the ‘conventional’ RT-PCR test took 20 hours longer when median turnaround times were compared. The EoU scores for the ‘real-time PCRs’ were ‘25’ and ‘26’, indicating PCRs to be ‘complex’ in terms of EoU and requirements. Our observations of turnaround times and EoU scores indicate that the ‘real-time’ RT-PCR method should be used in preference to the ‘conventional’ RT-PCR test, assuming that both methods are otherwise comparable in terms of sensitivity, specificity and cost. However, it is acknowledged that the technology is evolving rapidly, so what is complex now, with multiple steps, is gradually being automated so as to reduce the skill requirements, and improve turnaround times. The EoU score for the conventional cell culture diagnostic test was ‘26’, which reflects its general complexity and requirements in performance and interpretation. As judged by turnaround times, our findings indicate that consideration is given to replacement of conventional virus culture with an alternative test. However, virus culture remains of value in providing specimens for antigenic analysis and antiviral susceptibility, and guiding strain selection for vaccine production.
In the UK, treatment of ILI with NIs is influenced by illness duration and virus activity established through surveillance, rather than POCTs. Similarly, initial therapy of CAP with antibiotics is guided by severity scores – such as CURB-65 – and cannot await ‘early’ microbiology results, although ‘early’ results might subsequently influence the spectrum of antibiotics that are used.
For pneumococcal infection, only the BinaxNOW S. pneumoniae test provided results rapidly – the test had a median turnaround time of < 30 minutes, and 85% of results were reported within half-an-hour of specimen collection. The BinaxNOW S. pneumoniae POCT had an EoU score of 11 (i.e. component scores of ‘1’ for each of the 11 criteria for categorisation), indicating that it was as equally straightforward and undemanding to use as the Quidel QuickVUE Influenza A&B POCT. Like the QuickVUE Influenza A&B test, the BinaxNOW test can be used with minimum levels of training, knowledge and technical skills, can be stored at room temperature, and is readily transferable and convenient to use. The blood and sputum culture results were much slower in comparison. The median TtR growth of S. pneumoniae was 84.4 hours, although a provisional report of the growth of an organism was reported after a median of 36.8 hours. Antimicrobial sensitivity data did not become available for a median of 84.4 hours after specimen collection. Similarly, growth of S. pneumoniae from sputum was reported a median of 71.4 hours after collection, and the median TtR antimicrobial sensitivity was 133 hours.
As noted in the review of cases that tested positive in the pneumococcal POCT (see Chapter 4 ), the much shorter turnaround time of the urinary antigen test for pneumococcal antigen compared with blood and sputum culture, did not lead to any step-down in antimicrobial treatment within 24 hours of the test result, nor did a positive test result lead to the release any single-room accommodation. Similarly, the substantially faster turnaround time of the POCT for influenza compared with virus culture had no impact on the use of NIs, antibiotics or single-room accommodation but the number of patients with positive results was very small, and the data should therefore be interpreted with caution.
Chapter 8 Process outcomes and cost-effectiveness
Introduction
This chapter reports the results of a trial-based cost-effectiveness analysis. Resource-use data were collected prospectively during the time patients were in hospital via the CRFs, and retrospectively from discharge until day 28 via the 28-day follow-up. UK unit costs obtained from a variety of sources were then applied to the resource use data to estimate total cost per patient. As well as describing the distribution of total costs for the three diagnostic strategy groups, an incremental cost-effectiveness analysis was undertaken with cost per quality-adjusted life-year (QALY) (estimated using the EQ-5D and mortality data reported in Chapter 4 via an AUC approach218) being estimated when appropriate, together with cost per (correct) case detected (using the diagnostic data reported in Chapter 5 ). To simultaneously allow for the potential for correlation between cost and outcome, as well as their inherent uncertainty, together with the fact that some data were missing for both resource use (and therefore cost) and EQ-5D (see Chapter 4 for details), a Bayesian approach was adopted219–221 using Markov chain Monte Carlo (MCMC) methods, implemented in WinBUGS version 1.4.3 (MRC Biostatistics Unit, Cambridge, UK) to estimate the uncertainty surrounding the outcome measures. 222 A series of sensitivity analyses were undertaken, both to assess the sensitivity to the MCMC methods used, and to the prices of the index tests (PCR and POCT), and also the model structure used to estimate cost-effectiveness. The perspective adopted was that of the NHS.
Methods
Resource use data and unit costs
Table 32 displays the unit costs, derived from a variety of sources,223–226 for the various major resource use components identified in 3WS. As well as the unit costs in Table 32 , all drugs prescribed in the index admission, and subsequently associated with the index admission, were costed using the British National Formulary (BNF). 227 The price year adopted was 2007–8, i.e. the final year of the study,228 and those unit prices that were not in this year were adjusted accordingly using the Hospital and Community Health Services (HCHS) Pay and Price Index. 229 No discounting of either costs or QALYs was applied to the cost-effectiveness analyses, as the time horizon over which patients were followed was 28 days. As well as the average total cost per patient for each of the diagnostic strategies, the main cost components, namely hospital stay (both non-ITU and ITU), index test costs (including staffing and materials), complications [myocardial infarction, stroke, urinary tract infection, wound infection, MRSA/Clostridium difficile), post-discharge visits (GP/nurse visits) and A&E attendance], additional investigations (electrocardiography, full blood count, blood gases, blood biochemistry, oxygen levels, ward-based urine analysis and chest radiography) and antibiotic prescribing (both for the index admission and post discharge) were also reported in terms of means and SDs. Owing to the relatively large sample size, both cost components and mean resource were formally compared between the diagnostic strategies using parametric methods – one-way ANOVA or Fisher’s exact test as appropriate. 230
| Cost component/resource | Unit cost (£) | Source |
|---|---|---|
| Hospital cost per day | ||
| Ward | 276.21 | HRG code and DH Reference Costs223 |
| ITU | 1410.54 | HRG code and DH Reference Costs223 |
| Oxygen therapy | 212.00 | HRG code and DH Reference Costs223 |
| Ward-based CPAP | 219.75 | HRG code and DH Reference Costs223 |
| Tests (including staffing and materials) | ||
| Quidel | 15.83 | UHL NHS Trust |
| Binax | 25.56 | UHL NHS Trust |
| Blood culture | 46.00 | UHL NHS Trust |
| Viral culture | 48.00 | UHL NHS Trust |
| Sputum culture | 38.00 | UHL NHS Trust |
| PCR | 48.00 | UHL NHS Trust |
| Complications (average costs for acute phase) | ||
| Myocardial infarction | 1138.64 | Bravo-Vergel et al. 224 |
| Stroke | 790.03 | Bravo-Vergel et al. 224 |
| Urinary tract infection | 32.08 | Turner et al. 225 |
| Wound infection | 148.00 | DH NHS Reference Costs223 |
| MRSA/C. difficile | 688.00 | DH NHS Reference Costs223 |
| Post-discharge visits | ||
| GP: surgery | 36.00 | PSSRU226 |
| GP: home | 58.00 | PSSRU226 |
| Practice nurse | 11.00 | PSSRU226 |
| District nurse | 26.00 | PSSRU226 |
| Home-care worker | 39.00 | PSSRU226 |
| A&E attendance | 111.00 | PSSRU226 |
| Additional investigations | ||
| Electrocardiography | 33.45 | DH NHS Reference Costs223 |
| Full blood count | 2.99 | DH NHS Reference Costs223 |
| Blood gases | 2.99 | DH NHS Reference Costs223 |
| Oxygen levels | 2.99 | DH NHS Reference Costs223 |
| Blood biochemistry | 1.34 | DH NHS Reference Costs223 |
| Ward urine analysis | 9.15 | DH NHS Reference Costs223 |
| Chest radiography | 16.00 | DH NHS Reference Costs223 |
Statistical model
Model A
In order to simultaneously allow for the potential for correlation between cost and outcome a bivariate model was assumed for the two outcomes. 220,231 Thus, if y 1 ij and y 2 ij represented the total costs and QALYs for the ith patient in the jth group, respectively, these were then assumed to come from a bivariate normal distribution (model A), such that:
where μ1j and μ2 j are the mean total costs and mean QALYs for the jth randomisation group respectively, and Σ j is the associated covariance matrix. Adopting a Bayesian approach to estimating the unknown parameters of model (1), i.e. μ1 j , μ2 j and Σ j , prior distributions, representing a priori beliefs, are required for these parameters. To represent vague or non-informative prior beliefs regarding μ1 j and μ2 j appropriately diffuse normal distributions centred at zero were used, i.e. N(0,108) and N(0,10–2), respectively. For the covariance matrix, a Wishart prior distribution is placed on the precision matrix, i.e. the inverse of the covariance matrix. Thus:
where A in (2) represents a priori beliefs regarding the corresponding covariance matrix, and by setting k to be the rank of A, in this case k = 2, a non-informative prior distribution is obtained. Initially, A is set such that:
Having specified the prior distributions for model A as above, the marginal posterior distributions for μ 1j and μ 2j are required to assess cost-effectiveness. Owing to the number of unknown parameters in model A (1), MCMC methods are used in which random samples from the conditional posterior distributions are obtained, but which under ergodic theory will converge to the required marginal posterior distributions providing that a sufficient number of samples (iterations) are obtained and that the sampling algorithm has converged to an equilibrium distribution. 232
Estimation of model parameters and software
The results were reported as posterior means and 95% CrIs, which are analogous to CIs. Primary results are based on a ‘burn-in’ of 20,000 iterations followed by a further sample of 50,000 iterations. Further sensitivity analyses were conducted to assess both convergence/mixing of the sampling algorithm and influence of prior distributions, which were all chosen to be non-informative. Appendix 8 displays a sample of the WinBUGS code used for model A. The model parameters were estimated in WinBUGS 1.4.3,222 and further post-estimation processing of the samples was undertaken in R. 233
Missing data
There were missing values for both total costs and QALYs for a number of patients (costs – traditional 8.4%, POCT 8.6% and PCR 8.4%; QALYs – traditional 51.1%, POCT 44.0% and PCR 48.1%) and these were treated as unknown parameters in the estimation of model A, and hence at each iteration values were sampled from their corresponding posterior predictive distribution, i.e. conditional upon both the data and the model parameters, in a manner similar to multiple imputation. 234 Thus, appropriate allowance for all uncertainty in the MCMC estimated mean total costs and QALYs has been made. 235
Assessment of cost-effectiveness
Having estimated the mean effects, i.e. QALYs and total costs per patient, and after allowing for the imputation of missing data, the posterior probability of each diagnostic strategy being the least costly and also having the highest gain in QALYs was estimated from the MCMC samples, i.e. at each iteration the strategies were ranked and the probability of each strategy being ranked first (for QALYs) and third (for costs) was calculated as the proportion of iterations for which this was the case. An incremental cost-effectiveness analysis was then undertaken in which the three diagnostic strategies were first ranked in terms of ascending cost, and then any strategies which were dominated, i.e. for which there was another strategy which produced a greater QALY gain at lower cost, or extendedly dominated, i.e. for which there was another combination of strategies which produced the same QALY gain at lower cost, were excluded from the incremental analysis. For the two or three strategies that remain, cost-effectiveness was then assessed by calculating the appropriate incremental cost-effectiveness ratios (ICERs) to obtain an estimate of cost per additional QALY. Thus, for two strategies, j and k, the ICER is obtained by:
where μ ¯ 1 j and μ ¯ 2 j are obtained by averaging over the MCMC samples in R outside WinBUGS. 221
The probability of cost-effectiveness at willingness-to-pay thresholds, λ, of £20,000 per QALY and £30,000 per QALY, the currently accepted thresholds for NICE,236 were estimated as the proportion of MCMC samples for which the corresponding monetary net-benefit (NB) function was positive. 220
Thus, for strategy j the NB function is given by
In addition, the probability of error for any strategy deemed to be cost-effective was also estimated, i.e. the probability that by adopting that strategy an incorrect adoption decision had been made. For the incremental cost per confirmed case of influenza analysis, the strategies were again ranked in terms of ascending average total cost per patient, and the mean total cost per case, confirmed using serology (see Chapter 5 ), estimated. 237
Sensitivity analyses
A series of one-way sensitivity analyses were undertaken to examine whether the MCMC methods (i.e. initial settings of burn-in, sample length and starting values), prior distributions, statistical model or price of PCR or POCT had an impact on the mean costs and QALYs, i.e. μ 1 j and μ 2 j , and therefore on the incremental cost-effectiveness analysis.
Markov chain Monte Carlo methods
To assess evidence of non-convergence, the history trace plots, i.e. the sampled values at each iteration plotted against iteration number, were examined for systematic movement in the chain and therefore evidence of non-convergence. In addition, the auto-correlation functions/plots were examined to identify poor mixing of the MCMC sampler, i.e. that successive sampled values were not entirely random, and therefore a longer sample length would be required.
To assess the sensitivity of the cost-effectiveness results to the length of burn-in, length of sample, initial starting values and prior distributions in using MCMC methods to estimate mean costs and QALYs, a number of one-way sensitivity analyses were undertaken. Specifically, two additional sets of length of burn-in/sample were used: 10,000/20,000 and 50,000/100,000 compared with the base-case analysis, which used 20,000/50,000. Alternative prior distributions for μ1 j and μ2 j were also used, i.e. N(0,105) and N(0,1), respectively, compared with N(0,108) and N(0,10–2) in the base case, and A was set such that:
Finally, two additional chains, i.e. runs of the MCMC sampler, were used with qualitatively very different initial starting values to those used in the base case, and Brooks–Gelman–Rubin plots were explored,238 as well as the separate summary statistics for μ1 j and μ2 j , to identify evidence that the chains did not produce qualitatively similar results. The Brooks–Gelman–Rubin plots compare the width of 80% CrIs of the pooled chains with the average width of 80% CrIs across chains – the two should be the same unless there is evidence of non-convergence. This also gives an informal way in which to check whether a longer period of burn-in is required.
Model B
Model A (1) assumes that y 1ij and y 2ij , representing the total costs and QALYs for the ith patient in the jth group, respectively, are assumed to come from a bivariate normal distribution. This theoretically means that both totals costs and QALYs could become negative. To assess the sensitivity of the base-case results to this modelling assumption an alternative statistical model (model B)221 was developed in which y 1 ij is assumed to follow a gamma distribution, i.e. so that costs can only be positive, and logit of y 2 ij , i.e. log(y 2 ij /(1 – y 2ij )) is assumed to follow a normal distribution but that these two distributions are interlinked so that correlation between costs and QALYs is allowed. Thus:
The unknown parameters in (7) and (8) are then given plausible yet vague or non-informative prior distributions. The means of the costs and logit QALYs, i.e. μ1 j and μ2 j , are given uniform(1,50000) and normal(0,1000) distributions, respectively, whereas the shape parameters of the gamma distribution in (5), η j are given uniform(1,100) prior distributions, and σ j , the SD of logit QALYs in each randomisation group, is given a half-normal prior distribution with a mean of zero and a SD of 1 truncated at zero, i.e. σ j ∼ N(0,1)I(0,). Finally, βj, the regression parameters that represent the degree of correlation between costs and (logit) QALYs, through λ i – the scale parameter of the gamma distribution in (7) – are given vague normal prior distributions, i.e. β j ∼ N(0,1000). Although the mean costs can be estimated directly from the MCMC samples, because the logit transformation is not linear it cannot be directly back transformed to provide estimates of mean QALY gain. However, it is possible to estimate mean QALYs on their natural scale using Monte Carlo integration within R (see Appendix 8 , section C, for R code used). 221
Price reductions for polymerase chain reaction and point-of-care test
To assess the impact of price of the index test in the different diagnostic strategies, further cost-effectiveness analyses were undertaken in which either the price of PCR or POCT, as these are evolving technologies, was reduced by either 20% or 50%.
Results
Table 33 displays the mean/frequency of resource use for the main constituents of the cost components for patients with no missing data, i.e. complete case analysis. As can be seen, the only statistically significant difference between the three strategies was in terms of use of a practice nurse (p = 0.03). However, this should be interpreted with caution owing to the number of hypothesis tests undertaken. Table 34 displays the mean (and SD) cost per patient for the cost components that make up the overall total cost, as well as the mean overall total cost itself, for each of the three diagnostic strategies. It can be seen that the means for each cost component, as well as for the total, are similar across the three strategies, although there is considerable uncertainty surrounding some of these. Formal comparison of the three strategies using one-way ANOVA did not identify any statistically significant differences between them (p = 0.3). For the observed QALYs, i.e. based on patients who did not have any missing data, POCT produced a mean QALY gain of 0.008137 (SD 0.007191), PCR 0.007724 (SD 0.005977) and traditional laboratory culture 0.007631 (SD 0.006244), which were not statistically significant from one another (p = 0.7).
| Cost component | Traditional (n = 419) | Near patient (n = 418) | Rapid molecular (n = 415) | p-valuea |
|---|---|---|---|---|
| Hospital stay | Mean (SD)b [n] c | Mean (SD)b [n] c | Mean (SD)b [n] c | |
| Non-ITU (days) | 7.11 (11.30) [419] | 6.70 (10.96) [418] | 6.35 (9.56) [415] | 0.6 |
| ITU (days) | 0.07 (0.84) [3] | 0.15 (2.06) [3] | 0.03 (0.45) [2] | 0.4 |
| Oxygen therapy (hours) | 38.75 (108.47) [160] | 48.64 (214.14) [177] | 51.25 (257.75) [169] | 0.6 |
| Ward-based CPAP (hours) | 0.59 (11.74) [5] | 0.38 (5.63) [5] | 0.06 (0.87) [2] | 0.6 |
| Complications | n (%) | n (%) | n (%) | |
| Myocardial infarction | 3 (0.72) | 1 (0.24) | 4 (0.96) | 0.4 |
| Stroke | 1 (0.24) | 0 (0.00) | 0 (0.00) | 0.9 |
| Urinary tract infection | 5 (1.19) | 3 (0.72) | 6 (1.45) | 0.6 |
| Wound infection | 1 (0.24) | 1 (0.24) | 2 (0.48) | 0.7 |
| C. difficile | 6 (1.43) | 2 (0.48) | 1 (0.24) | 0.1 |
| MRSA | 1 (0.24) | 3 (0.72) | 2 (0.48) | 0.6 |
| Additional investigations | Mean (SD)b [n] c | Mean (SD)b [n] c | Mean (SD)b [n] c | |
| Electrocardiography | 1.51 (1.52) [344] | 1.47 (1.64) [336] | 1.44 (1.74) [322] | 0.8 |
| Full blood count | 2.71 (3.19) [399] | 2.69 (3.35) [397] | 2.51 (3.14) [385] | 0.6 |
| Blood gases | 1.86 (8.13) [249] | 2.11 (12.55) [245] | 1.39 (4.11) [245] | 0.5 |
| Oxygen levels | 15.88 (20.16) [397] | 16.24 (31.85) [387] | 15.57 (21.41) [395] | 0.9 |
| Blood biochemistry | 3.23 (5.17) [403] | 2.99 (3.84) [397] | 2.81 (3.68) [387] | 0.4 |
| Ward urine analysis | 0.40 (0.64) [141] | 0.38 (0.61) [140] | 0.37 (0.61) [130] | 0.7 |
| Chest radiography | 1.22 (1.04) [397] | 1.22 (1.07) [392] | 1.20 (0.98) [385] | 0.9 |
| Post-discharge visits | Mean (SD)b [n] c | Mean (SD)b [n] c | Mean (SD)b [n] c | |
| GP: surgery | 0.36 (0.77) [100] | 0.40 (0.98) [91] | 0.35 (0.84) [91] | 0.7 |
| GP: home | 0.14 (0.53) [37] | 0.11 (0.42) [34] | 0.10 (0.38) [33] | 0.4 |
| Practice nurse | 0.01 (0.15) [1] | 0.04 (0.29) [9] | 0.06 (0.41) [12] | 0.03 |
| District nurse | 0.17 (1.54) [22] | 0.09 (0.69) [25] | 0.09 (0.69) | 0.5 |
| Home-care worker | 0.28 (2.60) [17] | 0.15 (1.12) [18] | 0.24 (2.03) [18] | 0.6 |
| A&E attendance | 0.02 (0.15) [6] | 0.03 (0.23) [8] | 0.01 (0.11) [2] | 0.3 |
| Cost component | Traditional mean (SD), £ | Near-patient mean (SD), £ | Rapid molecular mean (SD), £ | p-valuea |
|---|---|---|---|---|
| Hospital stay: | 2004 (3174) | 1972 (3753) | 1782 (2698) | 0.5 |
| Non-ITU | 1977 (3121) | 1855 (3030) | 1759 (2652) | 0.6 |
| ITUa | 27 (485) | 117 (1662) | 24 (482) | 0.3 |
| Index testsb | 94 | 41 | 112 | |
| Complicationsa | 22 (139) | 11 (94) | 16 (120) | 0.5 |
| Post-discharge visitsa | 56 (143) | 52 (126) | 50 (156) | 0.8 |
| Additional investigations | 137 (116) | 138 (151) | 130 (118) | 0.6 |
| Antibiotics | 26 (56) | 23 (58) | 23 (61) | 0.7 |
| Total costs | 2326 (3370) [172c] | 2158 (3254) [167c] | 1980 (2355) [121c] | 0.3 |
Figure 19 displays the distribution of observed total costs for the three diagnostic strategy groups. As can be seen, the distributions are skew, but are very similar to one another.
FIGURE 19.
Distribution of total costs for each of the diagnostic strategies. (a) POCT; (b) PCR; (c) laboratory culture.
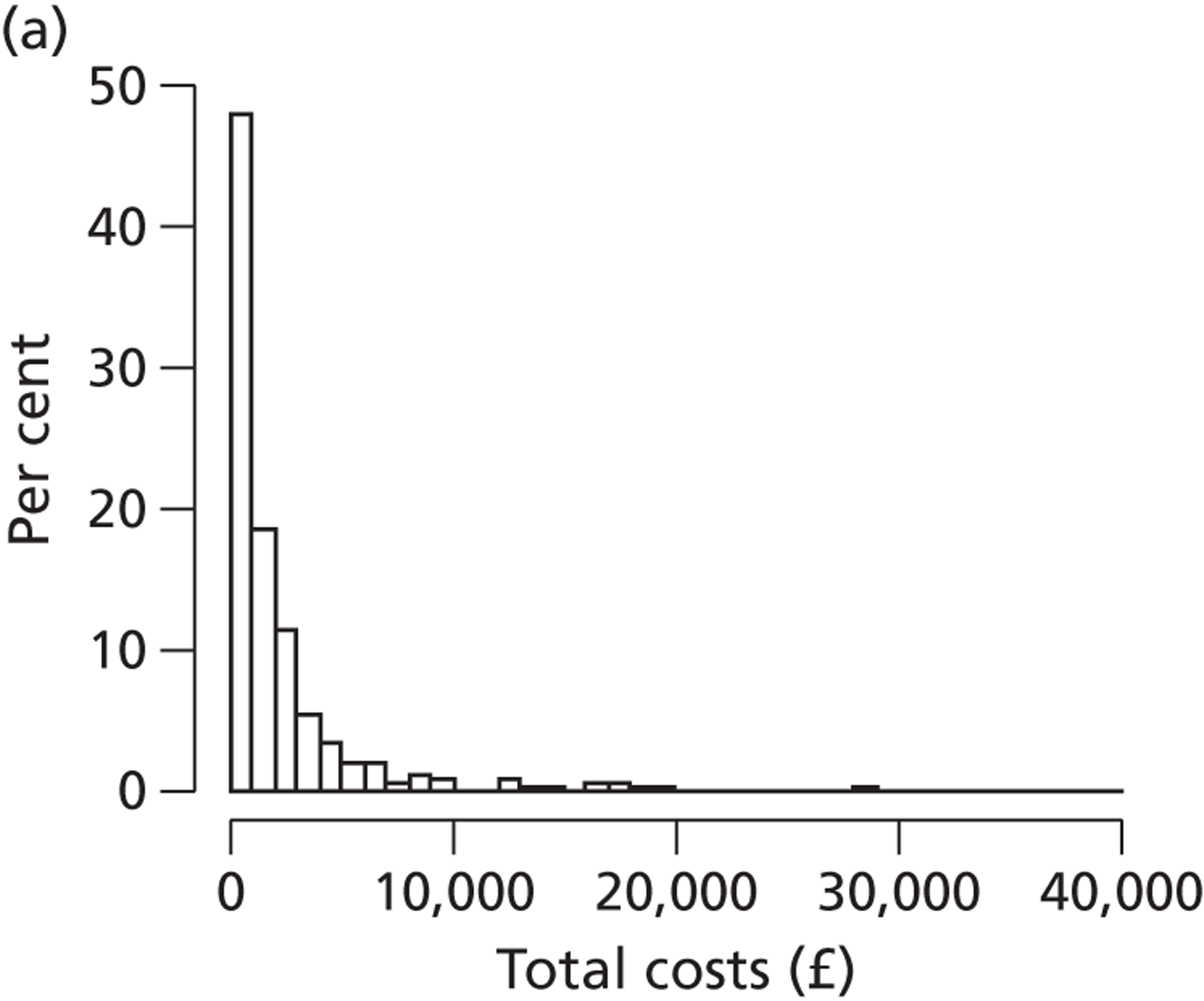
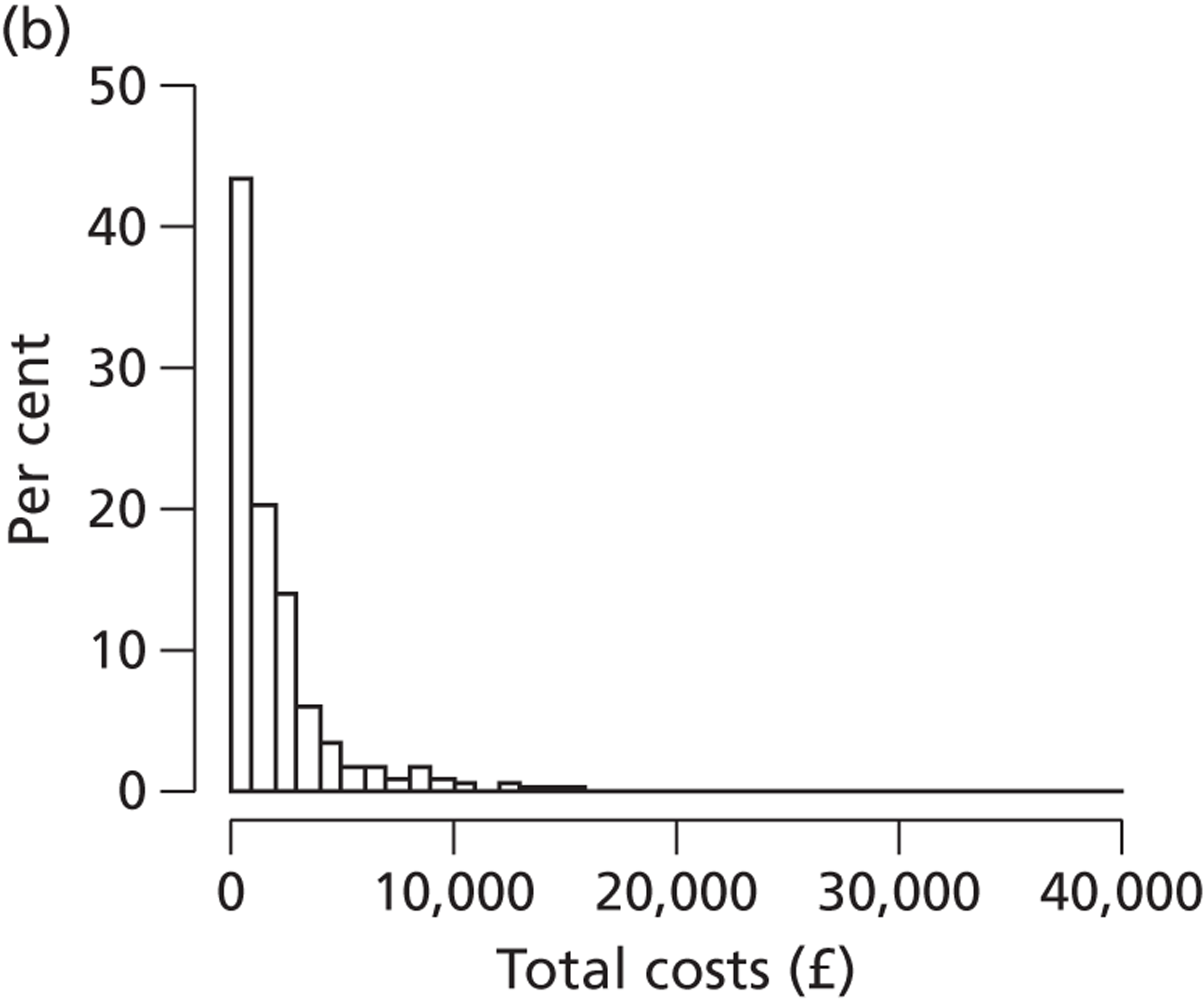
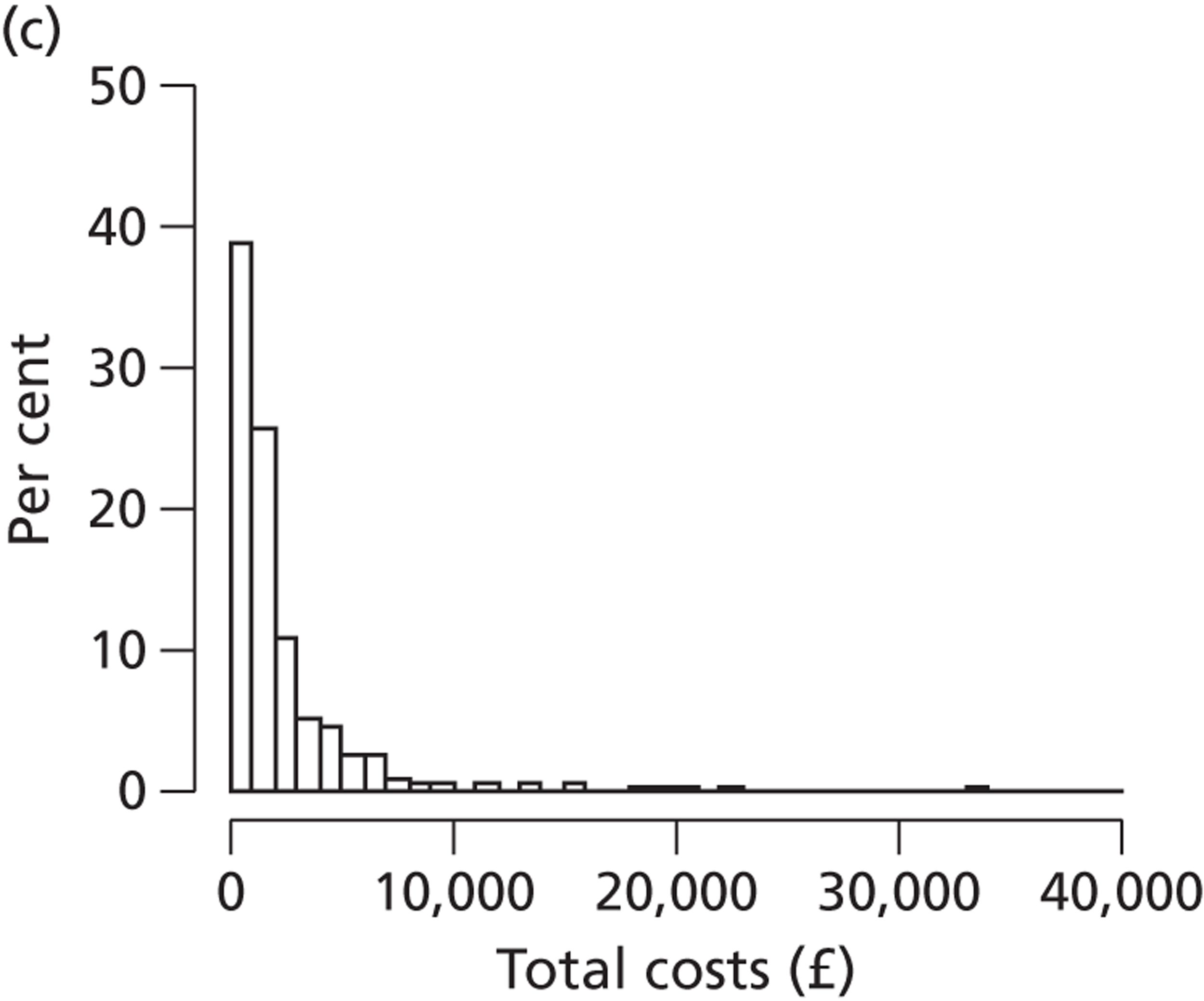
Table 35 displays the MCMC posterior mean estimates for the total costs and QALYs (gained during the 28 day follow-up period of the trial) associated with each of the three strategies, together with associated 95% CrIs, which can also be seen to very similar to one another. More formally, the posterior probabilities that each strategy has the highest QALY gain are all less than 50%, ranging from 21.1% for traditional laboratory culture to 48.9% for POCT, whilst the posterior probability that PCR is the least costly is only 78.6%. Figure 20 displays 50,000 samples for the mean total costs and QALYs gained for each of the three diagnostic strategies, and again demonstrates both the level of uncertainty in both costs and QALYs, and broad similarity between the strategies. Table 35 also shows a formal incremental analysis which indicates that traditional laboratory culture testing is dominated by POCT, in that it has the highest mean total costs but the smallest QALY gain, and is therefore excluded further from the incremental analysis. Although PCR has lower mean total cost than POCT (cost difference: +£181, 95% CrI: –£219 to +£587), POCT also has a larger QALY gain (QALY difference: +0.000256, 95% CrI: –0.001474 to +0.001978), and the associated ICER of POCT compared with PCR is £734,717.
| Mean total cost per patient, £ (95% CrI) | Mean QALYs gaineda (95% CrI) | Probability of being least costly | Probability of having highest QALY gain | Incremental mean total cost (95% CrI) | Incremental mean QALY gain, £ (95% CrI) | ICER, £ | Probability (of error) of being cost-effective at thresholds of: | |
|---|---|---|---|---|---|---|---|---|
| £20,000 per QALY | £30,000 per QALY | |||||||
| PCR | ||||||||
| 1978 (1743 to 2216) | 0.007779 (0.006555 to 0.008983) | 0.786 | 0.300 | –b | –b | –b | 0.783 (0.217) | 0.781 (0.219) |
| POCT | ||||||||
| 2159 (1828 to 2485) | 0.008035 (0.006772 to 0.009280) | 0.178 | 0.489 | +£181 (–£219 to +£587) | +0.000256 (–0.001474 to +0.001978) | 734,717 | 0.183 | 0.186 |
| Traditional | ||||||||
| 2327 (1989 to 2664) | 0.007588 (0.006334 to 0.008854) | 0.036 | 0.211 | Dominatedc | Dominatedc | Dominatedc | 0.034 | 0.033 |
FIGURE 20.
Mean costs and QALYs for ‘traditional’, ‘near-patient’ and ‘molecular’ diagnostic tests for 50,000 MCMC samples.
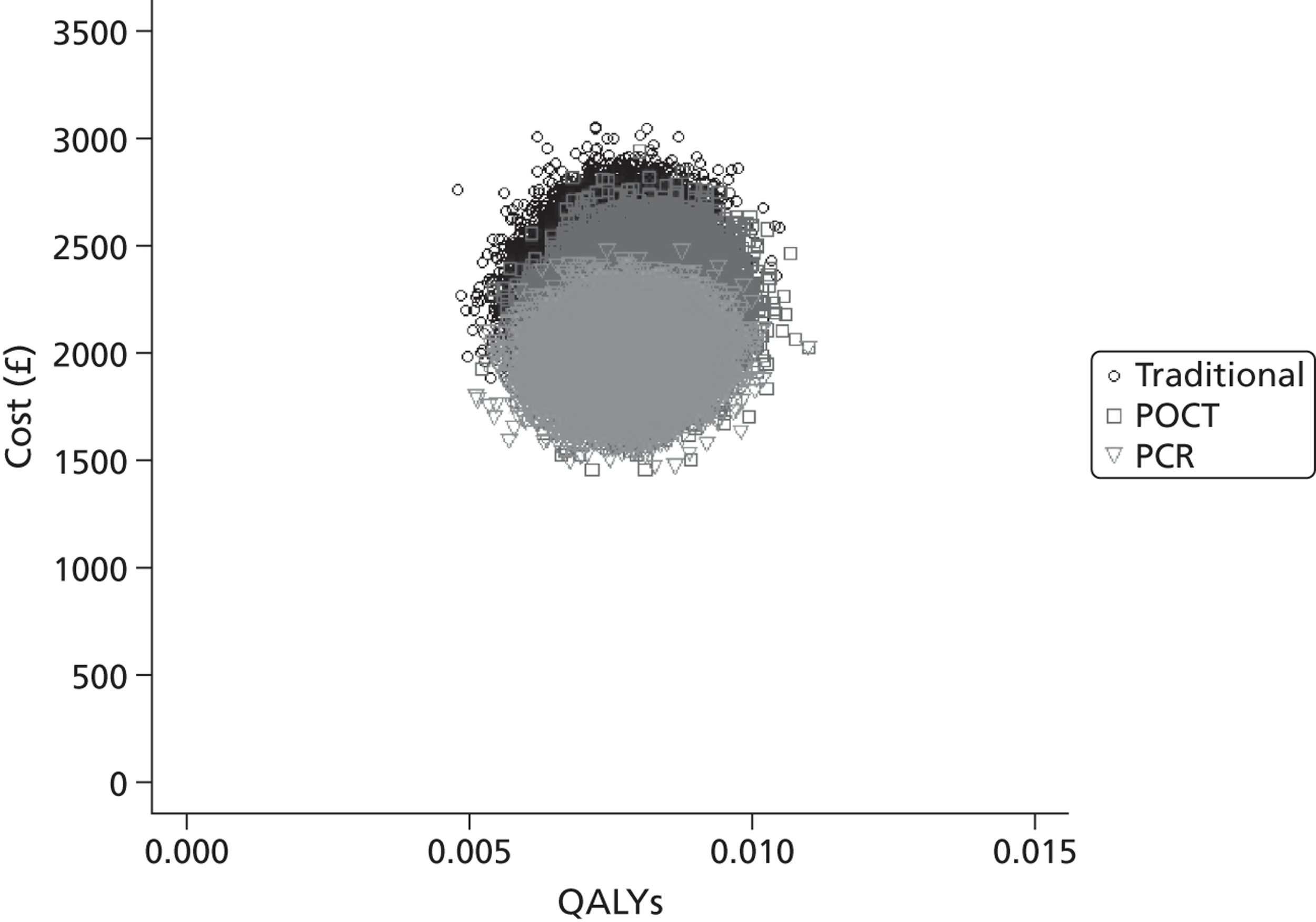
Figure 21 displays the cost-effectiveness plane, i.e. incremental costs plotted against incremental QALYs for POCT compared with PCR, together with a £20,000-per-QALY willingness-to-pay threshold. In fact, the probability of POCT being cost-effective at £20,000 threshold is 18.3%, and at £30,000 is 18.6%, reflecting the considerable uncertainty, whereas the probability of PCR being cost-effective at a £20,000 threshold is 78.3% and at £30,000 is 78.1%. The probability of error for adopting PCR, i.e. the probability of making an incorrect adoption decision, is 21.7% at a £20,000 threshold and 21.9% at £30,000. The corresponding cost-effectiveness acceptability curves (CEACs) for all three diagnostic strategies are displayed in Figure 22 .
FIGURE 21.
Cost-effectiveness plane, i.e. incremental costs plotted against incremental QALYs for POCT compared with PCR, using 50,000 MCMC samples together with a £20,000 per QALY willingness-to-pay threshold. Sampled values below the threshold line indicate that POCT should be adopted as the most cost-effective strategy at a willingness to pay of £20,000 per QALY.
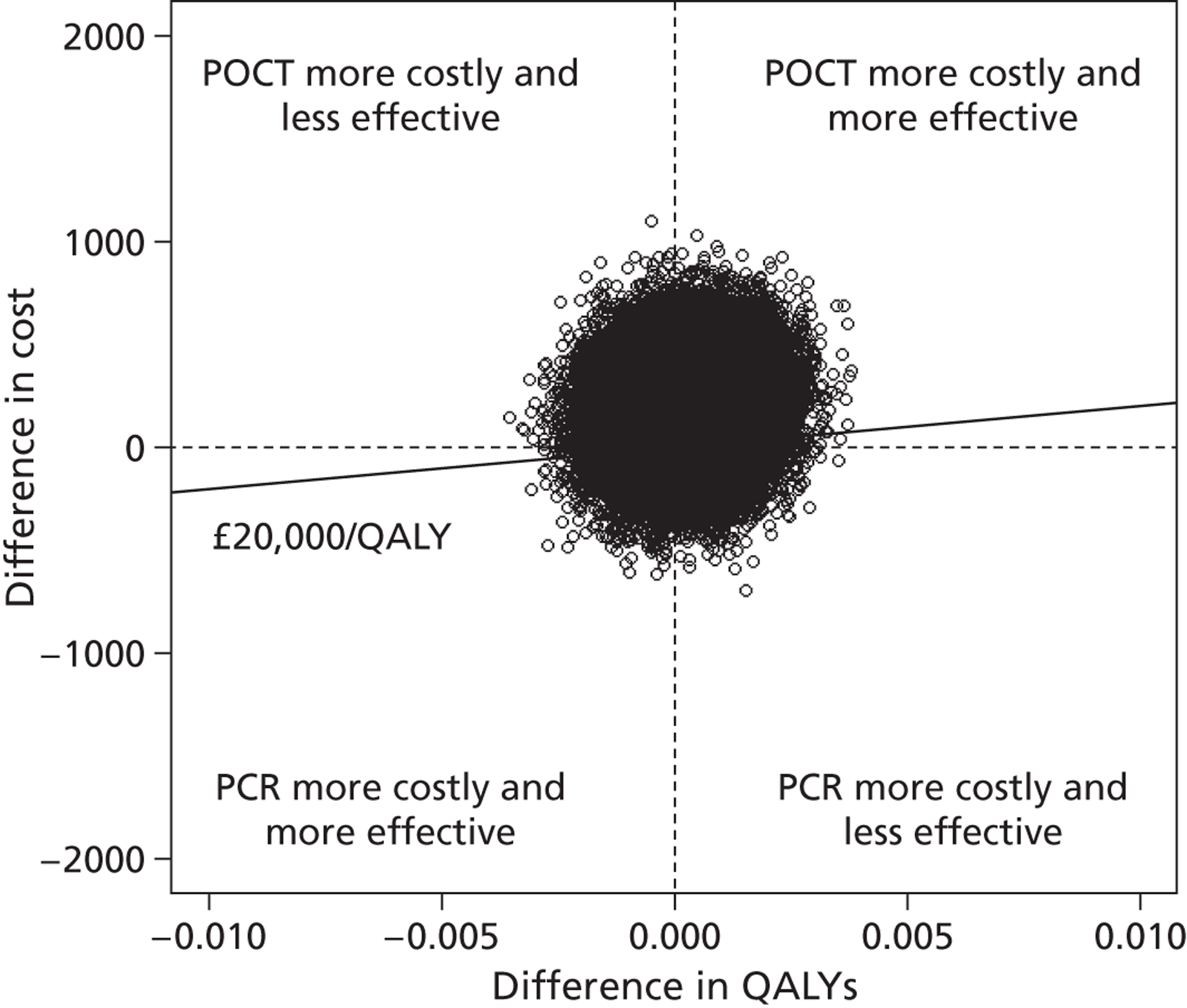
FIGURE 22.
Cost-effectiveness acceptability curve for the three diagnostic strategies, based on 50,000 MCMC samples together with a willingness-to-pay threshold of £20,000 per QALY.
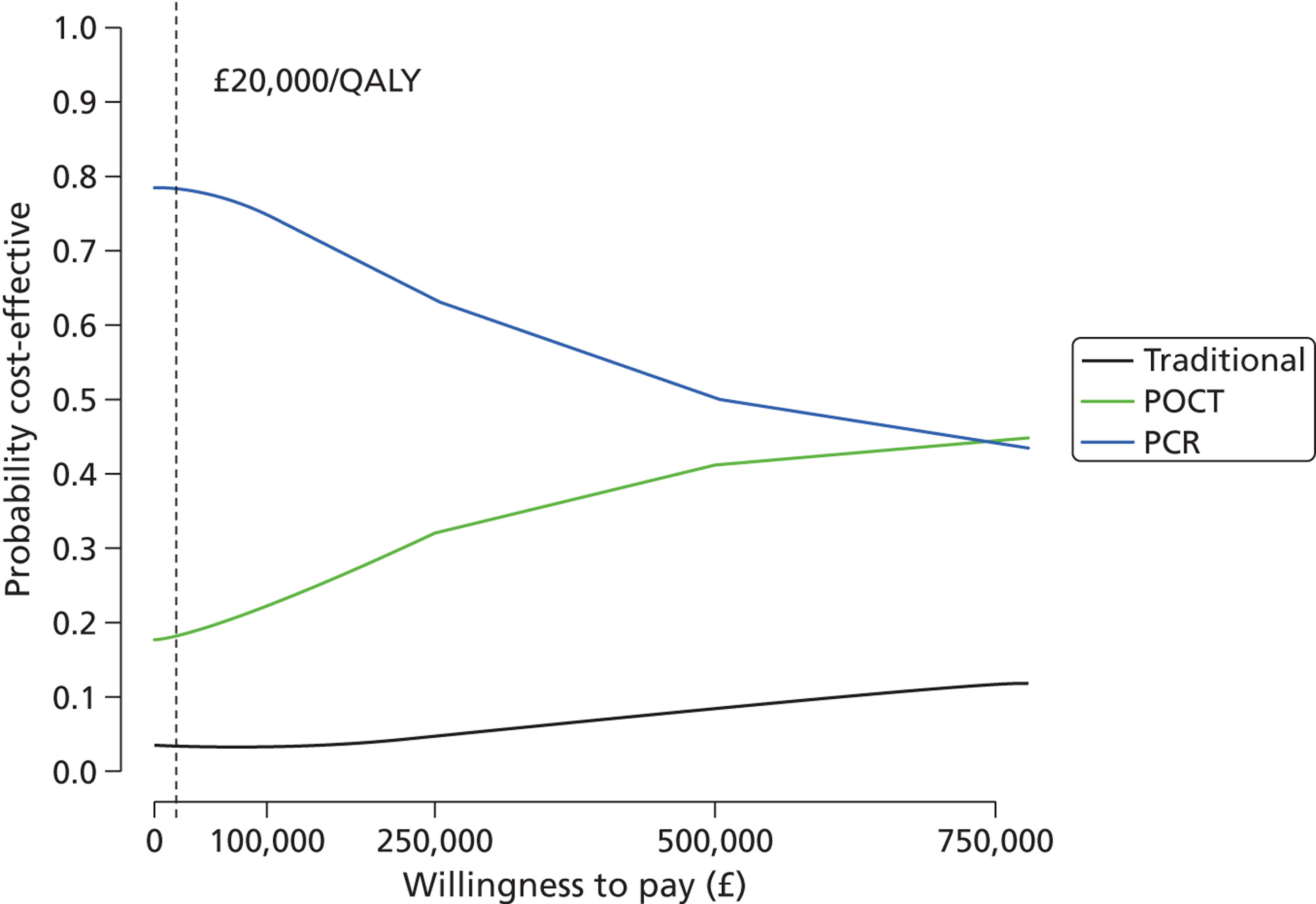
To assess the impact of price on the overall cost-effectiveness results, Table 36 displays the results of a series of one-way sensitivity analyses exploring the impact of 20% and 50% price reductions for POCT and PCR. It can be seen from Table 36 that all of the possible price reductions have relatively little impact on the estimated mean total costs and QALY gains for the three diagnostic strategies, for example even with a 50% price reduction for PCR the mean total costs for PCR are £1953 (95% CrI £1719 to £2189) compared with £1978 (95% CrI £1743 to £2165) in the base case, and similarly a 50% reduction in the price of POCT results in estimated mean total costs of £2139 (95% CrI £1813 to £2468) compared with £2159 (95% CrI £1828 to £2485) in the base case. In all cases not only are the estimated mean costs and QALYs very similar to the base case, but also the ranking of the treatments in terms of both costs and QALYs remains the same as in the base case.
| Models | Traditional | Rapid molecular | Near patient |
|---|---|---|---|
| Base case (model A) | |||
| Costs, average total cost (95% CrI) | 2327 (1989 to 2664) | 1978 (1743 to 2165) | 2159 (1828 to 2485) |
| QALYs | 0.007588 (0.006334 to 0.008854) | 0.007779 (0.006555 to 0.008983) | 0.008035 (0.006772 to 0.009280) |
| PCR, cost reduction of 20% | |||
| Costs, average total cost, £ (95% CrI) | 2327 (1989 to 2667) | 1967 (1733 to 2204) | 2160 (1834 to 2488) |
| QALYs | 0.007601 (0.006331 to 0.00889) | 0.007779 (0.006575 to 0.008985) | 0.008029 (0.006776 to 0.009286) |
| PCR, cost reduction of 50% | |||
| Costs, average total cost, £ (95% CrI) | 2327 (1989 to 2667) | 1953 (1719 to 2189) | 2160 (1834 to 2488) |
| QALYs | 0.007601 (0.006331 to 0.00889) | 0.007779 (0.006575 to 0.008985) | 0.008029 (0.006776 to 0.009286) |
| POCT, cost reduction of 20% | |||
| Costs, average total cost, £ (95% CrI) | 2327 (1989 to 2667) | 1977 (1743 to 2213) | 2152 (1825 to 2480) |
| QALYs | 0.007601 (0.006331 to 0.00889) | 0.007779 (0.006575 to 0.008985) | 0.008029 (0.006776 to 0.009286) |
| POCT, cost reduction of 50% | |||
| Costs, average total cost, £ (95% CrI) | 2327 (1989 to 2667) | 1977 (1743 to 2213) | 2139 (1813 to 2468) |
| QALYs | 0.007601 (0.006331 to 0.00889) | 0.007779 (0.006575 to 0.008985) | 0.008029 (0.006776 to 0.009286) |
| Alternative model (model B) | |||
| Costs, average total cost, £ (95% CrI) | 2349 (2131 to 2589) | 2007 (1819 to 2210) | 2189 (1976 to 2424) |
| QALYs | 0.008392 (0.007033 to 0.009750) | 0.008896 (0.007452 to 0.010341) | 0.009427 (0.007734 to 0.011120) |
Table 36 also presents the results obtained using an alternative statistical model (model B) in which costs are assumed to follow a gamma distribution and logit transformed QALYs a linked normal distribution. As anticipated, given the skewness of the original cost data presented in Figure 19 , the estimated mean costs for each of three diagnostic strategies are higher than in the base case, although for each diagnostic strategy the 95% CrIs for both mean costs and QALYs overlap, and the ranking of the treatments remains the same, with traditional laboratory culture being dominated by POCT, and the associated ICER for POCT compared with PCR, which still has the lowest estimated mean cost, being £342,750 per QALY, still considerably higher than the normal upper limit of £30,000 adopted by NICE in England and Wales.
Sensitivity analyses to the MCMC methods used are displayed in Appendix 8 , section B. Here, Table 47 displays the results of using different combinations of length of burn-in/sample, initial starting values and prior distributions. It can be seen that there appears to be little evidence of non-convergence of the MCMC estimation of model A, with the resulting estimated mean costs and QALYs being very similar across the different sensitivity analyses. This is further supported by Figures 23 – 25 , which display the history trace plots, the Brooks–Gelman–Rubin plots, and the autocorrelation function plots, respectively, for the estimated mean costs and QALYs for each diagnostic strategy, i.e. μ 1 j and μ 2 j . Figure 23 shows that the mean of the MCMC samples remains approximately constant over the 50,000 iterations with only random variation present, whereas Figure 24 shows that the two methods of calculating 80% CrIs (either by pooling the three chains, defined by the different sets of initial starting values, or by averaging over them) give very similar results in terms of the estimated mean costs and QALYs. Finally, Figure 25 indicates that the MCMC algorithm is efficiently sampling for the joint posterior distribution with relatively little autocorrelation present, i.e. successive samples appear to be virtually random, thus providing further evidence that the 20,000/50,000 burn-in/sample length combination used in the base case is sufficiently large.
FIGURE 23.
Markov chain Monte Carlo history plots for mean costs and QALYs.
FIGURE 24.
Brooks–Gelman–Rubin plots for chains 1, 2 and 3, representing three different sets of starting/initial values.
FIGURE 25.
Markov chain Monte Carlo autocorrelation plots for mean costs and QALYs.
Finally, Table 37 presents the results of an incremental cost per confirmed case of influenza analysis. In this analysis, potential cases of influenza identified by the three diagnostic strategies were confirmed by serology (see Chapter 5 ). It can be seen that on the basis of the diagnostic strategies the prevalence of influenza was low but confirmation with serology reduced this even further, with, in fact, no correctly identified cases being identified under the POCT strategy, thus hampering the calculation of a cost-per-case estimate. It should be noted that in Chapter 5 , as the aim was to estimate diagnostic performance, all 1252 admissions were used as each received all three diagnostic strategies but only those randomised to each strategy had their clinical management based on the strategy to which they randomised, and hence the absolute prevalence was considerably greater. Consequently, the cost per case for PCR was estimated as £164,570 (based on five cases) and for traditional laboratory culture £315,696 (based on three cases), with traditional laboratory culture being dominated by PCR.
| Mean total cost (£)/patient (95% CrI) | No. of patients with eligible serology | No. (%) of patients testing positive for influenza with index test | No. (%) of serology confirmed cases of influenza | Cost per confirmed case of influenza | Incremental cost per additional confirmed case of influenza detected |
|---|---|---|---|---|---|
| PCR | |||||
| 1978 (1743 to 2216) | 416 | 30 (7.2) | 5 (1.2) | 164,570 | –a |
| POCT | |||||
| 2159 (1828 to 2485) | 416 | 8 (1.9) | 0 (0) | –b | –a |
| Traditional | |||||
| 2327 (1989 to 2664) | 407 | 9 (2.2) | 3 (0.7) | 315,696 | Dominated |
Discussion
There is relatively little difference in the cost distributions or QALYs gained by each of the three diagnostic strategies. A strategy of using traditional laboratory culture led to an overall cost profile that is the most expensive but is also associated with the lowest gain in terms of QALYs, and is therefore dominated. Although POCT has the highest gain in terms of QALYs, this gain over PCR is not offset by its higher cost at current thresholds of willingness to pay.
The cost-per-case analysis is hampered by the relatively low prevalence (within each diagnostic strategy group) of influenza as diagnosed by serology. Indeed, as discussed in Chapter 5 and Chapter 6 , there are issues with the manner in which serology was undertaken in 3WS, although sensitivity analyses in Chapter 5 imply that as far possible this had little impact on the results, and in Chapter 6 the majority of other studies identified in fact used PCR as the ‘gold standard’. Thus, if the 30 cases of influenza identified in the PCR strategy group were in fact ‘true’ cases of influenza the cost per case detected would fall to £26,042 for that strategy compared with £164,570 when serology was used as a ‘gold standard’. If PCR was adopted as a ‘gold standard’ for the other two strategies, then five of the eight cases identified under the POCT strategy would be confirmed, rather than zero using serology, leading to a cost per case of £179,628, and all 9 cases identified by traditional laboratory culture would be confirmed leading to a cost per case of £105,232. Both POCT and traditional laboratory culture would however be dominated by PCR.
From a purely cost-effectiveness point of view, PCR would appear to be the most cost-effective diagnostic strategy, and, in fact, when viewed in light of the comparison with serology for influenza its superior diagnostic ability would reinforce that. However, set against this is the fact that there are no statistically significant differences between the three groups in terms of the main clinical and process outcomes. The fact that in the systematic review and meta-analysis, reported in Chapter 6 , the majority of studies identified used PCR as the ‘gold standard’ perhaps also underlines the fact PCR should be adopted as the de facto standard, but that POCT would appear to offer no clear benefit either in terms of cost or effectiveness.
As described in Chapter 2 , the 3WS was designed/powered to be able to estimate the diagnostic accuracy of the three strategies to within prespecified limits. In fact if the 3WS was designed with respect to clinical end points, for example length of stay, then the study would have been considerably smaller. However, with respect to cost-effectiveness or rather the differences in cost and effect, in terms of QALYs, a retrospective power analysis was undertaken. Assuming that cost-effectiveness was established using a willingness-to-pay threshold of £20,000 per QALY, the difference in mean costs and the difference in mean QALYs required to achieve such a threshold, providing the other outcome was held constant as estimated in Table 35 , the power of the study assuming a sample size of 1252 admissions was calculated. In terms of QALYs, the actual sample size of 3WS ensured over 90% power to detect a QALY difference that would ensure cost-effectiveness of one strategy over another. However, owing to the large variability in costs, as can be seen from Tables 34 and 35 for both the observed and estimated cost components and total costs, the 3WS as it was designed had < 50% power to be able to detect a minimum cost difference between any two strategies, assuming the QALY difference was as estimated in Table 35 . Whether 3WS could have ever been undertaken to ensure an appropriate level of power typically required, given the level of variability, is debatable, especially as its already relatively large sample size was driven by diagnostic accuracy rather than clinical effectiveness.
A possible major limitation of the cost-effectiveness analysis reported here is the fact that it is a purely trial-based analysis, with follow-up limited to 28 days post admission/randomisation, and that therefore further readmissions, for example, are not captured and costed appropriately. It could be argued that if any differences between the diagnostic strategies did exist then these would be expected to be seen within 28 days, and, in fact, within the index admission, and that if such a scenario did exist the key question would be whether any short-term benefit, for example if reduced length of stay was offset by a higher probability of readmission in the near future. However, as there are few differences, if any, between any of clinical outcomes, and in fact the clinical management of patients overall, then this would appear to be an unlikely scenario. A further limitation of the analyses presented is the level of missing data, approximately 50% for QALYs and 8.5% for costs, necessitating the use of MCMC predictive-based methods to impute missing values, conditional on the model and observed data. Although this approach is recommended for cost-effectiveness analyses,234 under the assumption that data were missing at random, a further sensitivity analysis using only complete cases was undertaken. The mean costs and QALYs for the three diagnostic strategies were broadly similar to those obtained using MCMC methods, and resulted in traditional laboratory culture remaining dominated, and POCT still having higher mean total cost than PCR, but with a smaller associated gain in QALYs, resulting in an ICER of £4,080,242 per QALY, and the probability of PCR being cost-effective rising to 93%. Finally, the perspective considered was that of the NHS, and it could be argued that there could be wider societal costs borne by patients or their informal carers, for example family members, which are not considered as part of the analyses presented here.
Chapter 9 Discussion
Main findings
Prescribing outcomes
-
lAntibiotic use was high in our cohort of patients. Altogether, 857 (73.1%) of all 1172 first admissions received at least one antibiotic for a median of 77 hours.
-
lOverall, 77.5% (96/120) of all first admissions with RSV or influenza received at least one antibiotic for a median of 77 hours – indicating that antibiotics are often prescribed for prolonged periods to patients with viral infections, many of whom are unlikely to derive clinical benefit. We found no evidence for an association between diagnostic group and time to final dose of antibiotics in patients infected with RSV or influenza.
-
lRegardless of the high level of antibiotic use, we found no evidence for any association between diagnostic group and three prescribing outcomes, specifically (1) time from admission to first narrow-spectrum antibiotic; (2) time from admission to first oral antibiotic; and (3) time from admission to cessation of antibiotics.
Clinical outcomes
-
Altogether, 48.5% (608/1252) of admissions had a raised temperature (≥ 37.3 °C) on admission. There was no evidence for any association between fever duration and diagnostic group for (1) all first admissions (regardless of any diagnosis) and (2) patients diagnosed with S. pneumoniae infection.
-
Overall, 43.6% (546/1252) of all admissions and 37.2% (436/1172) of first admissions received oxygen therapy during the admission. Time-to-event analysis for the 436 first admissions and for 40 out of 99 first admissions who were diagnosed with S. pneumoniae and were prescribed oxygen found no evidence of an association between diagnostic group and time to cessation of oxygen, in all patients, and in those diagnosed with S. pneumoniae.
-
CPAP dependence was infrequent in our patient cohort (1.2%, 14 out of 1172 first admissions). Time-to-event analysis by diagnostic group for all patients with a first admission found no evidence of an association between diagnostic group and time to cessation of CPAP.
-
Admissions to ITU were infrequent in this cohort (0.4%, 5 of 1172 first admissions). All five patients who were admitted to the ITU were ventilated. The small numbers of patients requiring ITU admission and ventilator support precluded comparison across the diagnostic groups for either end point.
-
We found no evidence for any association between duration of hospital stay and diagnostic group. Using a Cox proportional hazards model, we found that length of stay was comparable across the following groups (1) all first admissions who were discharged; (2) all first admissions discharged who were diagnosed with influenza; (3) all first admissions discharged who were diagnosed with RSV; and (4) all first admissions discharged who were diagnosed with S. pneumoniae infection.
-
Fifty deaths occurred within 28 days of admission. All 50 deaths occurred in first time admissions. We found no evidence to associate any diagnostic group with any increase or decrease in mortality for (1) ‘all’ patients, or for first admissions with a specific diagnosis of (2) influenza (n = 91); (3) RSV (n = 29); and (4) S. pneumoniae infection (n = 99).
-
Only 8.7% of first admissions (102/1172) were given single-room accommodation at some time during their admission. Hardly any (11.7%, 14/120) admissions with influenza or RSV received care in single-room accommodation. Owing to the small number of patients with RSV or influenza who were isolated, comparison across diagnostic groups was considered inappropriate. The majority of admissions with influenza and RSV were nursed in open areas of the hospital where they posed some degree of risk of nosocomial infection to vulnerable patients and staff. We found no evidence of any association with ‘containment’ across groups for infections with RSV, influenza, and S. pneumoniae.
-
We found no statistically significant differences between the three intervention groups in terms of EQ-5D scores on admission and subsequently. However, repeated measures analyses using a linear mixed-effects model revealed greater improvements in scores at both days 7 and 28 for the ‘near-patient’ and ‘rapid molecular’ groups compared with the ‘traditional’ group.
-
A review of data for eight patients in the ‘near-patient’ group with a positive test for influenza showed that none was eligible for treatment with a NI because they had illness duration of > 48 hours. None of the patients who remained in hospital after the result became available were isolated. There was no consistency in their subsequent treatment with antibiotics but the number of patients was too small to draw meaningful conclusions.
-
A review of data for 21 patients in the ‘near-patient’ group with a positive pneumococcal antigen test showed that none of the patients had a step-down in antibiotics within 24 hours of the result. The number eligible for treatment with a NI was small (4 out of 21), as assessed by duration of symptoms at presentation. There was no change in use of single-room accommodation following the diagnosis.
-
The median duration of symptoms before admission was 120 hours overall; few admissions were eligible for treatment with an NI as assessed by duration of symptoms.
Diagnostic accuracy
-
In comparison with PCR as the ‘gold standard’, the Quidel POCT had a sensitivity of 24.4% (95% CI 16% to 34.6%), a specificity of 99.7% (95% CI 99.2% to 99.9%), and PPVs and NPVs of 88.0% (95% CI 68.8% to 97.5%) and 94.4% (95% CI 93.0% to 95.7%), respectively.
-
In comparison with viral culture as the ‘gold standard’, the Quidel POCT had a sensitivity of 33.3% (95% CI 14.6% to 57.0%), a specificity of 98.6% (95% CI 97.7% to 99.2%), and PPVs and NPVs of 29.2% (95% CI 12.6% to 51.1%) and 98.8% (95% CI 98.0% to 99.3%), respectively.
-
In comparison with PCR as the ‘gold standard’, viral culture test had a sensitivity of 21.6%, (95% CI 13.5% to 31.6%), a specificity of 99.8% (95% CI 99.4% to 100%), and PPVs and NPVs of 90.5% (95% CI 69.6% to 98.8%) and 94.2% (95% CI 92.7% to 95.4%), respectively.
-
In comparison with viral culture as the ‘gold standard’, PCR had a sensitivity of 90.5%, (95% CI 69.6% to 98.8%), a specificity of 94.2% (95% CI 92.7% to 95.4%), and PPVs and NPVs of 21.6% (95% CI 13.5% to 31.6%) and 99.8% (95% CI 99.4% to 100.0%), respectively.
-
In comparison with serology as ‘gold standard’, PCR had the highest sensitivity (42.6%, 95% CI 28.3% to 57.8%). The sensitivity of viral culture and the Quidel POCT were low in comparison (culture: 13.3%, 95% CI 5.05% to 26.8%; POCT: 14.9%, 95% CI 6.2% to 28.3%).
-
In comparison with blood culture as the ‘gold standard’, the BinaxNOW pneumococcal POCT had a sensitivity of 57.1%, (95% CI 18.4% to 90.1%), a specificity of 92.5% (95% CI 90.6% to 94.1%), and PPVs and NPVs of 5.5% (95% CI 1.5% to 13.4%) and 99.6% (95% CI 99.0% to 99.9%), respectively.
-
In comparison with sputum culture as the ‘gold standard’, the BinaxNOW POCT had a sensitivity of 30.0% (95% CI 6.7% to 65.2%), a specificity of 92.0% (95% CI 88.1% to 95.0%), and PPVs and NPVs of 12.5% (95% CI 2.7% to 32.4%) and 97.2% (95% CI 94.3% to 98.9%), respectively.
-
In comparison with blood culture as the ‘gold standard’, sputum culture had a sensitivity of 100% (95% CI 2.5% to 100%), a specificity of 97.2% (95% CI 94.3% to 98.9%), and PPVs and NPVs of 12.5% (95% CI 0.3% to 52.7%) and 100% (95% CI 98.5% to 100%), respectively.
Systematic review and meta-analysis of point-of-care tests for influenza A and B
-
A bivariate mixed-effects meta-analysis model produced an overall estimate of sensitivity of 74% (95% CI 67% to 80%) and specificity of 99% (95% CI 98% to 99%). There was a high level of heterogeneity between the studies for both outcomes.
-
Exploration of between-study heterogeneity using bivariate mixed-effects meta-analysis showed that for some subgroup combinations the pooled estimate of sensitivity was considerably lower than that estimated for others:
-
Age Sensitivity of POCTs in children and adolescents was 86% (95% CI 75% to 93%) but 67% (95% CI 58% to 75%) in populations of ‘mixed’ age.
-
‘Gold standard’ Use of PCR as ‘gold standard’ produced an estimate of sensitivity of 51% (95% CI 38% to 64%), but the estimate of sensitivity was 86% (95% CI 77% to 92%) when virus culture was used as ‘gold standard’.
-
Target virus Sensitivity of POCTs for diagnosis of seasonal influenza was 84% (95% CI 74% to 90%) but 52% (95% CI 39% to 65%) for diagnosis of infection caused by 2009 pandemic H1N1 virus.
-
-
lComparison of estimates of sensitivity of POCTs produced by different manufacturers indicated that they were broadly similar – Directigen EZ: 85% (95% CI 71% to 93%); BinaxNOW: 69% (95% CI 58% to 79%); Quidel QuickVue: 66% (48–80%).
-
lComparison of estimates of sensitivity of POCTs for the detection of influenza A and B suggested that kits may detect influenza A more readily than influenza B – Influenza A: 81% (95% CI 64% to 91%); Influenza B: 59% (95% CI 47% to 70%).
-
lAnalyses of five studies152,182,198 that included both adults and children and compared the Quidel POCT with PCR produced an overall pooled sensitivity of 34% (95% CI 14% to 62%) and specificity of 99% (95% CI 97% to 100%), which are much more similar to those obtained in 3WS.
Speed of use
-
Quidel QuickVUE Influenza A&B POCT The median interval from specimen collection to the provision of a positive or negative result was 15 minutes (IQR 10–23 minutes).
-
BinaxNOW S. pneumoniae test: The median interval from specimen collection to the provision of a positive or negative result was 20 minutes (IQR 15–30 minutes).
-
Semi-nested multiplex PCR for influenza A subtypes H1 and H3, influenza B, and RSV A and B The median interval from specimen collection to the provision of a positive or negative result was 50.8 hours (IQR 44.3–92.6 hours).
-
One-step, quadriplex/multiplex, real-time RT-PCR for influenza, and one-step and two-step RSV and hMPV multiplex real-time PCR The median interval from specimen collection to the provision of a positive or negative result was 29.2 hours (IQR 26–46.9 hours). By 30 hours, results were available for 61.3% of 220 specimens that were tested.
-
Viral culture The median interval from specimen collection to the provision of a positive result for patients with influenza A or B was 629.6 hours (IQR 262.5–846.7 hours), which is approximately nine times greater than the median time to discharge or death (72.08 hours).
-
Blood culture: Eighty of the 973 blood cultures grew an organism (including contaminants), and 10 of the 80 grew S. pneumoniae with median time of 84.4 hours (IQR 70.7–137.8 hours); a provisional report was issued a median of 36.8 hours (IQR 22.7–48.75 hours) after specimen collection. Seven pneumococcal culture results were reported a median of 53.2 hours (IQR 30.4–58.1 hours) before the patients’ discharge. Three others were reported a median of 139.2 hours (IQR 27.9–163.4 hours) after death or discharge. Altogether 619 of 973 (63.6%) admissions were discharged or died before their blood culture results were reported.
-
Sputum culture The majority (941/1252: 75.2%, 95% CI 72.7% to 77.5%) of admissions with acute cardiopulmonary conditions were unable to produce sputum for culture. In total, 76 of the 296 sputum samples that were of acceptable quality for culture grew an organism; 10 of the 76 grew S. pneumoniae after a median interval from specimen collection of 71.4 hours (IQR 69.15–84.0 hours). The median TtR antimicrobial sensitivity was 133 hours (IQR 123.1–148.1 hours) for 8 out of the 10 isolates of S. pneumoniae. Altogether 129 of 294 (43.9%) patients were discharged before their sputum cultures were reported.
Ease of use
-
A scoring system was devised to compare the EoU of the different tests, based on 11 criteria, each assigned a score of 1, 2 or 3. A high score (maximum possible 33) reflects a high level of complexity, requiring specific expertise, equipment and facilities, etc. A score of ‘11’ identified test procedures as being straightforward and undemanding to use; scores of ‘12–22’ identified tests as being of moderate complexity and requirements; and scores of ‘23 and higher’ as being ‘complex’.
-
The QuickVUE Influenza A&B POCT and the BinaxNOW S. pneumoniae POCT were both rated as straightforward and undemanding to use. Blood culture was rated moderately complex. All other tests were rated as complex, with PCR testing being the most complex.
Process outcomes and cost-effectiveness
-
There were no statistically significant differences in the distributions of total costs, or QALYs gained, for the three diagnostic strategy groups, whether based on observed costs and effects, i.e. complete case analysis or after allowing for missing data using MCMC methods.
-
Average total NHS costs for the three diagnostic groups are relatively similar to one another (near-patient group, £2159; ‘molecular’ group £1978; ‘traditional’ group £2327), with the probability that any one strategy is the least costly not exceeding 79%.
-
The overall cost profile of traditional laboratory culture is the most expensive and is associated with the lowest gain in terms of QALYs.
-
The ‘near-patient’ group has the highest gain in terms of QALYs but this gain in QALYs is not offset by its higher cost at current thresholds of willingness to pay, with an ICER of £734,717.
-
In terms of cost per correct case of influenza detected, and verified by serology, both PCR and POCTs have lower estimates of cost than traditional laboratory culture but there is considerable uncertainty surrounding each. Overall, the cost per case detected was lowest by PCR.
Conclusions
-
The Quidel and BinaxNOW POCTs for influenza A and B and S. pneumoniae are straightforward and undemanding, and do not require laboratory facilities.
-
The Quidel and BinaxNOW POCTs for influenza A and B and S. pneumoniae provide results within minutes. Their speed of use provides opportunities to influence treatment decisions well within the median time to hospital discharge or death.
-
As judged by our trial, both the Quidel POCT for influenza A and B and traditional viral culture have low sensitivity when compared with PCR or serology as gold standard.
-
Meta-analysis of studies that included both adults and children and compared the Quidel POCT for influenza A and B with PCR produced an overall pooled sensitivity of 34%, which is similar to the observed sensitivity in our clinical trial.
-
The PCR tests are considered complex, requiring specialised equipment, reagents and expertise. The median TtR real-time PCR results for influenza A and B (≈29 hours) limits the usefulness of PCR in guiding treatment with NIs.
-
PCR has greater sensitivity than viral culture or the Quidel POCT for influenza A and B using serology as gold standard.
-
Conventional viral culture is demanding, requiring specialist equipment, reagents and skills. The time taken to report positive culture in our study was several-fold longer than the median duration of hospitalisation. Conventional viral culture results cannot provide results soon enough to influence clinical management or control of infection.
-
The BinaxNOW pneumococcal POCT has suboptimal sensitivity compared with blood culture. It cannot be used to rule out pneumococcal pneumonia.
-
Sputum culture is moderately demanding in terms of test site and expertise. The clinical usefulness of sputum culture is limited by the inability to produce sputum (75% were unable to produce sputum in our study) and TtR positive cultures (median ≈71 hours for S. pneumoniae).
-
Blood culture is moderately demanding in terms of equipment, test site and expertise. Blood cultures are often negative when the BinaxNOW pneumococcal POCT is positive. The median times to issuing a provisional report (≈37 hours) and identifying the organism as S. pneumoniae and its antimicrobial sensitivity (≈84 hours) further limit the usefulness of blood culture in guiding antimicrobial therapy.
-
All diagnostic tests that we evaluated had limitations, including suboptimal sensitivity, complexity, test requirements, or long turnaround time.
-
Few people in our study were admitted within 48 hours of illness onset. Virus shedding declines within days of illness onset, so diagnostic tests for influenza must be done early after illness onset and have very low limits of virus detection.
-
Few patients were prescribed NIs. The reason(s) remains speculative but delay between illness onset and admission, the knowledge base of junior doctors (i.e. unfamiliarity with viral diagnostic tests, NIs, and influenza and its complications) and the sensitivity and/or speed of use of the diagnostic tests are likely factors.
-
Many patients in this study were febrile on admission suggesting infection as a likely cause. The infrequent use of single-room accommodation across all admissions for acute cardiopulmonary illness (8.9%) and patients with influenza or RSV (11.7%) risks nosocomial transmission of infection.
-
We found no evidence that POCTs for influenza or S. pneumoniae infection, or PCR tests for influenza A and B and RSV A and B, influenced prescribing of antibiotics or NIs by clinicians providing care, or influenced clinical outcomes (including the duration of fever, requirement for supplemental oxygen, oxygen delivery by CPAP, admissions to ITU, ventilator support, deaths, or duration of hospitalisation) and use of single-room accommodation.
-
The total costs and QALYs for each diagnostic strategy were similar, although incrementally PCR was the most cost-effective strategy with a probability of being so of 78.3% at a willingness-to-pay threshold of £20,000 per QALY. Results of sensitivity analyses indicated that this conclusion appeared to be warranted. In terms of cost per correct case of influenza detected, traditional viral culture was the most expensive diagnostic strategy, and PCR was the least expensive.
-
Overall, there was a high level of consistency between the different facets of the study. The sensitivity of the Quidel POCT for influenza A and B was low and was in keeping with the results of our meta-analysis when adjustments were made for the ‘gold standard’, manufacturer and age distribution of those tested. The BinaxNOW pneumococcal test had suboptimal sensitivity that was comparable to sensitivities reported by others. Our study identified other factors that hamper patient care, notably the interval between illness onset and hospital admission, the limited availability of sputum, the infrequency of positive blood and sputum cultures, and the tardiness of individual tests (apart from the POCTs). We found no evidence that diagnostic strategy influenced clinical outcomes, or total costs and QALYs, findings that are consistent with the various limitations outlined above.
-
Our studies do not support routine use of POCTs for either influenza or pneumococcal antigen for adults hospitalised with acute cardiopulmonary conditions.
-
Our findings support the replacement of traditional viral culture by PCR – for reasons of greater sensitivity, speed, and cost-effectiveness.
Strengths and limitations of the study
Strengths
-
Compared with other studies identified in our meta-analysis, the 3WS is one of the largest undertaken on POCTs – only Ruest et al. ,152 Nougairede et al. 183 and Lucas et al. 142 had larger sample sizes, but clinicians in our study were blind to the nature of tests for clinical management.
-
Our study was powered to enable both clinical effectiveness (in terms of length of stay) and diagnostic performance to be evaluated with sufficient power/precision.
-
In terms of conduct, the prospective nature of 3WS meant that clinicians were blind to the results of the test strategies to which patients were not randomised. Thus, it enabled an unbiased assessment of the impact of the strategy to which they were randomised on the clinical management of eligible patients.
-
The research nurses who carried out the study spent most of their time on the admissions unit of the two hospitals providing care for acute medical admissions. They interacted directly with the teams of doctors and nurses providing initial care for the patient volunteers, particularly when patients were recruited, and also when the results of the POCTs were entered into the case notes and when the patients were followed up. Positive blood culture results were telephoned to the team in the usual manner, and results of microbiological tests were uploaded on to the pathology department results’ database when they became available. We ensured that bacteriology and virology results were made available to clinicians as soon as possible. Members of the research team and hospital microbiologists were available to answer any queries regarding the pathogens, the tests and the results of individual tests.
Limitations
-
The accuracy study for influenza and pneumococcal infection was limited primarily by the lack of an adequate reference standard. The serological tests for influenza were relatively unhelpful because the period between collection of acute and convalescent sera either coincided with annual vaccination against seasonal influenza or seasonal influenza activity. Blood culture is regarded as the gold standard for pneumococcal infection but cultures are often negative. This impacted on the cost-per-case analysis, and, as such, the findings of this analysis, should be considered exploratory.
-
It could be argued that the length of follow-up in 3WS, i.e. 28 days, was insufficient to detect any potential longer term consequences of the diagnostic strategies on the longer-term management and clinical outcomes of these patients. However, given the age distribution and clinical presentation of the patients in 3WS, 28 days should have enabled an appropriate assessment and evaluation of the index admission. The main driver in terms of patient outcome (and in fact of resource use, and therefore cost) was length of stay, and only 53 (4.2%) patient admissions had a length of stay beyond 28 days. The main outcome that further follow-up would have enabled to be assessed was readmission, although as 3WS covered the two acute hospitals in Leicestershire, readmissions pertaining to the index admission would have been identified, and, indeed, of the 1172 unique patients in 3WS, only 67 (5.7%) were readmitted (and rerandomised). Of course, there may have been other patients who were also readmitted but who declined to take any further part in 3WS. It also further brings into question the value of conducting further decision modelling over a longer time horizon, especially given that such an exercise would be dependent upon adequate resource use and outcome beyond that provided by 3WS.
-
This study was purposefully carried out during autumn through spring, a period that was specifically selected because it embraced seasonal influenza, RSV activity and the distinct winter seasonality when invasive pneumococcal disease reaches a peak. Clearly, if the study found no benefit from near-patient or molecular diagnostic tests when these pathogens peak (as occurred in this study) then it would be extremely unlikely to be of benefit during the summer months when cases of invasive pneumococcal disease are fewer, and those of RSV and influenza effectively absent. However, had the study shown a clear advantage of either the NPTs, or rapid molecular diagnosis, then there would be uncertainty of the value (both clinical effectiveness and cost-effectiveness) of such a programme when disease activity is much lower.
-
The molecular tests that were used in the study evolved during the study and were updated and implemented by a technician with considerable experience in molecular diagnostic tests, with back-up and training from the HPA. Although it could be argued that the time to report the PCR test results could have been shortened with a longer period of embedding the new developments into clinical practice, the complexity and turnaround times of PCR techniques actually decreased throughout the study. It could be argued that the study should have awaited newer developments and reconfiguration of the laboratory, as it is now possible to produce same day results. Indeed, this was implemented successfully in Leicester during the 2009 H1N1 pandemic; PCR results were available within 6 hours when specimens were delivered to the laboratory by 10 am.
-
Traditional virus culture technology was used as the comparator for PCR and the Quidel Influenza A + B POCT in this study. More rapid cell culture diagnostic techniques have been developed. Their cost, EoU, sensitivity and specificity, and clinical effectiveness and cost-effectiveness were not assessed in this study.
-
The cost-effectiveness analysis was subject to a variety of forms of uncertainty. There were a significant number of missing data, which, although allowed for in the MCMC-based analysis, was a concern. However, a series of sensitivity analyses has established that the conclusions of this report are, apparently, robust to both the inherent uncertainty in the data and methodological uncertainty induced by potentially different analytical approaches.
Implications for practice
-
Our findings do not support the routine use of POCTs for pneumococcal disease for all acute admissions with acute cardiopulmonary conditions. Indeed, the information provided by the POCT for pneumococcal antigen is considered, on the basis of our results, to be of questionable value even in patients with CAP. We note that at least three investigators have used the results of pneumococcal antigen test (exclusively) prospectively to target narrow-spectrum β-lactam treatment in CAP, with inconsistent results. 161,239,240 Guchev et al. 239 did a non-randomised study that evaluated a targeted approach to antibiotic therapy, based on the results of the pneumococcal urinary antigen test in young military recruits patients with non-severe pneumonia. Twenty-two per cent of patients with CAP had positive urinary tests and all were treated with oral amoxicillin. Treatment failures occurred in 5 out of 48 (10.4%) patients. Falguera et al. 240 treated 177 patients empirically and then randomised them into two arms when clinically stable; 89 patients were randomised to empiric treatment, and 88 to a target treatment study. Of the 88, 25 had positive urinary antigen tests and 63 had negative tests, i.e. 28.4% of the 88 patients were antigen positive. The 25 patients assigned to targeted treatment showed a statistically significant higher risk of clinical relapse compared with the remaining population (12% vs. 3%, p = 0.04). However, Sordé et al. 161 reduced the spectrum of antibiotics in 41 patients with positive antigen tests; pneumonia was cured in all patients. Factors for further consideration include the proportion of patients with CAP who have S. pneumoniae infection, the sensitivity and costs of the test, failure rate of ‘optimised’ antimicrobial therapy (i.e. step-down treatment) and whether or not the period of hospitalisation is cut by targeted treatment. The BTS, in its 2009 guidelines,17 recommended pneumococcal antigen testing in all patients with moderate- or high-severity CAP.
-
Our findings do not support the routine use of POCTs for influenza A and B throughout winter for adults presenting with acute cardiopulmonary conditions for three main reasons: first, the suboptimal sensitivity of these tests, particularly with the new lineage of H1N1 virus; second, evidence that the majority of patients present with illness duration exceeding 48 hours; and third, the lack of evidence that those identified as influenza positive had any change in treatment or isolation status. However, if further research substantiates preliminary evidence of benefit from NI treatment of patients who begin treatment in hospital > 48 hours after illness onset, there may be a place for using POCTs for influenza A and B in hospitals during periods of heightened influenza activity. The data accrued from our study and the 2009 H1N1 pandemic should facilitate modelling to identify whether this strategy might be cost-effective.
-
Our findings on the sensitivity, turnaround times, cost, and EoU of conventional diagnostic virology suggest that this technology should be replaced by PCR. The improved performance of PCR over conventional cell culture technology – in terms of test turnaround times and sensitivity – make PCR the preferred option, but it is acknowledged that PCR technology remains complex and demanding in terms of expertise and resources. We believe that laboratories should invest in developing molecular diagnostic processes and quality systems in preparation for forthcoming automation and molecular ‘black box technology’.
-
Our research identified as a sizeable problem the number of admissions with febrile acute respiratory illness who are nursed in open wards. This practice is likely to result in nosocomial transmission of influenza and other respiratory viruses that can be life-threatening in people with underlying heart and lung disease. Consideration must be given to one or more of the following: improving the design of future hospitals to include more single-room accommodation; managing single-room accommodation better than now; ensuring that all patients with acute respiratory illness follow ‘respiratory etiquette’; and use of quantitative PCR to support the flow of patients between single-roomed accommodation and an open-ward environment.
-
We are concerned that current NICE guidance70 is based on the results of double-blind RCTs that were designed and powered to establish whether NIs ameliorate symptomatic ILI. These studies were not powered to establish whether NI treatment prevents or ameliorates complications of influenza. Moreover, most RCTs were done in young otherwise healthy people who are not representative of patients who are hospitalised with influenza complications, including acute exacerbations of COPD, asthma, cystic fibrosis or heart failure. Recent publications indicate that (1) NIs may benefit patients when given > 48 hours after illness onset and (2) NIs ameliorate/prevent life-threatening complications. These observations were not generated by RCTs, rather by observational studies and risk being ignored.
Recommendations for research
-
We recommend a systematic review and meta-analysis of published data relating to the treatment of patients hospitalised with influenza with NIs to assess the evidence in support of treatment of patients hospitalised with influenza complications at > 48 hours after symptom onset.
-
Most patients with influenza complications in this study were unable to receive antiviral therapy because of delayed presentation. Patients risk serious outcomes from acute respiratory illness unless they are seen sooner. Research is needed to determine how widespread delayed presentation is, why it occurs and whether it can be reduced.
-
Because of the high specificity of POCTs for influenza, research is needed to determine their effectiveness in GP surgeries (or a commercial pharmacy setting) for people at risk of serious complications due to age and chronic ill-heath, during declared outbreaks.
-
There is good evidence that influenza virus exacerbates asthma, COPD, cystic fibrosis and is causally associated with CAP and acute bronchitis. Uncertainty about the role of NI treatment of patients presenting with these complaints during influenza outbreaks will remain until trials have shown clear benefits.
-
Controversy about the benefits of treatment with neuraminidase of patients presenting to hospital at > 48 hours (up to 6 days) after onset of symptoms will remain until clinical trials have established clear benefits. We recommend that this research include assessments of quantitative viral shedding and biomarkers to evaluate their role in guiding patient management.
Acknowledgements
KR Abrams is partially supported by the National Institute for Health Research (NIHR) as a Senior Investigator (NI-SI-0508–10061).
This study could not have been carried out without the help and support of research nurses Terry Smith, Aldona Kirkham, Annie Walkden, Sarah Bowrey and Hilary Pateman, who consented and recruited patients to the study, collected data and undertook the near-patient diagnostic tests. We are also grateful to others who contributed in various ways to the project, including the staff in the Microbiology Department, in general, for their effort. We have listed below particular individuals and their contributions.
Advice on original proposal:
-
Robert George, HPA, Centre for Infections, London.
-
Timothy Harrison, HPA, Centre for Infections, London.
-
David Turner, Principal Research Fellow in Health Economics, Wessex Institute, University of Southampton, Southampton.
Facilitation of trial recruitment:
-
Michael Steiner, University Hospitals of Leicester NHS Trust, Leicester.
-
Iain Stephenson, University Hospitals of Leicester NHS Trust, Leicester.
Contribution to laboratory analysis:
-
Catherine Thompson, HPA, Centre for Infections, London.
Contribution to the Trial Steering and Data Monitoring Committee:
-
Piers Lawford, University Hospitals Coventry and Warwickshire NHS Trust, Respiratory Medicine, Walsgrave Hospital, Coventry.
Design and maintenance of the database:
-
Hasmukh Patel, University of Leicester, Leicester.
Help to support the statistical analyses:
-
Chris Nelson at the Department of Health Sciences, University of Leicester, Leicester.
Contributions of authors
Karl G Nicholson was jointly responsible for the original proposal. He designed the study, drafted the protocol, contributed to the design of the data collection forms, led the process of integrating the various components into the final draft and drafted the final report.
Keith R Abrams was jointly responsible for the original proposal, for all of the statistical analyses and cost-effectiveness studies, and contributed to the drafting of the final report.
Sally Batham was trial manager and was involved in consent, data collection, patient contact, data entry and management of the research nurses, and contributed to the drafting of the final report.
Marie Jo Medina was responsible for the molecular diagnostics assays, collated data for all laboratory assays and contributed to the drafting of the final report. She also collated the evidence for the systematic review and meta-analysis of the POCTs for influenza.
Fiona C Warren performed statistical analyses regarding patient outcomes and contributed to the drafting of the final report.
Mike Barer was jointly responsible for the original proposal and oversaw the trial accounts.
Alison Bermingham advised on the original proposal and helped establish the multiplex PCR for influenza A and B, and RSV A and B, in Leicester. She also was responsible for the serological tests and interpretation of the results, and contributed to the drafting of the final report.
Tristan W Clark was responsible for coding the spectrum of the antibiotics that were prescribed to patients in this study and calculating the duration of their administration.
Nicholas Latimer advised on the collection/extraction of resource use data, identified unit cost sources and data, and advised on the analysis/reporting of the cost-effectiveness analyses.
Maria Fraser advised on the original proposal and, as consultant virologist at the University Hospitals of Leicester NHS Trust, supervised the virus isolation from clinical samples.
Nelun Perera was responsible for the conventional diagnostic bacteriology and for coordinating medical validation and reporting of laboratory results of conventional diagnostic tests to the clinicians responsible for the care of patients in this study, and contributed to the drafting of the final report.
K Rajakumar helped establish the multiplex PCR for influenza A and B and RSV A and B, in Leicester.
Maria Zambon advised on the original proposal and facilitated the initial transfer of the multiplex PCR to Leicester and the serological tests at the HPA Centre for Infections, Colindale, and contributed to the drafting of the final report.
The report was drafted by Karl Nicholson (principal investigator and Professor of Infectious Diseases), Keith Abrams (Professor of Medical Statistics), Sally Batham (trial manager), Marie Jo Medina, Fiona Warren and Maria Zambon. These authors take responsibility for the content of the report. All authors approved the final draft. Keith Abrams and Karl Nicholson are guarantors for the report.
Disclaimers
This report presents independent research funded by the National Institute for Health Research (NIHR). The views and opinions expressed by authors in this publication are those of the authors and do not necessarily reflect those of the NHS, the NIHR, NETSCC, the HTA programme or the Department of Health.
Publication
Bafadhel M, Clark TW, Reid C, Medina M, Batham S Barer M, et al. Procalcitonin and C reactive protein in hospitalised adult patients with community acquired pneumonia, exacerbation of asthma and chronic obstructive pulmonary disease [published online ahead of print October 28 2010]. Chest 2011;139:1410–18. DOI: 10.1378/chest.10-1747.
References
- National Statistics News Release . UK Population Set to Increase to 65 Million over the Next Ten Years 2009. www.statistics.gov.uk/pdfdir/pproj1007.pdf (accessed 11 February 2009).
- NHS under pressure as the number of elderly patients rises. The Times 2010.
- HESonline . Hospital Episode Statistics Headline Figures 2009–10 n.d. www.hesonline.nhs.uk/Ease/servlet/ContentServer?siteID = 1937&categoryID = 193 (accessed 15 August 2013).
- HESonline . Hospital Episode Statistics Primary Diagnosis: Summary 2009–10 n.d. www.hesonline.nhs.uk/Ease/servlet/ContentServer?siteID = 1937&categoryID = 202 (accessed 15 August 2013).
- McCormick A, Fleming D, Charlton J. Morbidity Statistics from General Practice – Fourth National Study 1991/92. London: HMSO; 1995.
- DeFrances CJ, Hall MJ, Podgornik MN. 2003 National Hospital Discharge Survey 2005;359:1-20. www.cdc.gov/nchs/data/ad/ad359.pdf (accessed 15 August 2013).
- Health, United States, 2009, with special feature on medical technology. Table 102 (page 1 of 3). Average length of stay in nonfederal short-stay hospitals, by sex, age, and selected first-listed diagnosis: United States, 1990, 2000, and 2006 n.d.:365-7. www.cdc.gov/nchs/data/hus/hus09.pdf#102oryID=202 (accessed 15 August 2013).
- Falsey AR, Cunningham CK, Barker WH, . Respiratory syncytial virus and influenza A infections in the hospitalised elderly. J Infect Dis 1995;172:389-94.
- Mufson MA, Chang V, Gill V, . The role of viruses, mycoplasmas and bacteria in acute pneumonia in civilian adults. Am J Epidemiol 1967;86:526-44.
- Fransen H, Heigl Z, Wolontis S, . Infections with viruses in patients hospitalised with acute respiratory illness. Scand J Infect Dis 1969;1:127-36.
- Dowell SF, Anderson LJ, Gary HE, . Respiratory syncytial virus is an important cause of community-acquired lower respiratory infection among hospitalized adults. J Infect Dis 1996;174:456-62. http://dx.doi.org/10.1093/infdis/174.3.456.
- Glezen WP, Greenberg SB, Atmar RL, Piedra PA, Couch RB. Impact of respiratory virus infections on persons with chronic underlying conditions. JAMA 2000;283:499-505. http://dx.doi.org/10.1001/jama.283.4.499.
- Falsey AR, Walsh EE, Hayden FG. Rhinovirus and coronavirus infection-associated hospitalizations among older adults. J Infect Dis 2002;185:1338-41. http://dx.doi.org/10.1086/339881.
- Macfarlane J, Mayon-White RT. The clinical impact of pneumococcal disease. London: Royal Society of Medicine Press; 1995.
- Lieberman D, Schlaeffer F, Boldur I, . Multiple pathogens in adult patients admitted with community-acquired pneumonia: a one year prospective study of 346 consecutive patients. Thorax 1996;51:179-84. http://dx.doi.org/10.1136/thx.51.2.179.
- Fine MJ, Smith MA, Carson CA, . Prognosis and outcomes of patients with community-acquired pneumonia: a meta-analysis. JAMA 1996;275:134-41. http://dx.doi.org/10.1001/jama.275.2.134.
- British Thoracic Society Community Acquired Pneumonia in Adults Guideline Group . Guidelines for the management of community acquired pneumonia in adults: update 2009. Thorax 2009;64:1-55. www.brit-thoracic.org.uk/Portals/0/Clinical%20Information/Pneumonia/Guidelines/CAPGuideline-full.pdfntimicrob.
- Nicholson KG, Wood JM, Zambon M. Influenza. Lancet 2003;362:1733-45. http://dx.doi.org/10.1016/S0140-6736(03)14854-4.
- Nguyen-Van-Tam JS, Openshaw PJM, Hashim A, Gadd EM, Lim WS, Semple MG, et al. Risk factors for hospitalisation and poor outcome with pandemic A/H1N1 influenza: United Kingdom first wave (May–September 2009). Thorax 2010;65:645-51. http://dx.doi.org/10.1136/thx.2010.135210.
- Nicholson KG. Impact of influenza and respiratory syncytial virus on mortality in England and Wales from January 1975 to December 1990. Epidemiol Infect 1996;116:51-63. http://dx.doi.org/10.1017/S0950268800058957.
- Meier CR, Napalkov PN, Wegmüller Y, Jefferson T, Jick H. Population-based study on incidence, risk factors, clinical complications and drug utilization associated with influenzas in the United Kingdom. Eur J Clin Microbiol Infect Dis 2000;19:834-42.
- Nicholson KG, McNally T, Silverman M, Simons P, Stockton JD, Zambon MC. Rates of hospitalisation for influenza, respiratory syncytial virus and human metapneumovirus among infants and young children. Vaccine 2006;24:102-8. http://dx.doi.org/10.1016/j.vaccine.2005.02.004.
- Nicholson KG, Baker DJ, Farquhar A, Hurd D, Kent J, Smith SH. Acute upper respiratory tract viral illness and influenza immunization in homes for the elderly. Epidemiol Infect 1990;105:609-18. http://dx.doi.org/10.1017/S0950268800048251.
- Nicholson KG, Kent J, Hammersley V, Cancio E. Acute viral infections of upper respiratory tract in elderly people living in the community: comparative, prospective, population based study of disease burden. BMJ 1997;315:456-62. http://dx.doi.org/10.1136/bmj.315.7115.1060.
- Kim PE, Musher DM, Glezen WP, Rodriguez-Barradas MC, Nahm WK, Wright CE. Association of invasive pneumococcal disease with season, atmospheric conditions, air pollution, and the isolation of respiratory viruses. Clin Infect Dis 1996;22:100-6. http://dx.doi.org/10.1093/clinids/22.1.100.
- Peltola V, Heikkinen T, Ruuskanen O, Jartti T, Hovi T, Kilpi T, et al. Temporal association between rhinovirus circulation in the community and invasive pneumococcal disease in children. Pediatr Infect Dis J 2011;30:456-61.
- Murdoch DR, Jennings LC. Association of respiratory virus activity and environmental factors with the incidence of invasive pneumococcal disease. J Infect 2009;58:37-46. http://dx.doi.org/10.1016/j.jinf.2008.10.011.
- Campbell SG, Marrie TJ, Anstey R, . The contribution of blood cultures to the clinical management of adult patients admitted to the hospital with community-acquired pneumonia. A prospective observational study. Chest 2003;123:1142-50. http://dx.doi.org/10.1378/chest.123.4.1142.
- Porath A, Schlaeffer F, Pick N, . Pneumococcal community-acquired pneumonia in 148 hospitalized adult patients. Eur J Clin Microbiol Infect Dis 1997;16:863-70. http://dx.doi.org/10.1007/BF01700551.
- Kauppinen M, Saikku P, Kujala P, . Clinical picture of community-acquired Chlamydia pneumoniae requiring hospital treatment: a comparison between chlamydial and pneumococcal pneumonia. Thorax 1996;51:185-9. http://dx.doi.org/10.1136/thx.51.2.185.
- Oosterheert JJ, Bonten MJ, Hak E, . How good is the evidence for the recommended empirical antimicrobial treatment of patients hospitalized because of community-acquired pneumonia? A systematic review. J Antimicrob Chemother 2003;52:555-63. http://dx.doi.org/10.1093/jac/dkg413.
- Woo PCY, Chiu SS, Seto W-H, . Cost-effectiveness of rapid diagnosis of viral respiratory tract infections in pediatric patients. J Clin Microbiol 1997;35:1579-81.
- Zambon MC, Hays J, Webster A, . Diagnosis of influenza in the community. Relationship of clinical diagnosis to confirmed virological, serologic, or molecular detection of influenza A. Arch Intern Med 2001;161:2116-22. http://dx.doi.org/10.1001/archinte.161.17.2116.
- Noyola DE, Clark B, O’Donnell FT, . Comparison of a new neuraminidase detection assay with an enzyme immunoassay, immunofluorescence, and culture for rapid detection of influenza A and B viruses in nasal wash specimens. J Clin Microbiol 2000;38:1161-5.
- Ellis JS, Fleming DM, Zambon MC. Multiplex RT-PCR for surveillance of influenza in England 1995/1996. J Clin Microbiol 1997;35:2076-82.
- Stockton J, Ellis JS, Saville M, Clewley JP, Zambon MC. Multiplex PCR for typing and subtyping influenza and respiratory syncytial viruses. J Clin Microbiol 1998;36:2990-5.
- Falsey AR, McCann RM, Hall WJ, Criddle MM. Evaluation of four methods for the diagnosis of respiratory syncytial virus infection in older adults. J Am Geriatr Soc 1996;44:71-3.
- Aida E, Casiano-Colón AE, Hulbert BB, Mayer TK, Walsh EE, Falsey AR. Lack of sensitivity of rapid antigen tests for the diagnosis of respiratory syncytial virus infection in adults. J Clin Virol 2003;28:169-74.
- Zambon MC, Stockton JD, Clewley JP, Fleming DM. Contribution of influenza and respiratory syncytial virus to community cases of influenza-like illness: an observational study. Lancet 2001;358:1410-16. http://dx.doi.org/10.1016/S0140-6736(01)06528-X.
- Ewig S, Schlochtermeier M, Goke N, . Applying sputum as a diagnostic tool in pneumonia. Chest 2002;121:1486-92. http://dx.doi.org/10.1378/chest.121.5.1486.
- Campbell SG, Marrie TJ, Anstey R, . The contribution of blood cultures to the clinical management of adult patients admitted to the hospital with community-acquired pneumonia. A prospective observational study. Chest 2003;123:1142-50. http://dx.doi.org/10.1378/chest.123.4.1142.
- Mandell LA, Marrie TJ, Grossman RF, . Canadian guidelines for the initial management of community-acquired pneumonia: an evidence based update by the Canadian Infectious Diseases Society and the Canadian Thoracic Society. Clin Infect Dis 2000;31:383-421. http://dx.doi.org/10.1086/313959.
- Moine P, Vercken J-B, Chevret C, . Severe community-acquired pneumonia: etiology, epidemiology and prognosis factors. Chest 1994;105:1487-95. http://dx.doi.org/10.1378/chest.105.5.1487.
- Chalasani NP, Valdecanaas MAL, Gopal AK, . Clinical utility of blood cultures in adult patients with community-acquired pneumonia without defined underlying risks. Chest 1995;108:932-8. http://dx.doi.org/10.1378/chest.108.4.932.
- Sanyal S, Smith PR, Saha AC, . Initial microbiologic studies did not affect outcome in patients hospitalised with community-acquired pneumonia. Am J Respir Crit Care Med 1999;160:346-8.
- Waterer GW, Jennings SG, Wunderink RG. The impact of blood cultures on antibiotic therapy in pneumococcal pneumonia. Chest 1999;116:1278-81. http://dx.doi.org/10.1378/chest.116.5.1278.
- Musher DM, Meddiwala MR, Phan HM, . Non-specificity of assaying for IgG antibody to pneumolysin in circulating immune complexes as a means to diagnose pneumococcal pneumonia. Clin Infect Dis 2001;32:534-8.
- Murdoch DR, Anderson TP, Beynon KA, . Evaluation of a PCR assay for detection of Streptococcus pneumoniae in respiratory and non-respiratory samples from adults with community-acquired pneumonia. J Clin Microbiol 2003;41:63-6.
- Smith MD, Sheppard CL, Hogan A, Harrison TG, Dance DAB, Derrington P, et al. Diagnosis of Streptococcus pneumoniae infections in adults with bacteremia and community-acquired pneumonia: clinical comparison of pneumococcal PCR and urinary antigen detection. J Clin Microbiol 2009;47:1046-9. http://dx.doi.org/10.1128/JCM.01480-08.
- Murdoch DR, Laing RTR, Mills GD, . Evaluation of a rapid immunochromatographic test for detection of Streptococcus pneumoniae antigen in urine samples from adults with community-acquired pneumonia. J Clin Microbiol 2001;39:3495-8. http://dx.doi.org/10.1128/JCM.39.10.3495-3498.2001.
- Farina C, Arosio M, Vailati F, . Urinary detection of Streptococcus pneumoniae antigen for diagnosis of pneumonia. New Microbiol 2002;25:259-63.
- Smith MD, Derrington P, Evans R, . Rapid diagnosis of bacteremic pneumococcus infections in adults by using the BinaxNOW Streptococcus pneumoniae urinary antigen test: a prospective controlled clinical evaluation. J Clin Microbiol 2003;41:2810-13. http://dx.doi.org/10.1128/JCM.41.7.2810-2813.2003.
- Roson B, Fernandez-Sabe N, Carratala J, Verdaguer R, Dorca J, Manresa F, et al. Contribution of a urinary antigen assay (BinaxNOW) to the early diagnosis of pneumococcal pneumonia. Clin Infect Dis 2004;38:222-6.
- Dominguez J, Blanco S, Rodrigo C, . Usefulness of urinary antigen detection by an immunochromatographic test for diagnosis of pneumococcal pneumonia in children. J Clin Microbiol 2003;41:2161-3. http://dx.doi.org/10.1128/JCM.41.5.2161-2163.2003.
- Kind P, Spiker B. Quality of Life and Pharmacoeconomics in Clinical Trials. Philadelphia, PA: Lippincott-Raven; 1996.
- The EuroQol Group . EuroQol: a new facility for the measurement of health-related quality of life. Health Policy 1990;16:199-208.
- Brooks R. EuroQol: the current state of play. Health Policy 1996;37:53-72. http://dx.doi.org/10.1016/0168-8510(96)00822-6.
- Guide to the Methods of Health Technology Appraisal. London: NICE; 2004.
- Dolan P, Gudex C, Kind P. A Social Tariff for EuroQoL: Results from a UK General Population Survey. York: University of York; 1995.
- Boom R, Sol CJA, Salimans MMM, Jansen CL, Wertheim-van Dillen PME, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol 1990;28:495-503.
- Ellis JS, Curran MD. Simultaneous molecular detection and confirmation of influenza AH5, with internal control. Methods Mol Biol 2011;665:161-81.
- Stephenson I, Democratis J, Lackenby A, McNally T, Smith J, Pareek M, et al. Neuraminidase inhibitor resistance after oseltamivir treatment of acute influenza A and B in children. Clin Infect Dis 2009;48:389-96. http://dx.doi.org/10.1086/596311.
- Wang K, Chalker V, Bermingham A, Harrison T, Mant D, Harnden A. Mycoplasma pneumoniae and respiratory virus infections in children with persistent cough in England: a retrospective analysis. Pediatr Infect Dis J 2011;30:1047-51. http://dx.doi.org/10.1097/INF.0b013e31822db5e2.
- Bryden AS. Isolation of influenza virus A and B in PLC/PRF/5 cells. Br J Biomed Sci 1996;53:93-5.
- Mézière A, Mollat C, Lapied R, Billaudel S, Courtieu AL. Detection of respiratory syncytial virus antigen after seventy-two hours of culture. J Med Virol 1990;31:241-4. http://dx.doi.org/10.1002/jmv.1890310312.
- Dolan P, Roberts J. Modelling values for EQ-5D health states: an alternative model using differences in valuations. Med Care 2002;40:442-6.
- Prieto L, Sacristan JA. What is the value of social values? The uselessness of assessing health-related quality of life through preference measures. BMC Med Res Meth 2004;4.
- Falsey AR, Murata Y, Walsh E. Impact of rapid diagnosis on management of adults hospitalised with influenza. Arch Intern Med 2007;167:354-60.
- D’Heilly J, Janoff EN, Nichol P, Nichol KL. Rapid diagnosis of influenza infection in older adults: influence on clinical care in a routine clinical setting. J Clin Virol 2008;42:124-8. http://dx.doi.org/10.1016/j.jcv.2007.12.014.
- National Institute for Health and Clinical Excellence (NICE) . Amantadine, Oseltamivir and Zanamivir for the Treatment of Influenza n.d. http://guidance.nice.org.uk/TA168/Guidance/pdf/English (accessed 25 February 2009).
- McGeer A, Green KA, Plevneshi A, Shigayeva A, Siddiqi N, Raboud J, et al. Antiviral therapy and outcomes of influenza requiring hospitalization in Ontario, Canada. Clin Infect Dis 2007;45:1568-75. http://dx.doi.org/10.1086/523584.
- Lee N, Cockram CS, Chan PKS, Hui DSC, Choi KW, Sung JJY. Antiviral treatment for patients hospitalized with severe influenza infection may affect clinical outcomes. Clin Infect Dis 2008;46:1323-4. http://dx.doi.org/10.1086/533477.
- Hanshaoworakul W, Simmerman JM, Narueponjirakul U, Sanasuttipun W, Shinde V, Kaewchana S, et al. Severe human influenza infections in Thailand: oseltamivir treatment and risk factors for fatal outcome. PLOS ONE 2009;4. http://dx.doi.org/10.1371/journal.pone.0006051.
- Yu H, Liao Q, Yuan Y, Zhou L, Xiang N, Huai Y, et al. Effectiveness of oseltamivir on disease progression and viral RNA shedding in patients with mild pandemic 2009 influenza A H1N1: opportunistic retrospective study of medical charts in China. BMJ 2010;341. http://dx.doi.org/10.1136/bmj.c4779.
- Jain S, Kamimoto L, Bramley AM, Schmitz AM, Benoit SR, Louie J, et al. Hospitalized patients with 2009 H1N1 influenza in the United States, April–June 2009. N Engl J Med 2009;361:1935-44. http://dx.doi.org/10.1056/NEJMoa0906695.
- Zarychanski R, Stuart TL, Kumar A, Doucette S, Elliott L, Kettner J, et al. Correlates of severe disease in patients with 2009 pandemic influenza (H1N1) virus infection. CMAJ 2010;182:257-64. http://dx.doi.org/10.1503/cmaj.091884.
- Lee N, Choi KW, Chan PKS, Hui DSC, Lui GCY, Ong BCK, et al. Outcomes of adults hospitalised with severe influenza. Thorax 2010;65:510-15. http://dx.doi.org/10.1136/thx.2009.130799.
- Dominguez-Cherit G, Lapinsky SE, Macias AE, Pinto R, Espinosa-Perez L, de la Torre A, et al. Critically ill patients with 2009 influenza A(H1N1) in Mexico. JAMA 2009;302:1880-7. http://dx.doi.org/10.1001/jama.2009.1536.
- Higuera Iglesias AL, Kudo K, Manabe T, Corcho Berdugo AE, Corrales Baeza A, Alfaro Ramos L, et al. Reducing occurrence and severity of pneumonia due to pandemic H1N1 2009 by early oseltamivir administration: a retrospective study in Mexico. PLOS ONE 2011;6.
- Siston AM, Rasmussen SA, Honein MA, Fry AM, Seib, K, Callaghan WM, et al. Pandemic 2009 Influenza A(H1N1) virus illness among pregnant women in the United States. JAMA 2010;303:1517-25. http://dx.doi.org/10.1001/jama.2010.479.
- Adisasmito W, Chan PKS, Lee N, Oner AF, Gasimov V, Aghayev F, et al. Effectiveness of antiviral treatment in human influenza A(H5N1) infections: analysis of a global patient registry. J Infect Dis 2010;202:1154-60. http://dx.doi.org/10.1086/656316.
- Ross AM, Kai J, Salter R, Ross J, Fleming DM. Presentation with influenza-like illness in general practice: implications for use of neuraminidase inhibitors. Community Dis Public Health 2000;3:256-60.
- Nilsson AC, Alemo B, Björkman P, Dillner L, Melhus Å, Nilsson B, et al. Around-the-clock, rapid diagnosis of influenza by means of membrane chromatography antigen testing confirmed by polymerase chain reaction. Infect Control Hosp Epidemiol 2008;29:177-9. http://dx.doi.org/10.1086/526446.
- Stripeli F, Sakkou Z, Papadopoulos N, Georgiou V, Gratsia P, Christodopolou I, et al. Performance of rapid influenza testing in hospitalized children. Eur J Clin Microbiol Infect Dis 2010;29:683-8. http://dx.doi.org/10.1007/s10096-010-0914-2.
- Vasoo S, Stevens J, Singh K. Rapid antigen tests for diagnosis of pandemic (swine) influenza A/H1N1. Clin Infect Dis 2009;49:1090-3.
- Heinonen S, Silvennoinen H, Lehtinen P, Vainionpää R, Heikkinen T. Feasibility of diagnosing influenza within 24 hours of symptom onset in children 1–3 years of age. Eur J Clin Microbiol Infect Dis 2011;30:387-92.
- Cazacu AC, Greer J, Taherivand M, Demmler GJ. Comparison of lateral-flow immunoassay and enzyme immunoassay with viral culture for rapid detection of influenza virus in nasal wash specimens from children. J Clin Microbiol 2003;41:2132-4. http://dx.doi.org/10.1128/JCM.41.5.2132-2134.2003.
- Hurt AC, Alexander R, Hibbert J, Deed N, Barr IG. Performance of six influenza rapid tests in detecting human influenza in clinical specimens. J Clin Virol 2007;39:132-5. http://dx.doi.org/10.1016/j.jcv.2007.03.002.
- Cruz AT, Cazacu AC, Greer JM, Demmler GJ. Rapid assays for the diagnosis of influenza A and B viruses in patients evaluated at a large tertiary care children’s hospital during two consecutive winter seasons. J Clin Virol 2008;41:143-7. http://dx.doi.org/10.1016/j.jcv.2007.11.006.
- Scansen KA, Bonsu BK, Stoner E, Mack K, Salamon D, Leber A, et al. Comparison of polyurethane foam to nylon flocked swabs for collection of secretions from the anterior nares in performance of a rapid influenza virus antigen test in a pediatric emergency department. J Clin Mirobiol 2010;48:852-6. http://dx.doi.org/10.1128/JCM.01897-09.
- Agoritsas K, Mack K, Bonsu BK, Goodman D, Salamon D, Marcon MJ. Evaluation of the Quidel QuickVue test for detection of influenza A and B viruses in the pediatric emergency medicine setting by use of three specimen collection methods. J Clin Microbiol 2006;44:2638-41. http://dx.doi.org/10.1128/JCM.02644-05.
- Walsh P, Overmyer CL, Pham K, Michaelson S, Gofman L, DeSalvia L, et al. Comparison of respiratory virus detection rates for infants and toddlers by use of flocked swabs, saline aspirates, and saline aspirates mixed in universal transport medium for room temperature storage and shipping. J Clin Microbiol 2008;46:2374-6. http://dx.doi.org/10.1128/JCM.00714-08.
- Schmid ML, Kudesia G, Wake S, Read RC. Prospective comparative study of culture specimens and methods in diagnosing influenza in adults. BMJ 1998;316. http://dx.doi.org/10.1136/bmj.316.7127.275.
- Smit M, Beynon KA, Murdoch DR, Jennings LC. Comparison of the NOW Influenza A & B, NOW Flu A, NOW Flu B, and Directigen Flu A + B assays, and immunofluorescence with viral culture for the detection of influenza A and B viruses. Diagn Microbiol Infect Dis 2007;57:67-70. http://dx.doi.org/10.1016/j.diagmicrobio.2006.11.003.
- Andre M, Odenholt I, Schwan A, Axelsson I, Eriksson M, Hoffman M. Upper respiratory tract infections in general practice: diagnosis, antibiotic prescribing, duration of symptoms and use of diagnostic tests. Scand J Infect Dis 2002;34:880-6.
- Molstad S. Reduction in antibiotic prescribing for respiratory tract infections is needed!. Scand J Prim Health Care 2003;21:196-8. http://dx.doi.org/10.1080/02813430310003273.
- Roumie CL, Halasa NB, Grijalva CG, Edwards KM, Zhu Y, Dittus RS, et al. Trends in antibiotic prescribing for adults in the United States: 1995 to 2002. J Gen Intern Med 2005;20:697-702. http://dx.doi.org/10.1111/j.1525-1497.2005.0148.x.
- Ashworth M, Latinovic R, Charlton J, Cox K, Rowlands G, Gulliford M. Why has antibiotic prescribing for respiratory illness declined in primary care? A longitudinal study using the General Practice Research Database. J Public Health 2004;26:268-74. http://dx.doi.org/10.1093/pubmed/fdh160.
- Smith SM, Fahey T, Smucny J, Becker LA. Antibiotics for acute bronchitis. Cochrane Database Syst Rev 2010;4. http://dx.doi.org/10.1002/14651858.CD000245.pub2.
- Graham VA, Milton AF, Knowles GK, Davies RJ. Routine antibiotics in hospital management of acute asthma. Lancet 1982;1:418-20. http://dx.doi.org/10.1016/S0140-6736(82)91619-1.
- National Asthma Education and Prevention Program Expert Panel Report 3: Guidelines for the Diagnosis and Management of Asthma. Bethesda, MD: National Heart, Lung, and Blood Institute; 2007.
- Vanderweil SG, Chu-Lin Tsai CL, Pelletier AJ, Espinola JA, Ashley F, Sullivan AF, et al. Inappropriate use of antibiotics for acute asthma in United States emergency departments. Acad Emerg Med 2008;15:736-43. http://dx.doi.org/10.1111/j.1553-2712.2008.00167.x.
- Puhan MA, Vollenweider D, Latshang T, Steurer J, Steurer-Stey C. Exacerbations of chronic obstructive pulmonary disease: when are antibiotics indicated? A systematic review. Respir Res 2007;4. http://dx.doi.org/10.1186/1465-9921-8-30.
- Ram FS, Rodriguez-Roisin R, Granados-Navarrete A, Garcia-Aymerich J, Barnes NC. Antibiotics for exacerbations of chronic obstructive pulmonary disease. Cochrane Database Syst Rev 2006;19. http://dx.doi.org/10.1002/14651858.CD004403.
- Chronic Obstructive Pulmonary Disease: Management of Chronic Obstructive Pulmonary Disease in Adults in Primary and Secondary Care. London: National Clinical Guideline Centre; 2010.
- Hawkey PM, Jones AM. The changing epidemiology of resistance. J Antimicrob Chemother 2009;64:3-10. http://dx.doi.org/10.1093/jac/dkp256.
- Steinman MA, Gonzales R, Linder JA, Landefeld CS. Changing use of antibiotics in community-based outpatient practice, 1991–1999. Ann Intern Med 2003;138:525-33. http://dx.doi.org/10.7326/0003-4819-138-7-200304010-00008.
- Bronzwaer SL, Cars O, Buchholz U, . A European study on the relationship between antimicrobial use and antimicrobial resistance. Emerg Infect Dis 2002;8:278-82. http://dx.doi.org/10.3201/eid0803.010192.
- Paterson DL. Collateral damage from cephalosporin or quinolone antibiotic therapy. Clin Infect Dis 2004;38:341-5. http://dx.doi.org/10.1086/382690.
- Bartlett JG. Diagnostic tests for agents of community-acquired pneumonia. Clin Infect Dis 2011;52:S296-304. http://dx.doi.org/10.1093/cid/cir045.
- Strålin K. Usefulness of aetiological tests for guiding antibiotic therapy in community-acquired pneumonia. Int J Antimicrob Agents 2008;31:3-11. http://dx.doi.org/10.1016/j.ijantimicag.2007.06.037.
- Abe T, Tokuda Y, Ishimatsu S, Birrer RB. Usefulness of initial blood cultures in patients admitted with pneumonia from an emergency department in Japan. J Infect Chemother 2009;15:180-6. http://dx.doi.org/10.1007/s10156-009-0682-z.
- Scott JA, Hall AJ. The value and complications of percutaneous transthoracic lung aspiration for the etiologic diagnosis of community-acquired pneumonia. Chest 1999;116:1716-32. http://dx.doi.org/10.1378/chest.116.6.1716.
- Corbo J, Friedman B, Bijur P, Gallagher EJ. Limited usefulness of initial blood cultures in community acquired pneumonia. Emerg Med J 2004;21:446-8.
- Kennedy M, Bates DW, Wright SB, Ruiz R, Wolfe RE, Shapiro NI. Do emergency department blood cultures change practice in patients with pneumonia?. Ann Emerg Med 2005;46:393-400. http://dx.doi.org/10.1016/j.annemergmed.2005.05.025.
- Mountain D, Bailey PM, O’Brien D, Jelinek GA. Blood cultures ordered in the adult emergency department are rarely useful. Eur J Emerg Med 2006;13:76-9. http://dx.doi.org/10.1097/01.mej.0000188231.45109.ec.
- Ramanujam P, Rathlev NK. Blood cultures do not change management in hospitalized patients with community-acquired pneumonia. Acad Emerg Med 2006;13:740-5. http://dx.doi.org/10.1197/j.aem.2006.03.554.
- Moran GJ, Abrahamian FM. Blood cultures for community acquired pneumonia: can we hit the target without a shotgun?. Ann Emerg Med 2005;46:407-8. http://dx.doi.org/10.1016/j.annemergmed.2005.07.004.
- Davidson M, Tempest B, Palmer DL. Bacteriologic diagnosis of acute pneumonia: comparison of sputum, transtracheal aspirates, and lung aspirates. JAMA 1976;235:158-63. http://dx.doi.org/10.1001/jama.1976.03260280016018.
- Musher DM, Montoya R, Wanahita A. Diagnostic value of microscopic examination of gram-stained sputum and sputum cultures in patients with bacteremic pneumococcal pneumonia. Clin Infect Dis 2004;39:165-9. http://dx.doi.org/10.1086/421497.
- Selickman J, Paxos M, File TM, Seltzer R, Bonilla H. Performance measure of urinary antigen in patients with Streptococcus pneumoniae pneumonia. Diagn Microbiol Infect Dis 2010;67:129-33. http://dx.doi.org/10.1016/j.diagmicrobio.2010.01.005.
- Strålin K, Kaltoft MS, Konradsen HB, Olcén P, Holmberg H. Comparison of two urinary antigen tests for establishment of pneumococcal etiology of adult community-acquired pneumonia. J Clin Microbiol 2004;42:3620-5. http://dx.doi.org/10.1128/JCM.42.8.3620-3625.2004.
- Hayden FG, Fritz RS, Lobo MC, Alvord WG, Strober W, Straus SE. Local and systemic cytokine responses during experimental human influenza A virus infection. J Clin Invest 1998;101:643-9. http://dx.doi.org/10.1172/JCI1355.
- Carrat F, Vergu E, Ferguson NM, Lemaitre M, Cauchemez S, Leach S. Time lines of infection and disease in human influenza: a review of volunteer challenge studies. Am J Epidemiol 2008;167:775-85. http://dx.doi.org/10.1093/aje/kwm375.
- Lee N, Chan PK, Hui DS, Rainer TH, Wong E, Choi K-W, et al. Viral loads and duration of viral shedding in adult patients hospitalized with influenza. J Infect Dis 2009;200:492-500. http://dx.doi.org/10.1086/600383.
- Siegel JD, Rhinehart E, Jackson M, Chiarello L. and the Healthcare Infection Control Practices Advisory Committee . Guideline for Isolation Precautions: Preventing Transmission of Infectious Agents in Healthcare Settings 2007. www.cdc.gov/ncidod/dhqp/pdf/isolation2007.pdf (accessed 15 August 2013).
- Dulek DE, Peebles RS. Viruses and asthma. Biochim Biophys Acta 2011;1810:1080-90. http://dx.doi.org/10.1016/j.bbagen.2011.01.012.
- Wat D, Gelder C, Hibbitts S, Cafferty F, Bowler I, Pierrepoint M, et al. The role of respiratory viruses in cystic fibrosis. J Cyst Fibros 2008;7:320-8. http://dx.doi.org/10.1016/j.jcf.2007.12.002.
- Mohan A, Chandra S, Agarwal D, . Prevalence of viral infection detected by PCR and RT-PCR in patients with acute exacerbation of COPD: a systematic review. Respirology 2010;15:536-42. http://dx.doi.org/10.1111/j.1440-1843.2010.01722.x.
- Falsey AR, Hennessey PA, Formica MA, Cox C, Walsh EE. Respiratory syncytial virus infection in elderly and high-risk adults. N Engl J Med 2005;352:1749-59. http://dx.doi.org/10.1056/NEJMoa043951.
- Louie JK, Hacker JK, Gonzales R, . Characterization of viral agents causing acute respiratory infection in a San Francisco University Medical Center Clinic during the influenza season. Clin Infect Dis 2005;41:822-8. http://dx.doi.org/10.1086/432800.
- Salgado CD, Farr BM, Hall KK, Hayden FG. Influenza in the acute hospital setting. Lancet Infect Dis 2002;2:145-55. http://dx.doi.org/10.1016/S1473-3099(02)00221-9.
- Maltezou HC, Drancourt M. Nosocomial influenza in children. J Hosp Infect 2003;55:83-91. http://dx.doi.org/10.1016/S0195-6701(03)00262-7.
- Kapila R, Lintz DI, Tescon FT, Ziskin L, Louria DB. A nosocomial outbreak of influenza A. Chest 1977;71:576-9. http://dx.doi.org/10.1378/chest.71.5.576.
- Sartor C, Zandotti C, Romain F, Jacomo V, Simon S, Atlan-Gepner C, et al. Nosocomial influenza outbreak: disruption of services in an internal medicine unit. Infect Control Hosp Epidemiol 2002;23:615-19. http://dx.doi.org/10.1086/501981.
- Elder AG, O’Donnell B, McCruden EAB, Symington IS, Carman WF. Incidence and recall of influenza in a cohort of healthcare workers during the 1993–4 epidemic: results of serum testing and questionnaire. BMJ 1996;313:1241-2. http://dx.doi.org/10.1136/bmj.313.7067.1241.
- Enstone JE, Myles PR, Openshaw PJ, Gadd EM, Lim WS, Semple MG, et al. Nosocomial pandemic (H1N1) 2009, United Kingdom, 2009–2010. Emerg Infect Dis 2011;17:592-8. http://dx.doi.org/10.3201/eid1704.101679.
- Estenssoro E, Ríos FG, Apezteguía C, Reina R, Neira J, Ceraso DH, et al. Pandemic 2009 influenza A in Argentina: a study of 337 patients on mechanical ventilation. Am J Respir Crit Care Med 2010;182:41-8. http://dx.doi.org/10.1164/201001-0037OC.
- Martín-Loeches I, Sanchez-Corral A, Diaz E, Granada RM, Zaragoza R, Villavicencio C, et al. Community-acquired respiratory coinfection in critically ill patients with pandemic 2009 influenza A(H1N1) virus. Chest 2011;139:555-62. http://dx.doi.org/10.1378/chest.10-1396.
- Gill JR, Sheng ZM, Ely SF, Guinee DG, Beasley MB, Suh J, et al. Pulmonary pathologic findings of fatal 2009 pandemic influenza A/H1N1 viral infections. Arch Pathol Lab Med 2010;134:235-43.
- Kim HS, Kim JH, Shin SY, Kang YA, Lee HG, Kim JS, et al. Fatal cases of 2009 pandemic influenza A (H1N1) in Korea. J Korean Med Sci 2011;26:22-7. http://dx.doi.org/10.3346/jkms.2011.26.1.22.
- Lucas S. Predictive clinicopathological features derived from systematic autopsy examination of patients who died with A/H1N1 influenza infection in the UK 2009–10 pandemic. Health Technol Assess 2010;14.
- Mauad T, Hajjar LA, Callegari GD, da Silva LF, Schout D, Galas FR, et al. Lung pathology in fatal novel human influenza A (H1N1) infection. Am J Respir Crit Care Med 2010;181:72-9. http://dx.doi.org/10.1164/rccm.200909-1420OC.
- Shieh WJ, Blau DM, Denison AM, Deleon-Carnes M, Adem P, Bhatnagar J, et al. 2009 pandemic influenza A (H1N1): pathology and pathogenesis of 100 fatal cases in the United States. Am J Pathol 2010;177:166-75.
- Centers for Disease Control and Prevention (CDC) . Bacterial coinfections in lung tissue specimens from fatal cases of 2009 pandemic influenza A (H1N1), United States, May–August 2009. MMWR 2009;58:1071-4.
- Rebelo-de-Andrade H, Zambon MC. Different diagnostic methods for detection of influenza epidemics. Epidemiol Infect 2000;124:515-22. http://dx.doi.org/10.1017/S0950268899003751.
- Chan KH, Maldeis N, Pope W, Yup A, Ozinskas A, Gill J, et al. Evaluation of the Directigen Flu A+B test for rapid diagnosis of influenza virus type A and B infections. J Clin Microbiol 2002;40:1675-80. http://dx.doi.org/10.1128/JCM.40.5.1675-1680.2002.
- Cheng PK, Wong KKY, Mak GC, Wong AH, Ng AYY, Chow SYK, et al. Performance of laboratory diagnostics for the detection of influenza A(H1N1)v virus as correlated with the time after symptom onset and viral load. J Clin Virol 2010;47:182-5. http://dx.doi.org/10.1016/j.jcv.2009.11.022.
- Mehlmann M, Bonner AB, Williams JV, Dankbar DM, Moore CL, Kuchta RD, et al. Comparison of the MChip to viral culture, reverse transcription-PCR, and the QuickVue Influenza A+B test for rapid diagnosis of influenza. J Clin Microbiol 2007;45:1234-7. http://dx.doi.org/10.1128/JCM.02202-06.
- Poehling KA, Griffin MR, Dittus RS, Tang YW, Holland K, Li H, et al. Bedside diagnosis of influenza virus infections in hospitalized children. Pediatrics 2002;110:83-8.
- Rahman M, Vandermause MF, Kieke BA, Belongia EA. Performance of BinaxNOW Flu A and B and direct fluorescent assay in comparison with a composite of viral culture or reverse transcription polymerase chain reaction for detection of influenza infection during the 2006 to 2007 season. Diagn Microbiol Infect Dis 2008;62:162-6. http://dx.doi.org/10.1016/j.diagmicrobio.2007.10.012.
- Ruest A, Michaud S, Deslandes S, Frost EH. Comparison of the Directigen flu A+B test, the QuickVue influenza test, and clinical case definition to viral culture and reverse transcription-PCR for rapid diagnosis of influenza virus infection. J Clin Microbiol 2003;41:3487-93. http://dx.doi.org/10.1128/JCM.41.8.3487-3493.2003.
- Simmerman JM, Chittaganpitch M, Erdman D, Sawatwong P, Uyeki TM, Dowell SF. Field performance and new uses of rapid influenza testing in Thailand. Int J Infect Dis 2007;11:166-71. http://dx.doi.org/10.1016/j.ijid.2006.01.005.
- Reina J, Fernandez-Baca V, Blanco I, Munar M. Comparison of Madin-Darby Canine Kidney Cells (MDCK) with a green monkey continuous cell line (Vero) and human lung embryonated cells (MRC-5) in the isolation of influenza virus from nasopharyngeal aspirates by shell vial culture. J Clin Microbiol 1997;35:1900-1.
- Leekha S, Zitterkopf NL, Espy MJ, Smith TF, Thompson RL. Duration of influenza A virus shedding in hospitalized patients and implications for infection control. Infect Control Hosp Epidemiol 2007;28:1071-6. http://dx.doi.org/10.1086/520101.
- Segonds C, Le Goff A, Chabanon G. Assessment of the contribution of the immunochromatographic pneumococcal urinary antigen test to the etiological diagnosis of pneumonia in hospitalized adults. Pathol Biol 2010;58:117-22.
- Ishida T, Hashimoto T, Arita M, Tojo Y, Tachibana H, Jinnai M. A 3-year prospective study of a urinary antigen-detection test for Streptococcus pneumoniae in community-acquired pneumonia: utility and clinical impact on the reported etiology. J Infect Chemother 2004;10:359-63. http://dx.doi.org/10.1007/s10156-004-0351-1.
- Marcos MA, de Anta J, de la Bellacasa JP, Gonzalez J, Martinez E, Garcia E, et al. Rapid urinary antigen test for diagnosis of pneumococcal community-acquired pneumonia in adults. Eur Respir J 2003;21:209-14. http://dx.doi.org/10.1183/09031936.03.00058802.
- Porcel JM, Ruiz-González A, Falguera M, Nogués A, Galindo C, Carratalá J, et al. Contribution of a pleural antigen assay (BinaxNOW) to the diagnosis of pneumococcal pneumonia. Chest 2007;131:1442-7. http://dx.doi.org/10.1378/chest.06-1884.
- Dominguez J, Gali N, Blanco S, Pedroso P, Prat C, Matas L, et al. Detection of Streptococcus pneumoniae antigen by a rapid immunochromatographic assay in urine samples. Chest 2001;119:243-9. http://dx.doi.org/10.1378/chest.119.1.243.
- Sordé R, Falcó V, Lowak M, Domingo E, Ferrer A, Burgos J, et al. Current and potential usefulness of pneumococcal urinary antigen detection in hospitalized patients with community-acquired pneumonia to guide antimicrobial therapy. Arch Intern Med 2011;171:166-72. http://dx.doi.org/10.1001/archinternmed.2010.347.
- Kalin M. Bacteremic pneumococcal pneumonia: value of culture of nasopharyngeal specimens and examination of washed sputum specimens. Eur J Clin Microbiol 1982;1:394-6. http://dx.doi.org/10.1007/BF02019941.
- Torres JM, Cardenas O, Vasquez A, Schlossberg D. Streptococcus pneumoniae bacteremia in a community hospital. Chest 1998;113:387-90. http://dx.doi.org/10.1378/chest.113.2.387.
- Watanakunakorn C, Bailey TA. Adult bacteremic pneumococcal pneumonia in a community teaching hospital, 1992–1996: a detailed analysis of 108 cases. Arch Intern Med 1997;157:1965-71. http://dx.doi.org/10.1001/archinte.157.17.1965.
- Drew WL. Value of sputum culture in diagnosis of pneumococcal pneumonia. J Clin Microbiol 1977;6:62-5.
- Fiala M. A study of the combined role of viruses, mycoplasmas and bacteria in adult pneumonia. Am J Med Sci 1969;257:44-51. http://dx.doi.org/10.1097/00000441-196901000-00005.
- García-Vázquez E, Marcos MA, Mensa J, de Roux A, Puig J, Font C, et al. Assessment of the usefulness of sputum culture for diagnosis of community-acquired pneumonia using the PORT predictive scoring system. Arch Intern Med 2004;164:1807-11. http://dx.doi.org/10.1001/archinte.164.16.1807.
- Andresen DN, Kesson AM. High sensitivity of a rapid immunochromatographic test for detection of influenza A virus 2009 H1N1 in nasopharyngeal aspirates from young children. J Clin Microbiol 2010;48:2658-9. http://dx.doi.org/10.1128/JCM.00229-10.
- Drexler JF, Helmer A, Kirberg H, Reber U, Panning M, Müller M, et al. Poor clinical sensitivity of rapid antigen test for influenza A pandemic (H1N1) 2009 virus. Emerg Infect Dis 2009;15:1662-4. http://dx.doi.org/10.3201/eid1510.091186.
- Choi YJ, Kim HJ, Park JS, Oh MH, Nam HS, Kim YB, et al. Evaluation of new rapid antigen test for detection of pandemic influenza A/H1N1 2009 virus. J Clin Microbiol 2010;48:2260-2. http://dx.doi.org/10.1128/JCM.02392-09.
- Angoulvant F, Bellettre X, Houhou N, Dexpert JB, Morin L, Siriez JY, et al. Sensitivity and specificity of a rapid influenza diagnostic test in children and clinical utility during influenza A (H1N1) 2009 outbreak. Emerg Med J 2011;28:924-6. http://dx.doi.org/10.1136/emj.2010.098533.
- Choi YJ, Nam HS, Park JS, Kim HJ, Park KB, Jeon MH, et al. Comparative analysis of the multiple test methods for the detection of pandemic influenza A/H1N1 2009 virus. J Microbiol Biotechnol 2010;20:1450-6. http://dx.doi.org/10.4014/jmb.1004.04039.
- Whiting P, Rutjes AW, Reitsma JB, Bossuyt PN, Kleijnen J. The development of QUADAS: a tool for the quality assessment of studies of diagnostic accuracy included in systematic reviews. BMC Med Res Methodol 2003;3.
- Whiting PF, Weswood ME, Rutjes AW, Reitsma JB, Bossuyt PN, Kleijnen J. Evaluation of QUADAS, a tool for the quality assessment of diagnostic accuracy studies. BMC Med Res Methodol 2006;6.
- Westwood ME, Whiting PF, Kleijnen J. How does study quality affect the results of a diagnostic meta-analysis?. BMC Med Res Methodol 2005;5.
- Bossuyt PM, Reitsma JB, Bruns DE, Gatsonis CA, Glasziou PP, Irwig LM, et al. Towards complete and accurate reporting of studies of diagnostic accuracy: the STARD initiative. BMJ 2003;326:41-4. http://dx.doi.org/10.1136/bmj.326.7379.41.
- Moher D, Liberati A, Tetzlaff J, Altman DG. The PRISMA Group (2009) . Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement n.d. www.prisma-statement.org/statement.htm (accessed 15 August 2013).
- Reitsma JB, Glas AS, Rutjes AWS, Scholten RJPM, Bossuyt PM, Zwinderman AH. Bivariate analysis of sensitivity and specificity produces informative summary measures in diagnostic reviews. J Clin Epidemiol 2005;58:982-90. http://dx.doi.org/10.1016/j.jclinepi.2005.02.022.
- Rutter CM, Gatsonis CA. A hierarchical regression approach to meta-analysis of diagnostic test accuracy evaluations. Stat Med 2001;20:2865-84. http://dx.doi.org/10.1002/sim.942.
- Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ 2003;327:557-60. http://dx.doi.org/10.1136/bmj.327.7414.557.
- Chomel JJ, Remilleux MF, Marchand P, Aymard M. Rapid diagnosis of influenza A. Comparison with ELISA immunocapture and culture. J Virol Meth 1992;37:337-43. http://dx.doi.org/10.1016/0166-0934(92)90033-A.
- Rashid H, Shafi S, Haworth E, El Bashir H, Ali KA, Memish ZA, et al. Value of rapid testing for influenza among Hajj pilgrims. Travel Med Infect Dis 2007;5:310-13. http://dx.doi.org/10.1016/j.tmaid.2007.07.006.
- Nougairede A, Ninove L, Zandotti C, de Lamballerie X, Gazin C, Drancourt M, et al. Point of care strategy for rapid diagnosis of novel A/H1N1 influenza virus. PLOS ONE 2010;5. http://dx.doi.org/10.1371/journal.pone.0009215.
- Hawkes M, Richardson SE, Ipp M, Schuh S, Adachi D, Tran D. Sensitivity of rapid influenza diagnostic testing for swine-origin 2009 a (H1N1) influenza virus in children. Pediatrics 2010;125:e639-44. http://dx.doi.org/10.1542/peds.2009-2669.
- Rodriguez WJ, Schwartz RH, Thorne MM. Evaluation of diagnostic tests for influenza in a pediatric practice. Pediatr Infect Dis J 2002;21:193-6. http://dx.doi.org/10.1097/00006454-200203000-00006.
- Sabetta JR, Mardin JS, Burns L, Barry K, Baisley C, Mahoney T, et al. Performance of rapid influenza diagnostic tests during two school outbreaks of 2009 pandemic influenza A (H1N1) virus infection – Connecticut, 2009. MMWR 2009;58:1029-32.
- Hara M, Takao M, Fukuda S, Shimazu Y, Miyazaki K. Evaluation of three immunochromatographic kits for rapid detection of influenza virus A and B. LabMedicine 2008;39:603-6. http://dx.doi.org/10.1309/LM6ONMGY7K9ETMGF.
- Shoji M, Shoji S, Okamoto M, Ohmi A, Nishimura H. Re-evaluation of clinical utility of throat swab specimens for rapid influenza diagnostic test. Kansenshogaku Zasshi 2009;83:19-25.
- Bellei N, Benfica D, Perosa AH, Carlucci R, Barros M, Granato C. Evaluation of a rapid test (QuickVue) compared with the shell vial assay for detection of influenza virus clearance after antiviral treatment. J Virol Methods 2003;109:85-8. http://dx.doi.org/10.1016/S0166-0934(03)00050-8.
- Booth S, Baleriola C, Rawlinson WD. Comparison of two rapid influenza A/B test kits with reference methods showing high specificity and sensitivity for influenza A infection. J Med Virol 2006;78:619-22. http://dx.doi.org/10.1002/jmv.20584.
- Chen Y, Xu F, Gui X, Yang K, Wu X, Zheng Q, et al. A rapid test for the detection of influenza A virus including pandemic influenza A/H1N1 2009. J Virol Methods 2010;167:100-2. http://dx.doi.org/10.1016/j.jviromet.2010.02.001.
- Hamilton MS, Abel DM, Ballam YJ, Otto MK, Nickell AF, Pence LM, et al. Clinical evaluation of the ZstatFlu-II test: a chemiluminescent rapid diagnostic test for influenza virus. J Clin Microbiol 2002;40:2331-4. http://dx.doi.org/10.1128/JCM.40.7.2331-2334.2002.
- Covalciuc KA, Webb KH, Carlson CA. Comparison of four clinical specimen types for detection of influenza A and B viruses by optical immunoassay (FLU OIA test) and cell culture methods. J Clin Microbiol 1999;37:3971-4.
- Waner JL, Todd SJ, Shalaby H, Murphy P, Wall LV. Comparison of Directigen FLU-A with viral isolation and direct immunofluorescence for the rapid detection and identification of influenza A virus. J Clin Microbiol 1991;29:479-82.
- Diederen BM, Veenendaal D, Jansen R, Herpers BL, Ligtvoet EE, Ijzerman EP. Rapid antigen test for pandemic (H1N1) 2009 virus. Emerg Infect Dis 2010;16:897-8. http://dx.doi.org/10.3201/eid1605.091574.
- Rahman M, Kieke BA, Vandermause MF, Mitchell PD, Greenlee RT, Belongia EA. Performance of Directigen flu A+B enzyme immunoassay and direct fluorescent assay for detection of influenza infection during the 2004–2005 season. Diagn Microbiol Infect Dis 2007;58:413-18. http://dx.doi.org/10.1016/j.diagmicrobio.2007.03.011.
- Yoo Y, Sohn JW, Park DW, Kim JY, Shin HK, Lee Y, et al. Clinical evaluation of the SD Bioline influenza virus antigen test for rapid detection of influenza viruses A and B in children and adults during the influenza season. Clin Vaccine Immunol 2007;14:1050-2. http://dx.doi.org/10.1128/CVI.00465-06.
- Lucas PM, Morgan OW, Gibbons TF, Guerrero AC, Maupin GM, Butler JL. Diagnosis of 2009 pandemic influenza A (pH1N1) and seasonal influenza using rapid influenza antigen tests, San Antonio, Texas, April–June 2009. Clin Infect Dis 2011;52:116-22. http://dx.doi.org/10.1093/cid/ciq027.
- Quach C, Newby D, Daoust G, Rubin E, McDonald J. QuickVue influenza test for rapid detection of influenza A and B viruses in a pediatric population. Clin Diagn Lab Immunol 2002;9:925-6. http://dx.doi.org/10.1128/CDLI.9.4.925-926.2002.
- Cazacu AC, Chung SE, Greer J, Demmler GJ. Comparison of the directigen flu A+B membrane enzyme immunoassay with viral culture for rapid detection of influenza A and B viruses in respiratory specimens. J Clin Microbiol 2004;42:3707-10. http://dx.doi.org/10.1128/JCM.42.8.3707-3710.2004.
- Cazacu AC, Demmler GJ, Neuman MA, Forbes BA, Chung S, Greer J, et al. Comparison of a new lateral-flow chromatographic membrane immunoassay to viral culture for rapid detection and differentiation of influenza A and B viruses in respiratory specimens. J Clin Microbiol 2004;42:3661-4. http://dx.doi.org/10.1128/JCM.42.8.3661-3664.2004.
- Kim YK, Uh Y, Chun J, Kim C, Kim HY. Evaluation of new hemagglutinin-based rapid antigen test for influenza A pandemic (H1N1) 2009. J Clin Virol 2010;49:69-72. http://dx.doi.org/10.1016/j.jcv.2010.06.012.
- Smidt N, Rutjes AWS, Van der Windt DAWM, Ostelo RWJG, Reitsma JB, Bossuyt PM, et al. Quality of reporting of diagnostic accuracy studies. Radiology 2005;235:347-53. http://dx.doi.org/10.1148/radiol.2352040507.
- Uyeki TM. Influenza diagnosis and treatment in children: a review of studies on clinically useful tests and antiviral treatment for influenza. Pediatr Infect Dis J 2003;22:164-77. http://dx.doi.org/10.1097/01.inf.0000050458.35010.b6.
- Petrozzino JJ, Smith C, Atkinson MJ. Rapid diagnostic testing for seasonal influenza: an evidence based review and comparison with unaided clinical diagnosis. J Emerg Med 2010;39:476-90. http://dx.doi.org/10.1016/j.jemermed.2009.11.031.
- Babin SM, Hsieh Y-H, Rothman RE, Gaydos CE. A meta-analysis of point-of-care laboratory tests in the diagnosis of novel 2009 swine-lineage pandemic influenza A (H1N1). Diagn Microbiol Infect Dis 2011;69:410-18. http://dx.doi.org/10.1016/j.diagmicrobio.2010.10.009.
- Cals JW, Hopstaken RM, Butler CC, Hood K, Severens JL, Dinant G. Improving management of patients with acute cough by C-reactive protein point of care testing and communication training (IMPAC3T): study protocol of a cluster randomised controlled trial. BMC Fam Pract 2007;8. http://dx.doi.org/10.1186/1471-2296-8-15.
- Holland CA, Kiechle FL. Point-of-care molecular diagnostic systems: past, present and future. Curr Opin Microbiol 2005;8:504-9. http://dx.doi.org/10.1016/j.mib.2005.08.001.
- McCord J, Nowak, RM, McCullough PA, Foreback C, Borzak S, Tokarski G, et al. Ninety-minute exclusion of acute myocardial infarction by use of quantitative point-of-care testing of myoglobin and troponin I. Circulation 2001;104:1483-8. http://dx.doi.org/10.1161/hc3801.096336.
- Karsh BT. Beyond usability: designing effective technology implementation systems to promote patient safety. Qual Saf Health Care 2004;13:388-94. http://dx.doi.org/10.1136/qhc.13.5.388.
- Aggelidis VP, Chatzoglou PD. Using a modified technology acceptance model in hospitals. Int J Med Inform 2009;78:115-26. http://dx.doi.org/10.1016/j.ijmedinf.2008.06.006.
- Gharabaghi F, Tellier R, Cheung R, Collins C, Broukhanski G, Drews SJ, et al. Comparison of a commercial qualitative real-time RT-PCR kit with direct immunofluorescence assay (DFA) and cell culture for detection of influenza A and B in children. J Clin Virol 2008;42:190-3. http://dx.doi.org/10.1016/j.jcv.2008.01.013.
- Jannes G, De Vos D. A review of current and future molecular diagnostic tests for use in the microbiology laboratory. Methods Mol Biol 2006;345:1-21.
- Leonardi GP, Leib H, Birkhead GS, Smith C, Costello P, Conron W. Comparison of rapid detection methods for influenza A virus and their value in health-care management of institutionalized geriatric patients. J Clin Microbiol 1994;32:70-4.
- US Food and Drug Administration (FDA) . CLIA Categorization Criteria n.d. www.fda.gov/MedicalDevices/DeviceRegulationandGuidance/IVDRegulatoryAssistance/ucm124208.htm (accessed 16 August 2013).
- Turner D, Wailoo A, Nicholson K, Cooper N, Sutton A, Abrams K. Systematic review and economic decision modelling for the prevention and treatment of influenza A and B. Health Technol Assess 2003;7.
- Burch J, Paulden M, Conti S, Stock C, Corbett M, Welton NJ, et al. Antiviral drugs for the treatment of influenza: a systematic review and economic evaluation. Health Technol Assess 2009;13.
- Morris S, Devlin N, Parkin D. Economic Analysis in Health Care. Chichester: John Wiley & Sons; 2007.
- O’Hagan A, Luce BR. A Primer on Bayesian Statistics in Health Economics and Outcomes Research. MEDTAP International; 2003.
- Spiegelhalter DJ, Abrams KR, Myles JP. Bayesian Approaches to Clinical Trials and Health-Care Evaluation. Chichester: Wiley and Sons; 2003.
- Baio G. Bayesian Methods in Health Economics. Boca Raton, FL: Chapman Hall, CRC; 2012.
- Spiegelhalter D, Thomas A, Best N, Lunn D. WinBUGS User Manual. Cambridge: MRC Biostatistics Unit; 2003.
- NHS Reference Costs 2008. London: DoH; 2008.
- Bravo-Vergel Y, Palmer S, Asseburg C, Fenwick E, de Belder M, Abrams K, et al. Is primary angioplasty cost effective in the UK? Results of a comprehensive decision analysis. Heart 2007;93:1238-43. http://dx.doi.org/10.1136/hrt.2006.111401.
- Turner D, Little P, Raftery J, Turner S, Smith H, Rumsby K, et al. Cost effectiveness of management strategies for urinary tract infections: results from randomised controlled trial. BMJ 2010;340. http://dx.doi.org/10.1136/bmj.c346.
- Curtis L. Unit Costs of Health and Social Care 2008. Canterbury: PSSRU; 2008.
- British National Formulary. No. 58, September 2009. London: BMA and RPS; 2009.
- Gray AM, Clarke PM, Wolstenholme JL, Wordsworth S. Applied Methods of Cost-Effectiveness Analysis in Healthcare. Oxford: Oxford University Press; 2011.
- Hospital and Community Health Services (HCHS) . Pay and Price Index n.d. www.doh.gov.uk/ (accessed 17 April 2013).
- Mihaylova B, Briggs A, O’Hagan A, Thompson SG. Review of statistical methods for analysing healthcare resources and costs. Health Econ 2011;20:897-916. http://dx.doi.org/10.1002/hec.1653.
- O’Hagan A, Stevens JW. A framework for cost-effectiveness analysis from clinical trial data. Health Econ 2001;10:303-15. http://dx.doi.org/10.1002/hec.617.
- Gilks WR, Richardson S, Spiegelhalter DJ. Markov Chain Monte Carlo in Practice. London: Chapman and Hall, UK; 1996.
- R: a Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2012.
- Burton A, Billingham LJ, Bryan S. Cost-effectiveness in clinical trials: using multiple imputation to deal with incomplete cost data. Clin Trials 2007;4:154-61. http://dx.doi.org/10.1177/1740774507076914.
- Lambert PC, Billingham LJ, Cooper NJ, Sutton AJ, Abrams KR. Estimating the cost-effectiveness of an intervention in a clinical trial when partial cost information is available: a Bayesian approach. Health Econ 2008;17:67-81. http://dx.doi.org/10.1002/hec.1243.
- Guide to the Methods of Technology Appraisal. London: NICE; 2008.
- Drummond MF, Sculpher MJ, O’Brien BJ, Torrance GW. Methods for the Economic Evaluation of Health Care Programs. Oxford: Oxford University Press; 2005.
- Brooks SP, Gelman A. Alternative methods for monitoring convergence of iterative simulations. J Comp Graph Stat 1998;7:434-55.
- Guchev IA, Yu VL, Sinopalnikov A, Klochkov OI, Kozlov RS, Stratchounski LS. Management of non-severe pneumonia in military trainees with the urinary antigen test for Streptococcus pneumoniae: an innovative approach to targeted therapy. Clin Infect Dis 2005;40:1608-16.
- Falguera M, Ruiz-Gonzalez A, Schoenenberger JA, Touzon C, Gazque I, Galindo C, et al. Prospective, randomised study to compare empirical treatment versus targeted treatment on the basis of the urine antigen results in hospitalised patients with community-acquired pneumonia. Thorax 2010;65:101-6. http://dx.doi.org/10.1136/thx.2009.118588.
- Balish A, Warnes CM, Wu K, Barnes N, Emery S, Berman L, et al. Evaluation of rapid influenza diagnostic tests for detection of novel influenza A (H1N1) Virus – United States, 2009. MMWR Morb Mortal Wkly Rep 2009;58:826-9.
- Bellman-Weiler R, Beilkircher B, Kurz K, Theurl I, Weiss G. Accuracy of bedside antigen tests in the diagnosis of new influenza A/H1N1v infection. Clin Microbiol Infect 2011;17:235-7. http://dx.doi.org/10.1111/j.1469-0691.2010.03235.x.
- Biggs C, Walsh P, Overmyer CL, Gonzalez D, Feola M, Mordechai E, et al. Performance of influenza rapid antigen testing in influenza in emergency department patients. Emerg Med J 2010;27:5-7. http://dx.doi.org/10.1136/emj.2009.078683.
- Blyth CC, Iredell JR, Dwyer DE. Rapid-test sensitivity for novel swine-origin influenza A (H1N1) virus in humans. N Engl J Med 2009;361. http://dx.doi.org/10.1056/NEJMc0909049.
- Cheng X, Yuan Q, Yue Q, Zheng Q, Ma Y, Yang B, et al. Evaluation of a new rapid influenza A diagnostic test for detection of pandemic (H1N1) 2009 and seasonal influenza A virus. J Clin Virol 2011;50:153-5. http://dx.doi.org/10.1016/j.jcv.2010.10.002.
- de la Tabla VO, Antequera P, Masia M, Ros P, Martin C, Gazquez G, et al. Clinical evaluation of rapid point-of-care testing for detection of novel influenza A (H1N1) virus in a population-based study in Spain. Clin Microbiol Infect 2010;16:1358-61.
- Faix DJ, Sherman SS, Waterman SH. Rapid-test sensitivity for novel swine-origin influenza A (H1N1) virus in humans. N Engl J Med 2009;361:728-9. http://dx.doi.org/10.1056/NEJMc0904264.
- Fernandez C, Cataleto M, Lee P, Feuerman M, Krilov L. Rapid influenza A testing for novel H1N1: point-of-care performance. Postgrad Med 2010;122:28-33. http://dx.doi.org/10.3810/pgm.2010.01.2096.
- Fuenzalida L, Blanco S, Prat C, Vivancos M, Dominguez MJ, Modol JM, et al. Utility of the rapid antigen detection BinaxNOW Influenza A&B test for detection of novel influenza A (H1N1) virus. Clin Microbiol Infect 2010;16:1574-6. http://dx.doi.org/10.1111/j.1469-0691.2010.03160.x.
- Ghebremedhin B, Engelmann I, Konig W, Konig B. Comparison of the performance of the rapid antigen detection actim Influenza A&B test and RT-PCR in different respiratory specimens. J Med Microbiol 2009;58:365-70. http://dx.doi.org/10.1099/jmm.0.004358-0.
- Gimeno C, Costa E, Navalpotro D, Bravo D, Ocete D, Clari MA, et al. Sensitivity of a marketed immunochromatographic assay specifically targeting the pandemic influenza A/H1N1 2009 virus. Diagn Microbiol Infect Dis 2010;68:80-2. http://dx.doi.org/10.1016/j.diagmicrobio.2010.05.005.
- Gordon A, Videa E, Saborio S, Lopez R, Kuan G, Reingold A, et al. Performance of an influenza rapid test in children in a primary healthcare setting in Nicaragua. PLOS ONE 2009;4. http://dx.doi.org/10.1371/journal.pone.0007907.
- Gordon A, Videa E, Saborio S, Lopez R, Kuan G, Balmaseda A, et al. Diagnostic accuracy of a rapid influenza test for pandemic influenza A H1N1. PLOS ONE 2010;5. http://dx.doi.org/10.1371/journal.pone.0010364.
- Herrmann B, Larsson C, Zweygberg BW. Simultaneous detection and typing of influenza viruses A and B by a nested reverse transcription-PCR: comparison to virus isolation and antigen detection by immunofluorescence and optical immunoassay (FLU OIA). J Clin Microbiol 2001;39:134-8. http://dx.doi.org/10.1128/JCM.39.1.134-138.2001.
- Kok J, Blyth CC, Foo H, Patterson J, Taylor J, McPhie K, et al. Comparison of a rapid antigen test with nucleic acid testing during cocirculation of pandemic influenza A/H1N1 2009 and seasonal influenza A/H3N2. J Clin Microbiol 2010;48:290-1. http://dx.doi.org/10.1128/JCM.01465-09.
- Landry ML, Cohen S, Ferguson D. Real-time PCR compared to BinaxNOW and cytospin-immunofluorescence for detection of influenza in hospitalized patients. J Clin Virol 2008;43:148-51. http://dx.doi.org/10.1016/j.jcv.2008.06.006.
- Lee GC, Jeon ES, Kim WS, Le DT, Yoo JH, Chong CK. Evaluation of a rapid diagnostic test, NanoSign(R) Influenza A/B Antigen, for detection of the 2009 pandemic influenza A/H1N1 viruses. Virol J 2010;7.
- Leveque N, Talmud D, Renois F, Barbe C, Andreoletti L. Evaluation of a new rapid test for the detection of influenza A and B viruses and pandemic (H1N1) 2009 virus subtyping in respiratory samples. J Med Microbiol 2011;60:1403-4. http://dx.doi.org/10.1099/jmm.0.030585-0.
- Louie JK, Guevara H, Boston E, Dahlke M, Nevarez M, Kong T, et al. Rapid influenza antigen test for diagnosis of pandemic (H1N1) 2009. Emerg Infect Dis 2010;16:824-6. http://dx.doi.org/10.3201/eid1605.091794.
- Poehling KA, Zhu Y, Tang YW, Edwards K. Accuracy and impact of a point-of-care rapid influenza test in young children with respiratory illnesses. Arch Pediatr Adolesc Med 2006;160:713-18. http://dx.doi.org/10.1001/archpedi.160.7.713.
- Pongthanapisith V, Sukasem C, Premchaiporn K, Srichantaratsamee C, Chantratita W. Clinical performance of three rapid diagnostic tests for influenza virus in nasopharyngeal specimens to detect novel swine-origin influenza viruses. Infection 2011;39:105-11. http://dx.doi.org/10.1007/s15010-011-0092-x.
- Pregliasco F, Puzelli S, Mensi C, Anselmi G, Marinello R, Tanzi ML, et al. Influenza virological surveillance in children: the use of the QuickVue rapid diagnostic test. J Med Virol 2004;73:269-73. http://dx.doi.org/10.1002/jmv.20086.
- Rouleau I, Charest H, Douville-Fradet M, Skowronski DM, De Serres G. Field performance of a rapid diagnostic test for influenza in an ambulatory setting. J Clin Microbiol 2009;47:2699-703. http://dx.doi.org/10.1128/JCM.00762-09.
- Suntarattiwong P, Jarman RG, Levy J, Baggett HC, Gibbons RV, Chotpitayasunondh T, et al. Clinical performance of a rapid influenza test and comparison of nasal versus throat swabs to detect 2009 pandemic influenza A (H1N1) infection in Thai children. Pediatr Infect Dis J 2010;29:366-7.
- Tsao K, Kuo Y, Huang C, Chau S, Chan E. Performance of rapid-test kits for the detection of the pandemic influenza A/H1N1 virus. J Virol Methods 2011;173:387-9. http://dx.doi.org/10.1016/j.jviromet.2011.02.009.
- Velasco JM, Montesa-Develos ML, Jarman RG, Lopez MN, Gibbons RV, Valderama MT, et al. Evaluation of QuickVue influenza A+B rapid test for detection of pandemic influenza A/H1N1 2009. J Clin Virol 2010;48:120-2. http://dx.doi.org/10.1016/j.jcv.2010.03.010.
- Watanabe M, Nakagawa N, Ito M, Ihara T. Sensitivity of rapid immunoassay for influenza A and B in the early phase of the disease. Pediatr Int 2009;51:211-15. http://dx.doi.org/10.1111/j.1442-200X.2008.02696.x.
- Watcharananan S, Kiertiburanakul S, Chantratita W. Rapid influenza diagnostic test during the outbreak of the novel influenza A/H1N1 2009 in Thailand: an experience with better test performance in resource limited setting. J Infect 2010;60:86-7. http://dx.doi.org/10.1016/j.jinf.2009.10.049.
- Weitzel T, Schnabel E, Dieckmann S, Borner U, Schweiger B. Evaluation of a new point-of-care test for influenza A and B virus in travellers with influenza-like symptoms. Clin Microbiol Infect 2007;13:665-9. http://dx.doi.org/10.1111/j.1469-0691.2007.01739.x.
- Yang J, Lo J, Ho Y, Wu H, Liu M. Pandemic H1N1 and seasonal H3N2 influenza infection in the human population show different distributions of viral loads, which substantially affect the performance of rapid influenza tests. Virus Res 2011;155:163-7. http://dx.doi.org/10.1016/j.virusres.2010.09.015.
Appendix 1 Summary of the protocol for the Three Winters Study
Study protocol
Randomised controlled trial to evaluate impact of diagnostic testing for influenza, respiratory syncytial virus and Streptococcus pneumoniae infection on the management of acute admissions in the elderly and high-risk 18–64-year-olds
Funder ref no. 03/39/18
Principal investigator Professor Karl G Nicholson,
Infectious & Tropical Diseases Unit,
Leicester Royal Infirmary,
Infirmary Square,
Leicester LE1 5WW, England
Telephone – 0116 2586164
Facsimile – 0116 2585067
Date 26/11/2007 Version 6
Protocol synopsis
Title
Randomised controlled trial to evaluate impact of diagnostic testing for influenza, respiratory syncytial virus and Streptococcus pneumoniae infection on the management of acute admissions in the elderly and high-risk 18–64-year-olds.
Sponsor
NHS R&D National Coordinating Centre for Health Technology Assessment.
Study population
Male or female elderly (> 65 years old) patients,
Male or female patients aged > 18 years with underlying heart or lung disease including asthma, or pneumonia/influenza type symptoms.
Study setting
Medical Admissions Units in the University Hospitals of Leicester NHS Trust (Leicester Royal Infirmary, Glenfield Hospital, Leicester General Hospital).
Duration
August 1 2005 – July 31 2008 (36m). Patient enrolment will occur during: September 1 2005 to June 30 2006;
September 1 2006 to June 30 2007; &
September 1 2007 to June 30 2008.
All patients will be followed up for 28 days.
Rationale
The purpose of this study is to determine the diagnostic accuracy and clinical- and cost-effectiveness of rapid molecular and near-patient diagnostic tests for influenza, RSV and S. pneumoniae infections in the elderly, and subjects aged > 18 years with chronic cardiopulmonary conditions, or pneumonia/influenza type symptoms, in comparison to traditional laboratory culture.
Objectives and hypotheses
Research objectives
-
To determine the diagnostic accuracy (sensitivity, specificity, positive and negative predictive values) of rapid molecular and near-patient diagnostic tests for influenza, RSV and S. pneumoniae infections in comparison to traditional laboratory culture.
-
To assess the potential benefits of ease of use, and speed of rapid molecular and near-patient diagnostic tests for influenza, RSV and S. pneumoniae infections, in comparison to traditional laboratory culture.
-
To determine whether rapid molecular and near-patient diagnostic tests for influenza, RSV and S. pneumoniae infections have any impact on the prescription of antimicrobials.
-
To determine whether rapid molecular and near-patient diagnostic tests for influenza, RSV and S. pneumoniae infections allow more appropriate use of isolation facilities, in comparison to traditional laboratory culture.
-
To compare the costs of performing rapid molecular and near-patient diagnostic tests for influenza, RSV and S. pneumoniae infections, in comparison to traditional laboratory culture.
-
To assess cost-savings associated with earlier use of narrow-spectrum antimicrobial therapy (or avoidance or discontinuation of antibiotics) in patients whose influenza, RSV and S. pneumoniae infections are diagnosed more rapidly by rapid molecular and near-patient diagnostic tests, in comparison to traditional laboratory culture.
-
To compare the outcome of patients whose influenza, RSV, and S. pneumoniae is diagnosed more rapidly by rapid molecular and near-patient diagnostic tests, compared with those who are diagnosed by traditional laboratory culture.
-
To assess the impact that rapid molecular and near-patient diagnostic tests have on the costs associated with an inpatient stay and on costs post-discharge up to a maximum of 28 days after admission.
-
To assess the impact that rapid molecular and near-patient diagnostic tests have on quality-of-life, as measured by the EuroQol, and to use this information to estimate the quality adjusted life years (QALYs) generated during the 28 days after admission.
-
To assess the cost-effectiveness of rapid molecular and near-patient diagnostic tests in comparison to traditional laboratory culture. This will be done on the basis of both cost per case detected and cost per QALY.
Hypotheses
-
The increased diagnostic accuracy of rapid molecular and near-patient tests over traditional laboratory methods improves patient management through better use of antimicrobials and isolation facilities. Rapid molecular and near-patient diagnostic tests for influenza, RSV and S. pneumoniae infections are more cost-effective than traditional laboratory diagnostic tests.
-
Rapid molecular and near-patient diagnostic tests for influenza, RSV and S. pneumoniae infections provide benefits in terms of (a) ease of use, and (b) more rapid results, in comparison to traditional laboratory culture.
-
Rapid molecular and near-patient diagnostic tests for influenza, RSV and S. pneumoniae infections result in earlier use of ‘narrow-spectrum’ antimicrobial therapy; an earlier switch from intravenous to oral therapy; and earlier discontinuation of antibiotics in patients infected with influenza and RSV – in comparison to traditional laboratory culture.
-
Rapid detection of influenza and RSV by rapid molecular and near-patient diagnostic tests leads to the appropriate isolation of patients, but only in hospitals/wards having an adequate provision of cubicles.
-
The costs of performing rapid molecular and near-patient diagnostic tests for influenza, RSV and S. pneumoniae infections, differ significantly from the cost of traditional laboratory culture.
-
The earlier use of narrow-spectrum antimicrobial therapy (or avoidance or discontinuation of antibiotics) in patients whose influenza, RSV and S. pneumoniae infections are diagnosed more rapidly by rapid molecular and near-patient diagnostic tests results in significant cost-savings, in comparison to traditional laboratory culture.
-
A streamlining of antimicrobial prescribing that may arise from more rapid diagnosis of influenza, RSV and S. pneumoniae infections by rapid molecular and near-patient diagnostic tests does not adversely affect patient outcome.
-
Any increase in costs incurred by rapid molecular and near-patient diagnostic tests, in comparison to traditional laboratory culture, are more than offset by savings that arise from either rational antimicrobial prescribing or earlier discharge from the hospital.
-
Rapid molecular and near-patient diagnostic tests result in an improvement in quality-of-life, as measured by the EuroQol, which arises from streamlining of antibiotics and earlier discharge into the community.
-
Rapid molecular and near-patient diagnostic tests are more cost-effective than traditional laboratory culture.
Observational objectives
-
To estimate the admission rates for influenza, RSV, and S. pneumoniae in the target population.
-
To compare the clinical characteristics and economic burden of influenza A and B and RSV A and B.
-
To review the implications of rapid diagnosis on isolation policy, and review alternate approaches to managing the infection control issues.
Hypotheses (observational)
-
The admission rates in the target population are higher for S. pneumoniae than influenza A and B. The admission rates for influenza A and B are similar to those for RSV A and B.
-
The clinical characteristics and economic burden of influenza A and B and RSV A and B are similar.
-
Rapid near-patient and/or molecular diagnostic tests will reveal more cases of influenza and RSV who require isolation than can be isolated. Alternate approaches to managing the infection control issues, such as the use of influenza neuraminidase inhibitors, may be pertinent.
Methodology
Study design
Prospective, randomised controlled trial of the impact of diagnostic testing [(i) Group 1: rapid near-patient diagnostic tests (influenza and pneumococcus), (ii) Group 2: rapid molecular tests (influenza and RSV), plus laboratory pneumococcal antigen testing, and (iii) Group 3: traditional ‘laboratory culture’ (influenza, RSV, and S. pneumoniae)] in elderly (> 65 years) and ‘high-risk’ patients, who present to Medical Admissions’ Units in Leicestershire with an acute cardio-pulmonary illness.
All tests will eventually be done on all patients who enter the study, but patients in Groups 1 and 2 will only be provided with rapid test results relating to their randomisation group.
Number of cases
Elderly Estimated 2752 cases of acute cardio-pulmonary illness in the elderly, of whom 664 will have pneumonia (J12.9-J18.9). An estimated 556 will have unspecified acute lower respiratory tract infections (J22.X). About one third of the pneumonia cases will have S. pneumoniae infections (n = 221); If ∼5% of all acute cardiopulmonary admissions have laboratory-confirmed RSV and ∼10% have influenza, then ∼138 cases of RSV and ∼275 cases of influenza would be studied in the elderly.
High-risk 18- to 64-year-old An estimated 83 cases of pneumonia, with one-third (n = 28) having S. pneumoniae infections. An estimated 93 cases of unspecified acute lower respiratory tract infections (J22.X); 29 acute unspecified URTI’s (J06.9); and 181 cases of COPD. If ∼5% of these admissions (n = 386) have laboratory-confirmed RSV and ∼10% have influenza, then ∼19 cases of RSV and ∼38 cases of influenza would be studied in 18 to 64 year-old high-risk patients.
Demographic data
-
Male or female elderly, aged > 65 years of age.
-
Male or female ‘high-risk’ patients with underlying heart or lung conditions, aged 18 to 64 years of age, or with pneumonia or influenza like symptoms.
Inclusion criteria
-
Able and willing to give written informed consent, OR a relative or carer is willing to give written informed assent for patients who are too debilitated to provide consent.
-
Age > 65 years, OR age > 18 years with underlying chronic heart or lung disease including asthma; OR with pneumonia or influenza like symptoms.
-
Have an acute exacerbation of chronic cardio-pulmonary illness of < 168 hours (7 days) duration, OR an acute cardio-pulmonary illness or influenza-like illness of < 7 days’ duration, including:
-
Pneumonia,
-
Influenza/influenza-like illness,
-
Exacerbations of chronic obstructive pulmonary disease (COPD),
-
Bronchitis,
-
Asthma,
-
Congestive heart failure,
-
Cardiac arrhythmia.
-
-
Able and willing to adhere to the procedures stated in the protocol.
-
Patients should have access to a telephone.
Exclusion criteria
-
Inclusion criteria not met.
-
Angina/suspected myocardial infarction.
-
Were recruited to this study within 28 days of the current admission.
-
Could not be recruited into the study within a 16-hour period of initial assessment by a doctor on the Medical Admissions Unit or a ward accepting acute medical admissions.
-
Enrolment in a study of antimicrobial therapy for the illness for which the patient was admitted.
Randomisation:
Patients in each centre will be randomly allocated to one of three diagnostic policy groups:
-
Group 1: near-patient tests (Quidel – influenza; BinaxNOW – pneumococcus).
-
Group 2: rapid molecular tests (‘flu & RSV plus laboratory testing of concentrated urine in the BinaxNOW assay); and
-
Group 3: traditional laboratory culture.
using computer generated randomisation codes stratified by centre.
Assessment methods:
Clinical
Impact of test result on prescribing, specifically:
-
Earlier use of ‘narrow-spectrum’ anti-microbial therapy.
-
Earlier switch from intravenous to oral therapy.
-
Avoidance or earlier discontinuation of antibiotics in patients infected with influenza and RSV, and
-
Prescriptions of influenza neuraminidase inhibitors.
Will be assessed in rapid near-patient (Group 1) and molecular diagnostic groups (Group 2) and compared with traditional laboratory culture (Group 3).
Clinical outcomes, specifically:
-
Length of hospital stay.
-
Fever duration.
-
Supplemental oxygen dependence and CPAP dependence.
-
Admissions to Intensive Care.
-
Ventilatory support, and
-
Deaths.
Will be assessed in rapid near-patient (Group 1) and molecular diagnostic groups (Group 2) in comparison to traditional laboratory culture (Group 3).
Duration of hospitalisation, until discharge or death, will be obtained from the UHL Leicester hospital activity analysis (i.e. from computerised records).
Fever duration The participants’ temperature charts will be monitored during the first 10 days of hospitalisation to identify when they first became apyrexial (temperature < 37.2 °C), and remained so for a period of at least 24 hours.
Supplemental oxygen dependence and CPAP dependence The participants will be monitored during the first 10 days of hospitalisation to identify when they no longer required oxygen for a period of at least 24 hours.
Admission to Intensive care, and Ventilatory support during the first 10 days of hospitalisation will be identified and documented by the study nurse in the Case Report Form.
Deaths that occur within a maximum of 28 days of hospitalisation will be identified and documented by the study nurse in the Case Report Form.
Quality of life, as measured by EuroQol, and quality adjusted life years generated during the 28 days after admission will be assessed in rapid near-patient (Group 1) and molecular diagnostic groups (Group 2) in comparison to traditional laboratory culture (Group 3).
Appropriate use of isolation facilities The time from admission to the Medical Admissions Unit to the time of admission into a single room (isolation cubicle) will be assessed in patients with confirmed influenza and RSV in the rapid near-patient (Group 1) and molecular diagnostic groups (Group 2) in comparison to traditional laboratory culture (Group 3).
The study nurse will document in the CRF where the patient was nursed throughout the first 7 days of admission.
Discharge diagnoses will be obtained from the UHL Leicester hospital activity analysis (i.e. from computerised records).
Financial
Costs of diagnostic tests, estimated by means of an ‘ingredients’ approach where all items needed to carry out the test are recorded and costed using appropriate local and national data, e.g., items to collect and transport specimens, media and reagents for the test, equipment to process specimens, technical support costs, etc. Costs will be identified for the following technologies:
-
Rapid near-patient test for influenza (Quidel).
-
Rapid near-patient test for pneumococcus (BinaxNOW).
-
Molecular (multiplex PCR) tests for influenza A and B and RSV A and B.
-
Culture (blood and sputum) for S. pneumoniae.
-
Gram staining of sputum samples.
-
Cell culture for influenza A and B.
-
Cell culture for RSV A and B.
-
Other tests that may be applied, e.g. immunofluorescence.
Care costs Cost of inpatient stay will be determined using information on length of stay and hospital costs to determine a ‘hotel’ cost of routine care. To this will be added the cost of any additional clinical care received such as diagnostic tests, drugs, etc. For patients who are discharged within 28 days of admission, health care resource use in the period after discharge will be recorded using a simple questionnaire administered in a telephone interview. These will be costed using appropriate national data, for example NHS reference costs unit costs compiled by the PSSRU at the University of Kent.
Cost-savings, which accrue from (i) a reduction in the use of resources; and (ii) earlier discharge from the hospital, will be identified by comparison of the costs for participants in Groups 1, and 2, compared with traditional laboratory culture (Group 3).
Economic evaluation of near-patient and rapid molecular diagnostic tests will be assessed by two main outcomes measures –
-
cost per case detected.
-
cost per QALY.
Laboratory
Diagnostic accuracy, (sensitivity, specificity, positive and negative predictive values) and discrepant analysis of near-patient and molecular diagnostic tests, will be estimated in comparison with traditional and other (e.g., serology) laboratory tests.
Ease of use of rapid near-patient and molecular tests The ease of use of molecular, near patient, and traditional laboratory culture will scored independently by three investigators in terms of whether they can be done:
On site:
-
Require special laboratory facilities.
-
Require special equipment.
-
The number of reagents required.
-
The number of steps.
-
Ease of disposal/decontamination of used equipment and reagents.
-
Technical competency required of the operator.
-
Training period required to reliably carry out the test, and any
-
Health and safety implications.
Speed of tests will be assessed in terms of the median time from specimen collection to result:
-
Appearing in the case notes.
-
Appearing on Pathology Department results’ database (APEX), and/or
-
Being phoned to the ward.
-
Being acted upon – in comparison to traditional laboratory culture.
Observational
Admission rates For influenza, RSV, and S. pneumoniae in the target population, taking into consideration the total population estimates, stratified by age, and the proportion of all patients by ICD code that were sampled.
Study procedures
Baseline (day 1)
Written informed consent from patient or assent from relative or carer, Inclusion/exclusion criteria,
-
Randomisation.
-
Basic demography.
-
Medical history/regular medication.
-
Presenting symptoms and the interval between their onset and admission.
-
Clinical findings.
-
Quality of Life assessment.
-
Investigations ordered by the admitting physicians.
-
Specimen collection (blood for antibody tests, sputum, nasopharyngeal specimen, and urine) for trial specific diagnostic tests.
-
Rapid near-patient diagnostic testing for influenza & pneumococcus (Group 1) on, or adjacent to, the ward. Results will be delivered to the nursing and/or medical team on the MAU and entered into the case notes. The time when the results were entered into the case notes will be recorded in the patient’s CRF together with the results.
-
Processing and transport of specimens to the laboratory of diagnostic specimens (Groups 1, 2, 3).
-
Antimicrobial and antiviral treatments prescribed/dose/frequency/route.
-
Isolation status.
-
Information relating to the above activities will be documented in the CRF.
Follow-up (days 2, 3, 4, 5, 6, 7, 8, 9, 10, and 28)
The following will be performed, updated and recorded in the CRF:
-
Time when diagnostic tests were made available to the nursing/medical staff.
-
Treatment, specifically the relationship between the timing/availability of diagnostic tests and changes in antimicrobial therapy. The nature of treatment given to all patients within 10 days of admission will be documented.
-
Isolation, specifically the relationship between the timing/availability of diagnostic tests and changes in isolation status. The isolation status of all patients throughout the first 7 days of admission will be carefully documented.
-
Admission to ITU and ventilatory support (within 28 days of hospitalisation).
-
Pyrexia – the timepoint when the patient first became apyrexial (< 37.2 °C), and remained so for > 24 hours (during days 1–10).
-
Oxygen requirement – the timepoint when supplemental oxygen was no longer required, and was not given for > 24 hours (during days 1–10).
-
Diagnostic studies (within 28 days of hospitalisation).
-
Duration of hospitalisation.
-
Deaths (within 28 days of hospitalisation).
-
EuroQol (days 7 and 28).
-
Discharge diagnosis.
-
Convalescent serum sample (days 10–90).
End points:
Clinical
Impact of test result on prescribing, specifically:
-
Time, from admission to MAU, to first administration of ‘narrow-spectrum’ antibiotics, for patients in Groups 1, 2, and 3, who are prescribed antibiotics.
-
Time, from admission to MAU, to first administration of oral antibiotics, for patients in Groups 1, 2, and 3, who are prescribed antibiotics.
-
Time (hours) from admission to MAU to prescription of ‘no antibiotics’ (oral or intravenous) administered to patients in Groups 1, 2, and 3, who have influenza or RSV, and
-
Proportion of patients with influenza in Groups 1, 2, and 3 who are prescribed neuraminidase inhibitors.
Clinical outcomes, specifically:
-
Length of hospital stay until discharge: First, for all patients in Groups 1, 2, and 3. Second, for all patients with (i) influenza; (ii) RSV; and (iii) S. pneumoniae infection in Groups 1, 2, and 3.
-
Fever duration (during the first 10 days of hospitalisation) Time from admission (hours) until the patient first became apyrexial (temperature < 37.2 °C), and remained so for a period of at least 24 hours: First, in all patients in Groups 1, and 2 in comparison to Group 3. Second, in patients with S. pneumoniae in Groups 1, and 2, in comparison to Group 3.
-
Supplemental oxygen dependence and CPAP dependence (during the first 10 days of hospitalisation) Times from admission (hours) until the patient required (i) no supplemental oxygen, and (ii) no CPAP, for a period of at least 24 hours: First, in all patients in Groups 1, and 2 in comparison to Group 3. Second, in patients with S. pneumoniae in Groups 1, and 2, in comparison to Group 3.
-
Admissions to Intensive Care (during the first 10 days of hospitalisation) First, the proportion of patients with S. pneumoniae infection in Groups 1, 2, and 3. Second, to better define the burden of influenza and RSV, the proportion of all patients with (i) influenza, and (ii) RSV who require ITU support.
-
Ventilatory support (during the first 10 days of hospitalisation): First, the proportion of patients with S. pneumoniae infection in Groups 1, 2, and 3. Second, to better define the burden of influenza and RSV, the proportion of all patients with (i) influenza and (ii) RSV who require ventilatory support.
-
Deaths (within 28 days of hospitalisation): First, the proportion of patients with S. pneumoniae infection in Groups 1, 2, and 3. Second, to better define the burden of influenza and RSV, the proportion of all patients with (i) influenza, and (ii) RSV, who die.
Quality of life, as measured by EuroQol, and quality adjusted life years generated during the 28 days after admission, will be assessed:
-
In all patients in Groups 1, and 2 in comparison to Group 3.
-
Second, in patients in patients with (i) influenza, (ii) RSV, and (iii) S. pneumoniae in Groups 1, and 2, in comparison to Group 3.
Use of isolation facilities:
-
The time from admission to the MAU to the time of admission to a single room (isolation cubicle) will be compared for patients with confirmed influenza or RSV in Groups 1, 2, and 3.
-
The proportion of patients with influenza or RSV in Groups 1, 2, and 3 who are isolated at any stage during the first 120 hours of the admission.
-
The proportion of patients with S. pneumoniae infection in Groups 1, 2, and 3 who are inappropriately isolated for > 12 hours.
Financial
Costs of diagnostic tests Costs will be identified for:
-
Rapid near-patient test for influenza (Quidel).
-
Rapid near-patient test for pneumococcus (BinaxNOW).
-
Molecular (multiplex PCR) tests for influenza A and B and RSV A and B.
-
Culture (blood and sputum) for S. pneumoniae.
-
Gram staining of sputum samples.
-
Cell culture for influenza A and B.
-
Cell culture for RSV A and B.
-
Other tests that may be applied, e.g., immunofluorescence.
Care costs of inpatient stay (+95% CI) will be determined for:
-
All patients in Groups 1, 2, and 3.
-
All patients (in all groups) with (i) influenza, (ii) RSV, and (iii) S. pneumoniae infection, and,
-
Patients with (i) influenza, (ii) RSV, and (iii) S. pneumoniae infection in Groups 1, 2, and 3.
For patients who are discharged within 28 days of admission, health care resource use in the period after discharge will be recorded using a simple questionnaire administered in a telephone interview or by post.
Cost-savings that accrue from earlier use of narrow-spectrum antibiotics, oral therapy, or avoidance, or discontinuation of antibiotics will be assessed in:
-
All patients in Groups 1, 2, and 3.
-
Patients with (i) influenza, (ii) RSV, and (iii) S. pneumoniae infection in Groups 1, 2, and 3.
Economic evaluation of near-patient and rapid molecular diagnostic tests will be assessed by two main outcomes measures:
-
Cost per case detected.
-
Cost per QALY.
Laboratory
Diagnostic accuracy:
-
Sensitivity.
-
Specificity.
-
Positive predictive value.
-
Negative predictive value.
and discrepant analysis of near-patient and molecular diagnostic tests, will be estimated in comparison with traditional and other (e.g. serology) laboratory tests.
Ease of use of rapid near-patient and molecular tests: molecular, near-patient, and traditional laboratory culture diagnostic tests will scored independently for ease of use by three investigators.
Speed of tests will be assessed in terms of the median time from specimen collection to result:
-
Appearing in the case-notes.
-
Appearing on Pathology Department results’ database (APEX).
-
Being phoned to the ward.
-
Being acted upon – in comparison to traditional laboratory culture.
Speed of tests will be determined for –
-
All patients in Groups 1, 2, and 3.
-
Patients with influenza; RSV, and S. pneumoniae infection in Groups 1, 2, and 3.
Observational
Admission rates, for:
-
Influenza.
-
RSV, and
-
S. pneumoniae.
Analysis
Sample size
The sample size is based on the admissions during 2002–03 and 2003–04 (September 1 – April 30) for elderly (> 65 years old) patients with acute cardio-pulmonary conditions, excluding angina and myocardial infarction. There were 2762 acute cardio-pulmonary admissions during September 1 – April 30 2002–03, and 2852 during 2003–04, i.e. an average of 11.57 admissions (> 65 years) per day.
The study will run for at 666 days (242 during each of Years 1 and 2, and 183 during Year 3), i.e. the number of eligible patients is estimated at 666 × 11.57 = 7705.6. We plan to recruit 5 days per week, which reduces the eligible number of patients to (5 ÷ 7) × 7705.6 = 5504. We understand that two-thirds are admitted during the period 09:00–21:00h, which reduces the evaluable pool to 3669. We estimate that three-quarters of eligible subjects will participate, i.e. we expect to recruit 2752 elderly (> 65 years old) patients with acute cardio-pulmonary conditions. Of these, 664 are expected to have ICD codes for pneumonia; 556 are expected to have unspecified acute lower respiratory tract infections; 683 are expected to have exacerbations of chronic obstructive pulmonary disease; and 735 are expected to be admitted with heart failure.
We estimated the number of admissions in ‘high-risk’ 18–64 year-olds by extrapolation using (i) national cardio-pulmonary hospital admission data for patients aged 15–59 years, 60–74 years, and 75 years and older and (ii) the number of cardio-pulmonary admissions aged > 65 years in Leicester.
We assumed that half of the patients admitted with pneumonia, unspecified lower respiratory tract infections, and exacerbations of COPD have underlying high-risk conditions. We expect to recruit 83 18–64-year old patients with pneumonia, 93 with unspecified lower respiratory tract infections, and 181 with COPD. These have not been included in the following estimates:
On the basis of historical data we expect that one-third of elderly patients with pneumonia have pneumococcal disease (i.e. 221) but expect the number identified by the pneumococcal antigen test to be higher. Of the 221, ∼73 should be randomly allocated to the rapid near-patient test (BinaxNOW) (Group 1); the remainder will be randomised to the group tested by traditional methods (Groups 2 & 3). However, as identical sample sets will be taken from each individual, diagnostic accuracy will be assessed in a minimum of 221 subjects.
We expect that 10% of the 2752 (elderly) patients will have influenza A or B. Of the 275, one third (∼91) will be allocated to Groups 1, 2, and 3. Identical sample sets will be taken from each individual, so the diagnostic accuracy of the tests will be assessed in all 275 subjects.
We expect that 5% of the 2752 (elderly) patients will have RSV A or B. Of the 137, one third (∼45) will be allocated to the rapid molecular group (Group 2); the remainder (∼90) will be allocated to the groups tested by traditional methods (Group 1 & 3). Identical sample sets will be taken from each individual, so the diagnostic accuracy of the tests will be assessed in all 137 subjects.
While the numbers of patients with influenza and RSV who are allocated to the ‘rapid’ near-patient or molecular tests are comparatively small, the impact of a ‘viral’ infection (RSV or influenza) infection on patient isolation, antimicrobial prescribing, and clinical outcomes may be compared using larger combined groups – i.e., rapid influenza & RSV (i.e. Quickview + molecular tests) n = (91 + 91 + 45) = 227 traditional influenza & RSV n = (91 + 45 + 45) = 182.
Statistical power:
This has been estimated for one laboratory and two clinical end points for the elderly population only.
Diagnostic accuracy
Assuming that the average sensitivity/specificity of the tests is 80%[90%] then allowing for a 20% dropout rate, a sample of 2752 (2000) i.e. only 2 winters) elderly (> 65 years) patients randomised into the trial would enable the sensitivity/specificity to be estimated to within, i.e. 2SE, 7.6% (8.9%) [5.7% (6.7%)] for a disease prevalence of 10%, and 5.4% (6.3%) [4.0% (4.7%)] for a disease prevalence of 5%.
Length of stay
2752 patients would enable a Minimum Clinically Significant Difference (MCSD) [between diagnostic policies] of 1 day in the mean length of stay (assuming SD = 6 days) to be detected at the 5% significance level with over 80% power, assuming a 20% dropout rate and adjusting for the fact that there are 3 groups.
Appropriate isolation levels: 2752 patients would also enable a Minimum Clinically Significant Difference (MCSD) [between diagnostic policies] of an improvement in appropriate use of isolation facilities from 5% to 15% to be detected at the 1% significance level with over 95% power, assuming a 20% dropout rate and adjusting for the fact that there are 3 groups.
Statistical methods:
All analyses for both process and clinical outcomes will be based on Intention to Treat (ITT) analyses.
Impact of test result on prescribing
The time to prescription of ‘narrow spectrum’, ‘oral antibiotics’ or ‘no antibiotics’ between the three groups will be assessed using survival analysis techniques, whilst the use of neuraminidase inhibitors in those with influenza will be assessed using chi-squared tests, together with 95% CIs. Further analyses to allow for potential differences in patient demographics and baseline clinical characteristics between the three diagnostic test groups, and not allowed by stratification, will make use of Cox proportional hazards regression modelling in the case of time to appropriate prescribing and logistic regression techniques in the case of neuraminidase inhibitors.
Length of hospital stay
Length of hospital stay in the three diagnostic groups will initially be compared using non-parametric methods. Further analyses to allow for potential differences in patient demographics and baseline clinical characteristics between the three diagnostic test groups, and not allowed by stratification, will make use of generalised linear models in order to accommodate any skewness in the data.
Mortality rates
Mortality rates between the three diagnostic testing groups will be compared by means of a Log-Rank Test and Kaplan–Meier survival curves. Adjustment for potential differences in patient demographics and baseline clinical characteristics between the three diagnostic test groups, and not allowed by stratification, will make use of Cox proportional hazards regression methods.
Diagnostic accuracy
Sensitivity, specificity, positive and negative predictive values of molecular and near-patient diagnostic tests in comparison to traditional laboratory methods will be calculated together with 95% CIs. Heterogeneity in the sensitivity and specificity with respect to patient demographics and baseline clinical characteristics will be explored as secondary analyses using patient defined sub-groups.
Admission to Intensive Care
The proportions of patients in the three groups who are admitted to intensive care within the first 10 days of admission will be compared using chi-squared tests, together with 95% CIs. Further analyses to allow for potential differences in patient demographics and baseline clinical characteristics between the three diagnostic test groups, and not allowed by stratification, will make use of logistic regression techniques.
Ventilatory support
The proportions of patients in the three groups who receive ventilatory support within the first 10 days of admission will be compared using chi-squared tests, together with 95% CIs. Further analyses to allow for potential differences in patient demographics and baseline clinical characteristics between the three diagnostic test groups, and not allowed by stratification, will make use of logistic regression techniques.
Appropriate use of isolation facilities
In patients with confirmed influenza or RSV the time taken from admission to the MAU to admission to a single room (isolation cubicle) will be compared between the three diagnostic groups by means of a Log-Rank Test and Kaplan–Meier survival curves. Adjustment for potential differences in patient demographics and baseline clinical characteristics between the three diagnostic test groups, and not allowed by stratification, will make use of Cox proportional hazards regression methods.
Quality of life (EQ-5D)
Quality of life in the three diagnostic groups will initially be compared using non-parametric methods. Further analyses to allow for potential differences in patient demographics and baseline clinical characteristics between the three diagnostic test groups, and not allowed by stratification, will make use of generalised linear models in order to accommodate any skewness in the data.
Speed of tests
Time taken to receive test results in the three diagnostic groups will initially be compared using non-parametric methods. Further analyses to allow for potential differences in patient demographics and baseline clinical characteristics between the three diagnostic test groups, and not allowed by stratification, will make use of generalised linear models in order to accommodate any skewness in the data.
Cost data
Cost data will be analysed using parametric and non-parametric statistical methods which explicitly allow for the censoring of (indirect & total) costs at 28 days, i.e. for those patients who are not discharged from hospital within 28 days and thus enable an unbiased assessment of potential cost differences between the three diagnostic groups to be made. Estimation of the cost distribution over longer timescales will make use of extrapolation techniques using time of discharge obtained from hospital information systems.
Trial plan and schedule of assessment
| Assessments | Study day | ||
|---|---|---|---|
| Admission (day 1) | Days 2, 3, 4, 7 (±1), & 10 (±1) | Day 28 (±2) | |
| Pre-study assessment by admitting medical team | ✓ | ||
| Note time of admission (CRF) | ✓ | ||
| Inclusion/Exclusion criteria check (CRF) | ✓ | ||
| Written informed Consent (or Assent) (CRF) | ✓ | ✓ (Consent upon recovery) | |
| Demography/immunisation status check (CRF) | ✓ | ||
| Medical history/concomitant medications check (CRF) | ✓ | ||
| Symptom assessment (CRF) | ✓ | ||
| Clinical findings, including body weight and temperature (CRF). Establish when patient first becomes apyrexial (≤ 37.2 °C), requires supplemental O2, and remains so for > 24 hours (CRF) | ✓ | ✓ (temperature, O2) | |
| EuroQol. Quality of Life assessment (CRF) | ✓ | ✓ (day 7) | ✓ |
| Diagnostic studies ordered by the admitting medical team (CRF) | ✓ | ✓ | |
| Collection of trial specific diagnostic specimens (CRF): | |||
| Nasopharyngeal (Quidel & virus culture) | ✓ | ||
| Sputum (Gram stain & culture) | ✓ | ||
| Urine (BinaxNOW) | ✓ | ||
| Blood (Blood cultures, serum antibodies) | ✓ | ✓ (antibody, day 10) | |
| Record time when diagnostic test results were made available to the ward nurses, and/or admitting team (CRF) | ✓ | ✓ | |
| Record antimicrobials & antivirals (identity, route, dose, time of administration) prescribed by admitting medical team (CRF) | ✓ | ||
| Note the time of any changes in antimicrobials & antivirals (identity, route, dose, time of administration) prescribed by admitting medical team (CRF) | ✓ | ||
| Bed location, i.e., record bay or a single room (CRF) | ✓ | ||
| Note time of any subsequent change in isolation status (CRF) | ✓ | ||
| ITU and ventilatory support: Document whether admitted or required ventilatory support (CRF) | ✓ | ✓ | |
| Date of hospital discharge/Death within 28 days of admission (CRF) | ✓ | ✓ | |
| Discharge diagnosis (CRF) | ✓ | ✓ | |
Appendix 2 BinaxNOW pneumococcal antigen test
Appendix 3 Collection and transport of nasopharyngeal specimens
The collection of nasopharyngeal specimens for this study followed guidance from the National Influenza Research Laboratory, HPA, Colindale, London, and the manufacturer of the POCT. A good specimen for the detection of influenza or RSV must contain a substantial number of respiratory epithelial cells, which are mainly obtained from the nasal swab. A throat swab alone will contain mainly squamous epithelial cells in which influenza does not replicate. In brief, collection of routine nasopharyngeal swabs was as follows:
-
a single swab with cotton wool bud is inserted in one nostril and rubbed against and above the nasal turbinates
-
a second swab is used to abrade the tonsils and pharynx
-
place both swabs in the same bijou bottle of virus transport medium
-
break off the swab sticks (scissors may be used)
-
screw lid tightly on to the bottle.
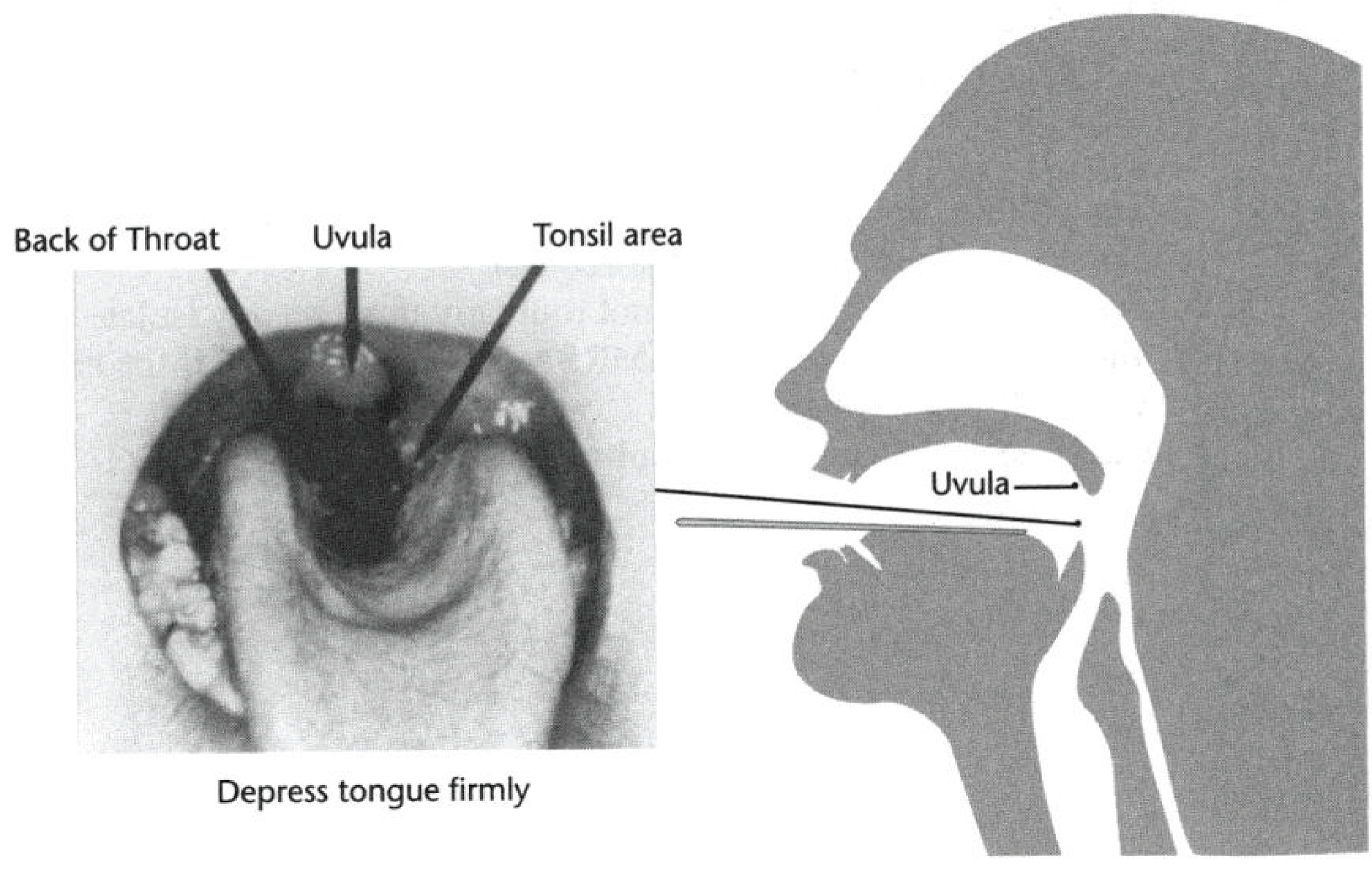
Trial-specific procedures were as follows.
‘Near-patient diagnostic group’ (QuickVue Influenza A + B test) and Deferred QuickVue test
-
The sterile sponge tipped swab provided in the QuickVue Influenza A + B test was inserted into the nostril that presented the most secretions under visual inspection, according to manufacturer’s instructions.
-
Using gentle rotation, the swab was gently pushed until resistance was met at the level of the turbinates (< 1 inch into the nostril). The swab was rotated a few times against the nasal wall.
-
Specimens collected from patients in the ‘near-patient diagnostic group’ were tested immediately for the presence of Influenza A and B according to the manufacturer’s instructions.
-
An identical specimen was collected from patients randomised to the molecular diagnostic group and conventional virus culture group. These specimens were collected into virus transport medium and were refrigerated at 4 °C until they were transported at the earliest opportunity by the hospital transport system to the microbiology laboratory. They were processed upon receipt during normal working hours by the QuickVue A + B test.
Deferred QuickVUE influenza A + B test, prompt and deferred molecular testing, and conventional virus culture
-
We collected nasal and pharyngeal swabs for molecular and conventional virus culture studies using the opposite nostril to that used for the ‘near-patient’ and ‘deferred’ QuickVue Influenza A + B tests.
-
Using gentle rotation, a dry swab with cotton wool bud was inserted into the nostril and rubbed against and above the nasal turbinates.
-
A second swab is used to abrade the tonsils and pharynx.
-
The swab tips from both swabs were placed into a single bijou bottle of virus transport medium, agitated and then cut off, or broken off, into the medium.
-
These specimens were refrigerated at 4 °C until they were transported to the Microbiology Laboratory at the earliest opportunity by the hospital transport system.
-
Specimens from subjects randomised to the ‘prompt’ molecular diagnostic group were processed upon receipt during normal working hours. Specimens for ‘deferred’ molecular diagnostic testing from subjects who were randomised to the ‘conventional’ diagnostic group were stored at ‒20 °C
-
Specimens from subjects randomised to the ‘prompt’ and ‘conventional’ diagnostic groups were processed upon receipt during normal working hours by conventional virus isolation tests.
Appendix 4 Quidel QuickVue Influenza A + B test
Appendix 5 Case report form, clinical study
Appendix 6 Publications that were screened for the meta-analysis
| Author | Location | Children | Adults | Influenza | Gold standard | POCT | Sensitivity (%) | Specificity (%) | PPV% | NPV% |
|---|---|---|---|---|---|---|---|---|---|---|
| Agoritsas, 200691 | USA | 2 weeks to 18 years | None | Seasonal | PCR and culture (R-mix) | QuickVue Influenza A + B | NPS: 85 (50/59) | NPS: 98 (62/63) | NPS: 98 (50/51) | NPS: 87 (62/71) |
| NPW: 69 (41/59) | NPW: 98 (62/63) | NPW: 98 (41/42) | NPW: 78 (62/80) | |||||||
| NS: 78 (46/59) | NS: 97 (61/63) | NS: 96 (46/48) | NS: 82 (61/74) | |||||||
| Andresen, 2010168 | Australia | 0–18 years | None | Pandemic | PCR | QuickVue Influenza A + B | NPA: 84 (122/145) | NPA: 98 | – | – |
| Swabs: 66 (47/71) | Swabs: 100 | |||||||||
| Balish, 2009241 | USA | Included, age not specified | Included, age not specified | Pandemic | PCR | Directigen EZ Flu A + B | 49 (21/43) | – | – | – |
| Included, age not specified | Included, age not specified | H1N1 | PCR | Directigen EZ Flu A + B | 75 (3/4) | – | – | – | ||
| Included, age not specified | Included, age not specified | H3N2 | PCR | Directigen EZ Flu A + B | 83 (10/12) | – | – | – | ||
| Included, age not specified | Included, age not specified | Pandemic | PCR | NOW Flu A + B | 40 (18/45) | – | – | – | ||
| Included, age not specified | Included, age not specified | H1N1 | PCR | NOW Flu A + B | 60 (3/5) | – | – | – | ||
| Included, age not specified | Included, age not specified | H3N2 | PCR | NOW Flu A + B | 80 (12/15) | – | – | – | ||
| Included, age not specified | Included, age not specified | Pandemic | PCR | QuickVue Influenza A + B | 69 (31/45) | – | – | – | ||
| Included, age not specified | Included, age not specified | H1N1 | PCR | QuickVue Influenza A + B | 80 (4/5) | – | – | – | ||
| Included, age not specified | Included, age not specified | H3N2 | PCR | QuickVue Influenza A + B | 80 (12/15) | – | – | – | ||
| Bellei, 2003189 | Brazil | None | 18–56 years | Seasonal | Culture on MDCK cells | QuickVue Influenza A + B | 74 (29/39) | 83 (48/58) | – | – |
| Bellman-Weiler, 2011242 | Austria | None | Included, age not specified | Pandemic | PCR | Directigen EZ Flu A + B | 32 (8/25) | 93 (66/71) | 62 (8/13) | 79.5 (66/83) |
| None | Included, age not specified | Pandemic | PCR | NOW Flu A + B | 28 (8/29) | 99 (77/78) | 89 (8/9) | 79 (77/98) | ||
| Biggs, 2010243 | USA | Included, age not specified | Included, age not specified | Seasonal | PCR | NOW Flu A + B | 75.3 (64/85) | 98 (458/469) | ||
| Blyth, 2009244 | Australia | Included, age not specified | Included, age not specified | Pandemic | PCR | QuickVue Influenza A + B | 25 (5/20) | |||
| Booth, 2006190 | Australia | 1–16 years | Included, age not specified | Seasonal, influenza A | Culture on MDCK cells and IFA | ImmunoCard Stat! Flu A&B | 80 (28/35) | 98 (185/189) | 87.5 (28/32) | 96 (185/192) |
| 1–16 years | Included, age not specified | Seasonal, influenza B | Culture on MDCK cells and IFA | ImmunoCard Stat! Flu A&B | 47 (7/15) | 100 (209/209) | 100 (7/7) | 96 (209/217) | ||
| 1–16 years | Included, age not specified | Seasonal, influenza A | Culture on MDCK cells and IFA | NOW Flu A + B | 80 (28/35) | 99 (188/189) | 97 (28/29) | 96 (188/195) | ||
| 1–16 years | Included, age not specified | Seasonal, influenza B | Culture on MDCK cells and IFA | NOW Flu A + B | 47 (7/15) | 100 (208/209) | 88 (7/8) | 96 (208/216) | ||
| Cazacu, 2004200 | USA | From ≥ 3 days | To 55.8 years | Seasonal | Culture on RMK cells | Directigen Flu A + B | 44 (96/219) | 100 (3863/3873) | 90.5 (96/106) | 97 (3863/3986) |
| Cazacu, 2004201 | Included, age not specified | Included, age not specified | Seasonal | Culture on RMK cells | XPECT Flu A + B | 94 (118/125) | 100 (275/275) | 100 (118/118) | 97.5 (275/282) | |
| Chan, 2002147 | USA | From < 2 years | To > 55 years | Seasonal, influenza A | Culture on MDCK cells | Directigen EZ Flu A + B | 100 (22/22) | 99 (225/228) | 88 (22/25) | 100 (225/225) |
| From < 2 years | To > 55 years | Seasonal, influenza B | Culture on MDCK cells | Directigen EZ Flu A + B | 88 (28/32) | 97 (211/218) | 80 (28/35) | 98 (211/215) | ||
| Chen, 2010191 | China | Included, age not specified | Included, age not specified | Influenza A | Culture on MDCK cells | Flu A Dot | 88.5 (69/78) | 99 (146/147) | 99 (69/70) | 94 (146/155) |
| Cheng, 2010148 | China | From 5 months | To 77 years | Pandemic | PCR and culture on MDCK cells | Fujirebio Espline Influenza A&B-N | 62 (37/60) | |||
| Cheng, 2011245 | From 6 months | To 86 years | Seasonal and pandemic | PCR | Directigen Flu A | 72 (169/235) | 100 (571/572) | 99 (169/170) | 90 (571/637) | |
| From 6 months | To 86 years | Seasonal and pandemic | PCR | Flu A Dot | 91 (214/235) | 100 (570/572) | 99 (214/216) | 96 (570/591) | ||
| Choi, 2010172 | Korea | From 5 months | To 78 years | Pandemic | PCR | NOW Flu A + B | 64.5 (91/141) ≥ 21 years: 47 Illness onset: 56.3 |
95 (107/113) | 94 (91/97) | 68 (107/157) |
| From 5 months | To 78 years | Pandemic | PCR | SD Bioline Influenza A/B/A(H1N1) Pandemic rapid test | 69.5 (98/141) ≥ 21 years: 57.9 Illness onset: 31.3 |
100 (113/113) | 100 (98/98) | 72 (113/156) | ||
| Choi, 2010170 | From 2 weeks | To 83 years | Pandemic | PCR | SD Bioline Influenza A/B/A(H1N1) Pandemic rapid test | 77 (241/313) | 100 (445/446) | |||
| Chomel, 1992181 | France | Included, age not specified | Included, age not specified | Seasonal, influenza A | ELISA | Directigen Flu A | 87.9 (87/99) | 97 (59/61) | 83 (59/71) | 97 (87/89) |
| Included, age not specified | Included, age not specified | Seasonal, influenza A | Culture on LLCMK2 cells | Directigen Flu A | 84 (21/25) | |||||
| Covalciuc, 1999193 | USA | From 2 months | To 76 years | Seasonal | Culture on RMK cells | BioStar OIA Flu A/B |
80.1 (121/151) | 73 (185/253) | ||
| de la Tabla, 2010246 | Spain | From 3 months | To 97 years | Pandemic | PCR | Clearview Exact Influenza A & B | 19 (55/297) | 100 | 100 | 75 |
| D’Heilly, 200869 | USA | None | 18–91 years | Seasonal | Culture | FLU OIA A/B | 65 (49/75) | 97 | 89 | 89 |
| Diederen, 2010195 | Netherlands | From birth | To 81 years | Pandemic | PCR | NOW Flu A + B | 47 (18/38) | 95 (92/97) | 78 (18/23) | 82 (92/112) |
| Drexler, 2009169 | Germany | Included, age not specified | Included, age not specified | Pandemic | PCR | NOW Flu A + B | 11.1 (16/144) | |||
| Faix, 2009247 | USA | Included, age not specified | Included, age not specified | Pandemic | PCR | QuickVue Influenza A + B | 51 (20/39) | 99 | ||
| Included, age not specified | Included, age not specified | Influenza A (H1N1) | PCR | QuickVue Influenza A + B | 63 (12/19) | 99 | ||||
| Included, age not specified | Included, age not specified | Influenza A (H1N1) | PCR | QuickVue Influenza A + B | 31 (6/19) | 99 | ||||
| Fernandez, 2010248 | USA | From < 2 years | To > 65 years | Seasonal and Pandemic | R-mix culture and DFA | QuickVue Influenza A + B | 77 (113/147) | 85 | 74 | 87 |
| Fuenzalida, 2010249 | Spain | From 4 days | To 87 years | Pandemic | PCR | NOW Flu A + B | 60.3 (137/227) | 94 | 88 | 75 |
| Ghebremedhin, 2009250 | Germany | 7 weeks to 6.5 years | None | Influenza A | PCR | Actim Influenza A&B | 69.2 (9/13) | 100 (460/460) | 100 (9/9) | 99 (460/464) |
| 7 weeks to 6.5 years | None | Influenza B | PCR | Actim Influenza A&B | 60 (6/10) | 100 (463/463) | 100 (6/6) | 98 (463/467) | ||
| 7 weeks to 6.5 years | None | Influenza A | PCR | NOW Flu A + B | 57 (4/7) | 100 | ||||
| 7 weeks to 6.5 years | None | Influenza A | PCR | NOW Flu A + B | 43 (3/7) | 100 | ||||
| Gimeno, 2010251 | Spain | From < 1 year | To 87 years | Pandemic | PCR | SD Bioline Influenza A/B/A(H1N1) Pandemic rapid test | 26.9 (24/49) NPS: 35 Oropharyngeal swabs: 13 |
|||
| Gordon, 2009252 | Nicaragua | 2–12 years | None | Seasonal | PCR | QuickVue Influenza A + B | 68.5 (246/359) | 98 | ||
| 2–12 years | None | Influenza A | PCR | QuickVue Influenza A + B | 65.2 (146/224) | |||||
| 2–12 years | None | Influenza B | PCR | QuickVue Influenza A + B | 74.1 (100/135) | |||||
| Gordon, 2010253 | 2–14 years | None | Pandemic | PCR | QuickVue Influenza A + B | 64.1 (59/92) | 98 | |||
| Hamilton, 2002192 | USA | 12 days to 19 years | None | Seasonal | Culture on RMK cells | Directigen Flu A + B | 75 (49/65) | 93 (218/235) | 74 (49/66) | 93 (218/234) |
| 12 days to 19 years | None | Seasonal | Culture on RMK cells | Zstat Flu-II | 88 (57/65) | 92 (216/235) | 75 (57/76) | 96 (216/224) | ||
| Hara, 2008187 | Japan | 4 months to 19 years | None | Seasonal, influenza A | Culture on MDCK cells | Directigen EZ Flu A + B | 100 (53/53) | 99 (438/441) | 100 (53/53) | 100 (441/441) |
| 4 months to 19 years | None | Seasonal, influenza B | Culture on MDCK cells | Directigen EZ Flu A + B | 76 (206/270) | 99 (221/224) | 99 (206/209) | 78 (221/285) | ||
| 4 months to 19 years | None | Seasonal, influenza A | Culture on MDCK cells | Fujirebio Espline Influenza A&B-N | 100 (53/53) | 100 (441/441) | 100 (53/53) | 100 (441/441) | ||
| 4 months to 19 years | None | Seasonal, influenza B | Culture on MDCK cells | Fujirebio Espline Influenza A&B-N | 86 (232/270) | 100 (224/224) | 100 (232/232) | 85 (224/262) | ||
| 4 months to 19 years | None | Seasonal, influenza A | Culture on MDCK cells | NOW Flu A + B | 100 (53/53) | 98 (430/441) | 100 (531/531) | 100 (441/441) | ||
| 4 months to 19 years | None | Seasonal, influenza B | Culture on MDCK cells | NOW Flu A + B | 78 (211/270) | 99 (221/224) | 99 (211/214) | 79 (221/280) | ||
| Hawkes, 2010184 | Canada | 1 month to 17 years | None | Pandemic | PCR | NOW Flu A + B | 62 (66/107) | 99 (70/71) | 99 (66/67) | 63 (70/111) |
| 1 month to 17 years | None | Influenza A | DFA | NOW Flu A + B | 69 (72/104) | 99 (567/575) | 90 (72/80) | 95 (567/599) | ||
| 1 month to 17 years | None | Influenza A | Culture and DFA | NOW Flu A + B | 68 (75/111) | 99 (590/596) | 93 (75/81) | 94 (590/626) | ||
| 1 month to 17 years | None | Influenza B | DFA | NOW Flu A + B | 72 (51/71) | 100 (605/608) | 93 (75/81) | 97 (605/625) | ||
| 1 month to 17 years | None | Influenza B | Culture and DFA | NOW Flu A + B | 68 (52/77) | 100 (628/630) | 96 (52/54) | 96 (628/653) | ||
| Heinonen, 201186 | Finland | 1–3 years | None | Influenza A | PCR, culture on MDCK and TR-FIA | Actim Influenza A&B | 90 (28/31) | 99 (127/128) | 97 (28/29) | 98 (127/130) |
| 1–3 years | None | Influenza B | PCR, culture on MDCK and TR-FIA | Actim Influenza A&B | 25 (2/8) | 100 (150/150) | 100 (2/2) | 96 (150/156) | ||
| Herrmann, 2011254 | Sweden | From 2 months | To 83 years | Seasonal | PCR | BioStar OIA Flu A/B |
56.5 (52/92) | 89.1 | ||
| Hurt, 200788 | Australia | From 4 days | To 64 years | Influenza A | RETCIF | Directigen EZ Flu A + B | 69 (34/49) | 100 (128/128) | 100 (34/34) | 90 (128/143) |
| From 4 days | To 64 years | Influenza B | RETCIF | Directigen EZ Flu A + B | 30 (3/10) | 100 (167/167) | 100 (3/3) | 96 (167/174) | ||
| From 4 days | To 64 years | Influenza A | RETCIF | Denka Seiken Quick Ex-Flu | 71 (35/49) | 100 (128/128) | 100 (35/35) | 90 (128/142) | ||
| From 4 days | To 64 years | Influenza B | RETCIF | Denka Seiken Quick Ex-Flu | 30 (3/10) | 100 (167/167) | 100 (3/3) | 96 (167/174) | ||
| From 4 days | To 64 years | Influenza A | RETCIF | Fujirebio Espline Influenza A&B-N | 67 (33/49) | 100 (128/128) | 100 (5/5) | 89 (128/144) | ||
| From 4 days | To 64 years | Influenza B | RETCIF | Fujirebio Espline Influenza A&B-N | 30 (3/10) | 100 (167/167) | 100 (3/3) | 96 (167/174) | ||
| From 4 days | To 64 years | Influenza A | RETCIF | NOW Flu A + B | 73 (36/49) | 99 (127/128) | 97 (36/37) | 91 (127/140) | ||
| From 4 days | To 64 years | Influenza B | RETCIF | NOW Flu A + B | 30 (3/10) | 100 (167/167) | 100 (3/3) | 96 (167/174) | ||
| From 4 days | To 64 years | Influenza A | RETCIF | QuickVue Influenza A + B | 67 (33/49) | 100 (12/128) | 100 (33/33) | 89 (128/144) | ||
| From 4 days | To 64 years | Influenza B | RETCIF | QuickVue Influenza A + B | 30 (3/10) | 100 (167/167) | 100 (3/3) | 96 (167/174) | ||
| From 4 days | To 64 years | Influenza B | RETCIF | Rockeby Influenza A Antigen Test | 10 (5/49) | 100 (128/128) | 100 (5/5) | 74 (128/172) | ||
| Kim, 2010202 | Korea | From 2 months | To 78 years | Pandemic | PCR | SD Bioline Influenza A/B/A(H1N1) Pandemic rapid test | 70 (182/260) | 98 (677/688) | 94 (182/193) | 90 (677/755) |
| Kok, 2010255 | Australia | Included, age not specified | Included, age not specified | Pandemic | PCR | QuickVue Influenza A + B | 53.4 (93/174) | 100 | 100 | 76 |
| Included, age not specified | Included, age not specified | Influenza A (H3N2) | PCR | QuickVue Influenza A + B | 77.2 (68/88) | 100 | 100 | 92 | ||
| Included, age not specified | Included, age not specified | Influenza A (H1N2) | PCR | QuickVue Influenza A + B | 74.2 (72/97) | 100 | 100 | 90 | ||
| Landry, 2008256 | USA | From 4 months | to 93 years | Seasonal | PCR | NOW Flu A + B | 53 (70/132) | 98 | 97 | 63 |
| Lee, 2010257 | Korea | Yang, 2011 | Taiwan | Pandemic | PCR | NanoSign Influenza A/B Ag | 79.4 (158/199) | 97 (801/824) | 87 (158/181) | 95 (801/842) |
| Leveque, 2011258 | France | Included, age not specified | Included, age not specified | Pandemic | PCR | SD Bioline Influenza A/B/A(H1N1) Pandemic rapid test | 39.3 (22/56) | 100 | 100 | 59.5 |
| Louie, 2010259 | USA | From < 1 year | To 80 years | Pandemic | PCR | QuickVue Influenza A + B | 66 (266/404) | 84 (250/299) | 84 (266/315) | 64 (250/388) |
| Lucas, 2011198 | USA | From < 5 years | To > 60 years | Influenza A | PCR | QuickVue Influenza A + B | 15 (2/13) | 99 (1464/1479) | 12 (2/17) | 99 (1464/1475) |
| From < 5 years | To > 60 years | Influenza B | PCR | QuickVue Influenza A + B | 31 (4/13) | 99 (610/615) | 44 (4/9) | 98 (610/619) | ||
| From < 5 years | To > 60 years | Pandemic | PCR | QuickVue Influenza A + B | 20 (6/30) | 99 (1464/1479) | 29 (6/21) | 98 (1076/1096) | ||
| Mehlmann, 2007149 | USA | From 3 months | To 86 years | Seasonal | Culture (Rmix) | QuickVue Influenza A + B | 93 (53/57) | 100 (49/49) | 100 (53/53) | 92 (49/53) |
| From 3 months | To 86 years | Seasonal | PCR | QuickVue Influenza A + B | 87 (53/61) | 97 | 98 | 92 | ||
| Nilsson, 200883 | Sweden | None | 19–86 years | Seasonal | PCR | NOW Flu A + B | 52 (63/120) | 100 | 73 | |
| Nougairede, 2010183 | France | Included, age not specified | Included, age not specified | Pandemic | PCR | Directigen EZ Flu A + B | 57.7 (64/111) | 100 (1910/1910) | 100 (64/64) | 97.5 (1910/1957) |
| Poehling, 2002150 | USA | 3 months to 18 years | None | Seasonal | PCR and culture on RMK cells | QuickVue Influenza A + B | 73.7 (14/19) | 98 | 74 | 98 |
| Poehling, 2006260 | 0–59 months | None | Seasonal | PCR and culture on RMK cells | QuickVue Influenza A + B | 82 (42/51) | 99 | 98 | 94 | |
| Pongthanapisith, 2011261 | Thailand | 6 months to 15 years | None | Pandemic and seasonal | PCR | Osom Influenza A&B | 69.5 (114/164) | 100 (46/46) | 100 (114/114) | 48 (46/96) |
| 6 months to 15 years | None | Pandemic and seasonal | PCR | QuickVue Influenza A + B | 58.5 (96/164) | 100 (46/46) | 100 (96/96) | 40 (46/114) | ||
| 6 months to 15 years | None | Pandemic and seasonal | PCR | SD Bioline Influenza A/B/A(H1N1) Pandemic rapid test | 45.1 (74/164) | 100 (46/46) | 100 (74/74) | 34 (46/136) | ||
| Pregliasco, 2004262 | Italy | 0–14 years | None | Seasonal | Culture on MDCK cells | QuickVue Influenza A + B | 54.5 (6/11) | 98.5 (323/328) | 54.5 (6/11) | 98.5 (323/328) |
| 0–14 years | None | Seasonal | PCR | QuickVue Influenza A + B | 58.3 (7/12) | 98.8 (323/327) | 63.6 (7/11) | 98.5 (323/328) | ||
| Quach, 2002199 | Canada | Included, age not specified | None | Seasonal | Culture on RMK and MDCK cells | QuickVue Influenza A + B | 79.2 (42/53) | 82.6 (204/247) | 49.4 (42/85) | 94.9 (204/215) |
| Rahman, 2007196 | USA | From 6 months | To ≥ 65 years | Seasonal | Culture on RMK cells | Directigen EZ Flu A + B | 42 (18/43) | 96 (72/75) | 86 (18/21) | 74 (72/97) |
| Rahman, 2008151 | USA | From 6 months | To ≥ 65 years | Seasonal | PCR and culture on RMK cells | NOW Flu A + B | 61 (11/18) | 100 (55/55) | 100 (11/11) | 89 (55/62) |
| Rashid, 2007182 | UK and Saudi Arabia | From 1 year | To 85 years | Seasonal | PCR | QuickVue Influenza A + B | 22 (13/58) | 99 (492/497) | 72 (13/18) | 92 (492/537) |
| Rodriguez, 2002185 | USA | From < 3 years | To > 21 years | Seasonal | Culture and DFA | BioStar OIA Flu A/B | 93 (54/58) | 82 (47/57) | 84 (54/64) | 92 (47/51) |
| From < 3 years | To > 21 years | Seasonal | Culture and DFA | Directigen EZ Flu A + B | 95 (55/58) | 84 (49/58) | 86 (55/64) | 94 (49/52) | ||
| From < 3 years | To > 21 years | Seasonal | Culture and DFA | QuickVue Influenza A + B | 95 (54/57) | 76 (42/55) | 81 (54/67) | 93 (42/45) | ||
| From < 3 years | To > 21 years | Seasonal | Culture and DFA | Zstat Flu AB | 72 (41/57) | 83 (48/58) | 80 (41/51) | 75 (48/64) | ||
| Rouleau, 2009263 | Canada | From < 1 year | To 82 years | Seasonal | PCR | QuickVue Influenza A + B | 19.5 (52/267) | 99.1 (219/221) | 96 (52/54) | 51 (219/434) |
| Ruest, 2003152 | Canada | From 1 day | To 98 years | Seasonal | Culture on MDCK cells | Directigen Flu A + B | 86 (57/66) | 94 (115/122) | 89 (57/64) | 92 (115/124) |
| From 1 day | To 98 years | Seasonal | PCR | Directigen Flu A + B | 80 (63/79) | 98 (121/123) | 97 (63/65) | 88 (121/137) | ||
| From 1 day | To 98 years | Seasonal | Culture on MDCK cells | QuickVue Influenza A + B | 91 (64/70) | 86 (105/122) | 79 (64/81) | 95 (105/111) | ||
| From 1 day | To 98 years | Seasonal | PCR | QuickVue Influenza A + B | 85 (72/84) | 92 (127/138) | 87 (72/83) | 90 (114/127) | ||
| Sabetta, 2009186 | USA | Included, age not specified | Included, age not specified | Pandemic | PCR | XPECT Flu A + B | 47 (23/49) | 86 (12/14) | 92 (23/25) | 32 (12/38) |
| Scansen, 201090 | USA | ≤ 17 years | None | Influenza A | PCR | QuickVue Influenza A + B | Swabs: | Swabs: | Swabs: | Swabs: |
| Foam 81 (30/37) | Foam 98 (62/63) | Foam 97 (30/31) | Foam 90 (81/90) | |||||||
| Flocked 59 (22/37) | Flocked 98 (62/63) | Flocked 96 (22/23) | Flocked 81 (62/77) | |||||||
| ≤ 17 years | None | Influenza B | PCR | QuickVue Influenza A + B | Swabs: | Swabs: | Swabs: | Swabs: | ||
| Foam 53 (10/19) | Foam 100 (81/81) | Foam 100 (10/10) | Foam 90 (81/90) | |||||||
| Flocked 42 (8/19) | Flocked 100 (81/81) | Flocked 100 (8/8) | Flocked 88 (81/92) | |||||||
| ≤ 17 years | None | Influenza A | Culture: R-Mix and DFA | QuickVue Influenza A + B | Swabs: | Swabs: | Swabs: | Swabs: | ||
| Foam 85 (29/34) | Foam 97 (64/66) | Foam 94 (29/31) | Foam 93 (64/69) | |||||||
| Flocked 65 (22/34) | Flocked 97 (64/66) | Flocked 92 (22/24) | Flocked 84 (64/76) | |||||||
| ≤ 17 years | None | Influenza B | Culture: R-Mix and DFA | QuickVue Influenza A + B | Swabs: | Swabs: | Swabs: | Swabs: |
||
| Foam 60 (9/15) | Foam 99 (84/85) | Foam 90 (9/10) | Foam 93 (84/90) | |||||||
| Flocked 53 (8/15) | Flocked 100 (85/85) | Flocked 100 (8/8) | Flocked 92 (85/92) | |||||||
| Simmerman, 2007153 | Thailand | From 1 month | To 86 years | Seasonal | Culture on MDCK cells | QuickVue Influenza A + B | 77 (158/205) | 96 | 82 | 95 |
| Stripeli, 201084 | Greece | 6 months to 14 years | None | Seasonal | PCR | QuickVue Influenza A + B | 67.5 (27/40) | 96 (169/176) | 79 (27/34) | 93 (169/182) |
| Suntarattiwong, 2010264 | Thailand | 6 months to 2 years | None | Pandemic | PCR | QuickVue Influenza A + B | 76.7 (23/30) | |||
| To 14 years | None | Pandemic | PCR | QuickVue Influenza A + B | 62.7 (89/142) | 99.2 (235/237) | 97.8 (89/91) | 81.6 (235/288) | ||
| To 14 years | None | Seasonal | PCR | QuickVue Influenza A + B | 69.2 (27/39) | |||||
| Shoji, 2009188 | Japan | From 1 year | To 88 years | Influenza A | Culture | QuickVue Influenza A + B | 87.8 (72/82) | 90.1 (322/357) | 67.3 (72/107) | 97 (322/332) |
| Influenza B | Culture | QuickVue Influenza A + B | 80.4 (176/219) | 95 (209/220) | 94.1 (176/187) | 83 (209/252) | ||||
| Tsao, 2011265 | Taiwan | Included, age not specified | Included, age not specified | Pandemic | PCR | Formosa One Sure Flu A/B Rapid Test | 53.2 (33/62) | |||
| Included, age not specified | Included, age not specified | Pandemic | PCR | QuickVue Influenza A + B | 45.2 (28/62) | 100 | ||||
| Vasoo, 200985 | USA | From 7 months | To 58 years | Pandemic | PCR | Directigen EZ Flu A + B | 46.7 (28/60) | 100 | 100 | 89.6 |
| From 7 months | To 58 years | Pandemic | PCR | NOW Flu A + B | 38.3 (23/60) | 100 | 100 | 88.2 | ||
| From 7 months | To 58 years | Pandemic | PCR | QuickVue Influenza A + B | 53.3 (32/60) | 100 | 100 | 90.8 | ||
| Velasco, 2010266 | Philippines | From 6 months | To 73 years | Pandemic | PCR | QuickVue Influenza A + B | 63 (142/226) | 96 (110/114) | 97 (142/146) | 57 (110/194) |
| Waner, 1991194 | USA | From < 2 months | None | Seasonal | Culture on RMK cells | Directigen Flu A | 100 (23/23) | 91.6 (153/167) | 62 (23/37) | 100 (153/153) |
| From < 2 months | None | Seasonal | IFA | Directigen Flu A | 100 (24/24) | 95 (152/160) | 75 (24/32) | 100 (152/152) | ||
| Watanabe, 2009267 | Japan | 1–13 years | None | Seasonal | Culture on MDCK cells | Capilia FluA, B | 76.9 (444/577) | |||
| Watcharaanan, 2010268 | Thailand | From 3 months | To 73 years | Pandemic | PCR | QuickVue Influenza A + B | 75 (9/12) | 80 (41/51) | 47 (9/19) | 93 (41/44) |
| From 3 months | To 73 years | Seasonal | PCR | QuickVue Influenza A + B | 50 (7/14) | 80 (41/51) | 41 (7/17) | 85 (41/48) | ||
| Weitzel, 2007269 | Germany | From 4 years | To 80 years | Seasonal | PCR and culture on MDCK cells | ImmunoCard Stat! Flu A&B | 67 (18/27) | 99 (175/176) | 95 (18/19) | 95 (175/184) |
| Yang, 2011270 | Taiwan | From < 1 year | To > 60 years | Pandemic | PCR | Fujirebio Espline Influenza A&B-N | 54.6 (425/778) | |||
| From < 1 year | To > 60 years | Influenza A H3N2 | PCR | Fujirebio Espline Influenza A&B-N | 72.3 (164/227) | |||||
| Yoo, 2007197 | Korea | 3 months to 15 years | 18.5–81.9 years | Seasonal | Culture (R-mix) | QuickVue Influenza A + B | 46.7 (35/75) | 94.5 (208/220) | 74.5 (35/47) | 83.9 (208/248) |
| 3 months to 15 years | 18.5–81.9 years | Influenza A | Culture (R-mix) | SD Bioline Influenza A/B/A(H1N1) Pandemic rapid test | 61.9 (26/42) | 96.8 (245/253) | 76.5 (26/34) | 93.9 (245/261) | ||
| 3 months to 15 years | 18.5–81.9 years | Influenza B | Culture (R-mix) | SD Bioline Influenza A/B/A(H1N1) Pandemic rapid test | 54.5 (18/33) | 100 (262/262) | 100 (18/18) | 94.6 (262/277) |
Appendix 7 Ease-of-use scores
| Categories | Criteria for categorisation | EoU score | ||
|---|---|---|---|---|
| Test site | Is a POCT | 1 | ||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| Requires purpose-built accommodation, ventilation systems and dedicated space | ||||
| Equipment | No specialised equipment required and is readily transferred between hospital facilities | 1 | ||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| Specialised equipment is essential and non-portable | ||||
| Materials and reagents | Are stable, pre-packaged and/or pre-measured, and require no special handling steps or storage | 1 | ||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| May be labile, requiring special storage or require special handling steps; their preparation requires manual steps, e.g. volumetric measurement | ||||
| Operational steps | Either automatically executed or easily controlled | 1 | ||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| Require close monitoring and control, and may require special preparation, precise temperature control or timing, accurate pipetting or extensive calculations | ||||
| Training, experience and knowledge | Require minimal scientific and technical knowledge and experience; and knowledge to perform test can be obtained through on-job instruction (commensurate with AFC Band 4, or less) | 1 | ||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| Specialised scientific and technical knowledge essential and substantial experience required (commensurate with AFC Band 7, or higher) | ||||
| Calibration and quality control | Calibration is either automatic or not required; quality control materials are included, or readily available, and are stable | 1 | ||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| Calibration and quality control materials, if available, may be labile or unavailable; or technical expertise is required for calibration | ||||
| Interpretation and judgement | Minimal interpretation and judgement are required, and problems require limited interpretation, judgement and decision-making | 1 | ||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| Extensive interpretation and judgement are required; resolution of problems requires extensive interpretation, judgement and decision-making | ||||
| Test system troubleshooting and equipment maintenance | Test system troubleshooting is automatic or self-correcting or clearly described or requires minimal judgement | 1 | ||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| Test system troubleshooting is not automatic or self-correcting and requires decision-making to resolve most problems; or equipment maintenance requires special knowledge, skills and abilities | ||||
| Time to reporting of results | Results are reported within 4 hours of collection of almost all specimens | 1 | ||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| Median time to reporting of results exceeds the median duration of hospitalisations/time to death | ||||
| Health and safety | The test is completed using low levels of personal protection, i.e. gloves | 1 | ||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| One or more operational steps requires bio-safety level Category 3, or higher | ||||
| Storage and disposal of waste test materials and reagents | Waste materials and reagents are stored and disposed using medium duty ‘Clinical waste’ plastic bags and ‘Sharps’ containers, or a sluice | 1 | ||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| Waste materials or reagents include hazardous materials that require special attention | ||||
| Total EoU score | 11 | |||
| Categories | Criteria for categorisation | EoU score | ||
|---|---|---|---|---|
| Test site | Is a POCT | 1 | ||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| Requires purpose-built accommodation, ventilation systems and dedicated space | ||||
| Equipment | No specialised equipment required and is readily transferred between hospital facilities | 1 | ||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| Specialised equipment is essential and non-portable | ||||
| Materials and reagents | Are stable, pre-packaged and/or pre-measured, and require no special handling steps or storage | 1 | ||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| May be labile, requiring special storage or require special handling steps; their preparation requires manual steps, e.g. volumetric measurement | ||||
| Operational steps | Either automatically executed or easily controlled | 1 | ||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| Require close monitoring and control, and may require special preparation, precise temperature control or timing, accurate pipetting or extensive calculations | ||||
| Training, experience and knowledge | Require minimal scientific and technical knowledge and experience; and knowledge to perform test can be obtained through on-job instruction (commensurate with AFC Band 4, or less) | 1 | ||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| Specialised scientific and technical knowledge essential and substantial experience required (commensurate with AFC Band 7, or higher) | ||||
| Calibration and quality control | Calibration is either automatic or not required; quality control materials are included, or readily available, and are stable | 1 | ||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| Calibration and quality control materials, if available, may be labile or unavailable; or technical expertise is required for calibration | ||||
| Interpretation and judgement | Minimal interpretation and judgement are required, and problems require limited interpretation, judgement and decision-making | 1 | ||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| Extensive interpretation and judgement are required; resolution of problems requires extensive interpretation, judgement and decision-making | ||||
| Test system troubleshooting and equipment maintenance | Test system troubleshooting is automatic or self-correcting or clearly described or requires minimal judgement | 1 | ||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| Test system troubleshooting is not automatic or self-correcting and requires decision-making to resolve most problems; or equipment maintenance requires special knowledge, skills and abilities | ||||
| Time to reporting of results | Results are reported within 4 hours of collection of almost all specimens | 1 | ||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| Median time to reporting of results exceeds the median duration of hospitalisations/time to death | ||||
| Health and safety | The test is completed using low levels of personal protection, i.e. gloves | 1 | ||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| One or more operational steps requires bio-safety level Category 3, or higher | ||||
| Storage and disposal of waste test materials and reagents | Waste materials and reagents are stored and disposed using medium duty ‘Clinical waste’ plastic bags and ‘Sharps’ containers, or a sluice | 1 | ||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| Waste materials or reagents include hazardous materials that require special attention | ||||
| Total EoU score | 11 | |||
| Categories | Criteria for categorisation | EoU score | ||
|---|---|---|---|---|
| Test site | Is a POCT | |||
| Intermediate between descriptions listed for scores of 1 and 3 | 2 | |||
| Requires purpose-built accommodation, ventilation systems and dedicated space | ||||
| Equipment | No specialised equipment required and is readily transferred between hospital facilities | |||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| Specialised equipment is essential and non-portable | 3 | |||
| Materials and reagents | Are stable, pre-packaged and/or pre-measured, and require no special handling steps or storage | |||
| Intermediate between descriptions listed for scores of 1 and 3 | 2 | |||
| May be labile, requiring special storage or require special handling steps; their preparation requires manual steps, e.g. volumetric measurement | ||||
| Operational steps | Either automatically executed or easily controlled | |||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| Require close monitoring and control, and may require special preparation, precise temperature control or timing, accurate pipetting or extensive calculations | 3 | |||
| Training, experience and knowledge | Require minimal scientific and technical knowledge and experience; and knowledge to perform test can be obtained through on-job instruction (commensurate with AFC Band 4, or less) | |||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| Specialised scientific and technical knowledge essential and substantial experience required (commensurate with AFC Band 7, or higher) | 3 | |||
| Calibration and quality control | Calibration is either automatic or not required; quality control materials are included, or readily available, and are stable | |||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| Calibration and quality control materials, if available, may be labile or unavailable; or technical expertise is required for calibration | 3 | |||
| Interpretation and judgement | Minimal interpretation and judgement are required, and problems require limited interpretation, judgement and decision-making | |||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| Extensive interpretation and judgement are required; resolution of problems requires extensive interpretation, judgement and decision-making | 3 | |||
| Test system troubleshooting and equipment maintenance | Test system troubleshooting is automatic or self-correcting or clearly described or requires minimal judgement | |||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| Test system troubleshooting is not automatic or self-correcting and requires decision-making to resolve most problems; or equipment maintenance requires special knowledge, skills, and abilities | 3 | |||
| Time to reporting of results | Results are reported within 4 hours of collection of almost all specimens | |||
| Intermediate between descriptions listed for scores of 1 and 3 | 2 | |||
| Median time to reporting of results exceeds the median duration of hospitalisations/time to death | ||||
| Health and safety | The test is completed using low levels of personal protection, i.e. gloves | |||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| One or more operational steps requires bio-safety level Category 3, or higher | 3 | |||
| Storage and disposal of waste test materials and reagents | Waste materials and reagents are stored and disposed using medium duty ‘Clinical waste’ plastic bags and ‘Sharps’ containers, or a sluice | |||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| Waste materials or reagents include hazardous materials that require special attention | 3 | |||
| Total EoU score | 30 | |||
| Categories | Criteria for categorisation | EoU score | ||
|---|---|---|---|---|
| Test site | Is a POCT | |||
| Intermediate between descriptions listed for scores of 1 and 3 | 2 | |||
| Requires purpose-built accommodation, ventilation systems and dedicated space | ||||
| Equipment | No specialised equipment required and is readily transferred between hospital facilities | |||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| Specialised equipment is essential and non-portable | 3 | |||
| Materials and reagents | Are stable, pre-packaged and/or pre-measured, and require no special handling steps or storage | 1 | ||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| May be labile requiring special storage or require special handling steps; their preparation requires manual steps, e.g. volumetric measurement | ||||
| Operational steps | Either automatically executed or easily controlled | |||
| Intermediate between descriptions listed for scores of 1 and 3 | 2 | |||
| Require close monitoring and control, and may require special preparation, precise temperature control or timing, accurate pipetting or extensive calculations | ||||
| Training, experience and knowledge | Require minimal scientific and technical knowledge and experience; and knowledge to perform test can be obtained through on-job instruction (commensurate with AFC Band 4, or less) | |||
| Intermediate between descriptions listed for scores of 1 and 3 | 2 | |||
| Specialised scientific and technical knowledge essential and substantial experience required (commensurate with AFC Band 7, or higher) | ||||
| Calibration and quality control | Calibration is either automatic or not required; quality control materials are included, or readily available, and are stable | |||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| Calibration and quality control materials, if available, may be labile or unavailable; or technical expertise is required for calibration | 3 | |||
| Interpretation and judgement | Minimal interpretation and judgement are required, and problems require limited interpretation, judgement and decision-making | |||
| Intermediate between descriptions listed for scores of 1 and 3 | 2 | |||
| Extensive interpretation and judgement are required; resolution of problems requires extensive interpretation, judgement and decision-making | ||||
| Test system troubleshooting and equipment maintenance | Test system troubleshooting is automatic or self-correcting or clearly described or requires minimal judgement | |||
| Intermediate between descriptions listed for scores of 1 and 3 | 2 | |||
| Test system troubleshooting is not automatic or self-correcting and requires decision-making to resolve most problems; or equipment maintenance requires special knowledge, skills and abilities | ||||
| Time to reporting of results | Results are reported within 4 hours of collection of almost all specimens | |||
| Intermediate between descriptions listed for scores of 1 and 3 | 2 | |||
| Median time to reporting of results exceeds the median duration of hospitalisations/time to death | ||||
| Health and safety | The test is completed using low levels of personal protection, i.e. gloves | |||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| One or more operational steps requires bio-safety level Category 3, or higher | 3 | |||
| Storage and disposal of waste test materials and reagents | Waste materials and reagents are stored and disposed using medium duty ‘Clinical waste’ plastic bags and ‘Sharps’ containers, or a sluice | |||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| Waste materials or reagents include hazardous materials that require special attention | 3 | |||
| Total EoU score | 25 | |||
| Categories | Criteria for categorisation | EoU score | ||
|---|---|---|---|---|
| Test site | Is a POCT | |||
| Intermediate between descriptions listed for scores of 1 and 3 | 2 | |||
| Requires purpose-built accommodation, ventilation systems and dedicated space | ||||
| Equipment | No specialised equipment required and is readily transferred between hospital facilities | |||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| Specialised equipment is essential and non-portable | 3 | |||
| Materials and reagents | Are stable, pre-packaged and/or pre-measured, and require no special handling steps or storage | 1 | ||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| May be labile requiring special storage or require special handling steps; their preparation requires manual steps, e.g. volumetric measurement | ||||
| Operational steps | Either automatically executed or easily controlled | |||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| Require close monitoring and control, and may require special preparation, precise temperature control or timing, accurate pipetting or extensive calculations | 3 | |||
| Training, experience and knowledge | Require minimal scientific and technical knowledge and experience; and knowledge to perform test can be obtained through on-job instruction (commensurate with AFC Band 4, or less) | |||
| Intermediate between descriptions listed for scores of 1 and 3 | 2 | |||
| Specialised scientific and technical knowledge essential and substantial experience required (commensurate with AFC Band 7, or higher) | ||||
| Calibration and quality control | Calibration is either automatic or not required; quality control materials are included, or readily available, and are stable | |||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| Calibration and quality control materials, if available, may be labile or unavailable; or technical expertise is required for calibration | 3 | |||
| Interpretation and judgement | Minimal interpretation and judgement are required, and problems require limited interpretation, judgement and decision-making | |||
| Intermediate between descriptions listed for scores of 1 and 3 | 2 | |||
| Extensive interpretation and judgement are required; resolution of problems requires extensive interpretation, judgement and decision-making | ||||
| Test system troubleshooting and equipment maintenance | Test system troubleshooting is automatic or self-correcting or clearly described or requires minimal judgement | |||
| Intermediate between descriptions listed for scores of 1 and 3 | 2 | |||
| Test system troubleshooting is not automatic or self-correcting and requires decision-making to resolve most problems; or equipment maintenance requires special knowledge, skills and abilities | ||||
| Time to reporting of results | Results are reported within 4 hours of collection of almost all specimens | |||
| Intermediate between descriptions listed for scores of 1 and 3 | 2 | |||
| Median time to reporting of results exceeds the median duration of hospitalisations/time to death | ||||
| Health and safety | The test is completed using low levels of personal protection, i.e. gloves | |||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| One or more operational steps requires bio-safety level Category 3, or higher | 3 | |||
| Storage and disposal of waste test materials and reagents | Waste materials and reagents are stored and disposed using medium duty ‘Clinical waste’ plastic bags and ‘Sharps’ containers, or a sluice | |||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| Waste materials or reagents include hazardous materials that require special attention | 3 | |||
| Total EoU score | 26 | |||
| Categories | Criteria for categorisation | EoU score | ||
|---|---|---|---|---|
| Test site | Is a POCT | |||
| Intermediate between descriptions listed for scores of 1 and 3 | 2 | |||
| Requires purpose-built accommodation, ventilation systems and dedicated space | ||||
| Equipment | No specialised equipment required and is readily transferred between hospital facilities | |||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| Specialised equipment is essential and non-portable | 3 | |||
| Materials and reagents | Are stable, pre-packaged and/or pre-measured, and require no special handling steps or storage | |||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| May be labile requiring special storage or require special handling steps; their preparation requires manual steps, e.g. volumetric measurement | 3 | |||
| Operational steps | Either automatically executed or easily controlled | |||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| Require close monitoring and control, and may require special preparation, precise temperature control or timing, accurate pipetting or extensive calculations | 3 | |||
| Training, experience and knowledge | Require minimal scientific and technical knowledge and experience; and knowledge to perform test can be obtained through on-job instruction (commensurate with AFC Band 4, or less) | |||
| Intermediate between descriptions listed for scores of 1 and 3 | 2 | |||
| Specialised scientific and technical knowledge essential and substantial experience required (commensurate with AFC Band 7, or higher) | ||||
| Calibration and quality control | Calibration is either automatic or not required; quality control materials are included, or readily available, and are stable | 1 | ||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| Calibration and quality control materials, if available, may be labile or unavailable; or technical expertise is required for calibration | ||||
| Interpretation and judgement | Minimal interpretation and judgement are required, and problems require limited interpretation, judgement and decision-making | |||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| Extensive interpretation and judgement are required; resolution of problems requires extensive interpretation, judgement and decision-making | 3 | |||
| Test system troubleshooting and equipment maintenance | Test system troubleshooting is automatic or self-correcting or clearly described or requires minimal judgement | |||
| Intermediate between descriptions listed for scores of 1 and 3 | 2 | |||
| Test system troubleshooting is not automatic or self-correcting and requires decision-making to resolve most problems; or equipment maintenance requires special knowledge, skills and abilities | ||||
| Time to reporting of results | Results are reported within 4 hours of collection of almost all specimens | |||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| Median time to reporting of results exceeds the median duration of hospitalisations/time to death | 3 | |||
| Health and safety | The test is completed using low levels of personal protection, i.e. gloves | |||
| Intermediate between descriptions listed for scores of 1 and 3 | 2 | |||
| One or more operational steps requires bio-safety level Category 3, or higher | ||||
| Storage and disposal of waste test materials and reagents | Waste materials and reagents are stored and disposed using medium duty ‘Clinical waste’ plastic bags and ‘Sharps’ containers, or a sluice | |||
| Intermediate between descriptions listed for scores of 1 and 3 | 2 | |||
| Waste materials or reagents include hazardous materials that require special attention | ||||
| Total EoU score | 26 | |||
| Categories | Criteria for categorisation | EoU score | ||
|---|---|---|---|---|
| Test site | Is a POCT | |||
| Intermediate between descriptions listed for scores of 1 and 3 | 2 | |||
| Requires purpose-built accommodation, ventilation systems and dedicated space | ||||
| Equipment | No specialised equipment required and is readily transferred between hospital facilities | |||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| Specialised equipment is essential and non-portable | 3 | |||
| Materials and reagents | Are stable, pre-packaged and/or pre-measured, and require no special handling steps or storage | 1 | ||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| May be labile requiring special storage or require special handling steps; their preparation requires manual steps, e.g. volumetric measurement | ||||
| Operational steps | Either automatically executed or easily controlled | 1 | ||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| Require close monitoring and control, and may require special preparation, precise temperature control or timing, accurate pipetting or extensive calculations | ||||
| Training, experience and knowledge | Require minimal scientific and technical knowledge and experience; and knowledge to perform test can be obtained through on-job instruction (commensurate with AFC Band 4, or less) | |||
| Intermediate between descriptions listed for scores of 1 and 3 | 2 | |||
| Specialised scientific and technical knowledge essential and substantial experience required (commensurate with AFC Band 7, or higher) | ||||
| Calibration and quality control | Calibration is either automatic or not required; quality control materials are included, or readily available, and are stable | |||
| Intermediate between descriptions listed for scores of 1 and 3 | 2 | |||
| Calibration and quality control materials, if available, may be labile or unavailable; or technical expertise is required for calibration | ||||
| Interpretation and judgement | Minimal interpretation and judgement are required, and problems require limited interpretation, judgement and decision-making | |||
| Intermediate between descriptions listed for scores of 1 and 3 | 2 | |||
| Extensive interpretation and judgement are required; resolution of problems requires extensive interpretation, judgement and decision-making | ||||
| Test system troubleshooting and equipment maintenance | Test system troubleshooting is automatic or self-correcting or clearly described or requires minimal judgement | 1 | ||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| Test system troubleshooting is not automatic or self-correcting and requires decision-making to resolve most problems; or equipment maintenance requires special knowledge, skills and abilities | ||||
| Time to reporting of results | Results are reported within 4 hours of collection of almost all specimens | |||
| Intermediate between descriptions listed for scores of 1 and 3 | 2 | |||
| Median time to reporting of results exceeds the median duration of hospitalisations/time to death | ||||
| Health and safety | The test is completed using low levels of personal protection, i.e. gloves | |||
| Intermediate between descriptions listed for scores of 1 and 3 | 2 | |||
| One or more operational steps requires bio-safety level Category 3, or higher | ||||
| Storage and disposal of waste test materials and reagents | Waste materials and reagents are stored and disposed using medium duty ‘Clinical waste’ plastic bags and ‘Sharps’ containers, or a sluice | |||
| Intermediate between descriptions listed for scores of 1 and 3 | 2 | |||
| Waste materials or reagents include hazardous materials that require special attention | ||||
| Total EoU score | 20 | |||
| Categories | Criteria for categorisation | EoU score | ||
|---|---|---|---|---|
| Test site | Is a POCT | |||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| Requires purpose-built accommodation, ventilation systems and dedicated space | 3 | |||
| Equipment | No specialised equipment required and is readily transferred between hospital facilities | |||
| Intermediate between descriptions listed for scores of 1 and 3 | 2 | |||
| Specialised equipment is essential and non-portable | ||||
| Materials and reagents | Are stable, pre-packaged and/or pre-measured, and require no special handling steps or storage | |||
| Intermediate between descriptions listed for scores of 1 and 3 | 2 | |||
| May be labile requiring special storage or require special handling steps; their preparation requires manual steps, e.g. volumetric measurement | ||||
| Operational steps | Either automatically executed or easily controlled | 1 | ||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| Require close monitoring and control, and may require special preparation, precise temperature control or timing, accurate pipetting or extensive calculations | ||||
| Training, experience and knowledge | Require minimal scientific and technical knowledge and experience; and knowledge to perform test can be obtained through on-job instruction (commensurate with AFC Band 4, or less) | |||
| Intermediate between descriptions listed for scores of 1 and 3 | 2 | |||
| Specialised scientific and technical knowledge essential and substantial experience required (commensurate with AFC Band 7, or higher) | ||||
| Calibration and quality control | Calibration is either automatic or not required; quality control materials are included, or readily available, and are stable | 1 | ||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| Calibration and quality control materials, if available, may be labile or unavailable; or technical expertise is required for calibration | ||||
| Interpretation and judgement | Minimal interpretation and judgement are required, and problems require limited interpretation, judgement and decision-making | |||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| Extensive interpretation and judgement are required; resolution of problems requires extensive interpretation, judgement and decision-making | 3 | |||
| Test system troubleshooting and equipment maintenance | Test system troubleshooting is automatic or self-correcting or clearly described or requires minimal judgement | |||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| Test system troubleshooting is not automatic or self-correcting and requires decision-making to resolve most problems; or equipment maintenance requires special knowledge, skills and abilities | 3 | |||
| Time to reporting of results | Results are reported within 4 hours of collection of almost all specimens | |||
| Intermediate between descriptions listed for scores of 1 and 3 | 2 | |||
| Median time to reporting of results exceeds the median duration of hospitalisations/time to death | ||||
| Health and safety | The test is completed using low levels of personal protection, i.e. gloves | |||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| One or more operational steps requires bio-safety level Category 3, or higher | 3 | |||
| Storage and disposal of waste test materials and reagents | Waste materials and reagents are stored and disposed using medium duty ‘Clinical waste’ plastic bags and ‘Sharps’ containers, or a sluice | |||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| Waste materials or reagents include hazardous materials that require special attention | 3 | |||
| Total EoU score | 25 | |||
| Categories | Criteria for categorisation | EoU score | ||
|---|---|---|---|---|
| Test site | Is a POCT | |||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| Requires purpose-built accommodation, ventilation systems and dedicated space | 3 | |||
| Equipment | No specialised equipment required and is readily transferred between hospital facilities | |||
| Intermediate between descriptions listed for scores of 1 and 3 | 2 | |||
| Specialised equipment is essential and non-portable | ||||
| Materials and reagents | Are stable, pre-packaged and/or pre-measured, and require no special handling steps or storage | |||
| Intermediate between descriptions listed for scores of 1 and 3 | 2 | |||
| May be labile requiring special storage or require special handling steps; their preparation requires manual steps, e.g. volumetric measurement | ||||
| Operational steps | Either automatically executed or easily controlled | |||
| Intermediate between descriptions listed for scores of 1 and 3 | 2 | |||
| Require close monitoring and control, and may require special preparation, precise temperature control or timing, accurate pipetting or extensive calculations | ||||
| Training, experience and knowledge | Require minimal scientific and technical knowledge and experience; and knowledge to perform test can be obtained through on-job instruction (commensurate with AFC Band 4, or less) | |||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| Specialised scientific and technical knowledge essential and substantial experience required (commensurate with AFC Band 7, or higher) | 3 | |||
| Calibration and quality control | Calibration is either automatic or not required; quality control materials are included, or readily available, and are stable | 1 | ||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| Calibration and quality control materials, if available, may be labile or unavailable; or technical expertise is required for calibration | ||||
| Interpretation and judgement | Minimal interpretation and judgement are required, and problems require limited interpretation, judgement and decision-making | |||
| Intermediate between descriptions listed for scores of 1 and 3 | 2 | |||
| Extensive interpretation and judgement are required; resolution of problems requires extensive interpretation, judgement and decision-making | ||||
| Test system troubleshooting and equipment maintenance | Test system troubleshooting is automatic or self-correcting or clearly described or requires minimal judgement | |||
| Intermediate between descriptions listed for scores of 1 and 3 | 2 | |||
| Test system troubleshooting is not automatic or self-correcting and requires decision-making to resolve most problems; or equipment maintenance requires special knowledge, skills and abilities | ||||
| Time to reporting of results | Results are reported within 4 hours of collection of almost all specimens | |||
| Intermediate between descriptions listed for scores of 1 and 3 | 2 | |||
| Median time to reporting of results exceeds the median duration of hospitalisations/time to death | ||||
| Health and safety | The test is completed using low levels of personal protection, i.e. gloves | |||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| One or more operational steps requires bio-safety level Category 3, or higher | 3 | |||
| Storage and disposal of waste test materials and reagents | Waste materials and reagents are stored and disposed using medium duty ‘Clinical waste’ plastic bags and ‘Sharps’ containers, or a sluice | 1 | ||
| Intermediate between descriptions listed for scores of 1 and 3 | ||||
| Waste materials or reagents include hazardous materials that require special attention | ||||
| Total EoU score | 23 | |||
Appendix 8 Sample of WinBUGS code used
A. WinBUGS code (model A)
Model
Data – original scale
Specifying prior distributions for Wishart distribution
B. Markov chain Monte Carlo sensitivity analyses
Table 47 displays the results of various sensitivity analyses exploring the impact of length of ‘burn-in’ and sample, different initial/starting values, and changing the prior distributions used in terms of their impact on the posterior mean costs and QALYs for the three patient groups.
| Parameter | Traditional | PCR | POCT |
|---|---|---|---|
| Base case (model A) | |||
| Costs (£) | 2327 (1989 to 2664) | 1978 (1743 to 2165) | 2159 (1828 to 2485) |
| QALYs | 0.007588 (0.006334 to 0.008854) | 0.007779 (0.006555 to 0.008983) | 0.008035 (0.006772 to 0.009280) |
| Burn-in/sample | |||
| 10k/20k | |||
| Costs (£) | 2327 (1992 to 2661) | 1977 (1739 to 2216) | 2160 (1831 to 2491) |
| QALYs | 0.007596 (0.006337 to 0.008882) | 0.007776 (0.006553 to 0.008990) | 0.008030 (0.006786 to 0.009286) |
| 50k/100k | |||
| Costs (£) | 2326 (1991 to 2663) | 1977 (1741 to 2215) | 2160 (1833 to 2487) |
| QALYs | 0.007599 (0.006330 to 0.008877) | 0.007776 (0.006563 to 0.008988) | 0.008033 (0.006789 to 0.009281) |
| Alternative priors | |||
| Costs (£) | 1786 (1478 to 2091) | 1724 (1500 to 1947) | 1682 (1384 to 1979) |
| QALYs | 0.007322 (0.004132 to 0.010550) | 0.007682 (0.004706 to 0.010670) | 0.007853 (0.004968 to 0.010750) |
| Alternative starting values | |||
| Chain 1 | |||
| Costs (£) | 2327 (1994 to 2664) | 1977 (1739 to 2213) | 2159 (1834 to 2485) |
| QALYs | 0.007597 (0.006310 to 0.008866) | 0.007771 (0.006557 to 0.008987) | 0.008037 (0.006787 to 0.009286) |
| Chain 2 | |||
| Costs (£) | 2327 (1989 to 2666) | 1977 (1741 to 2215) | 2160 (1837 to 2485) |
| QALYs | 0.007598 (0.006326 to 0.008882) | 0.007770 (0.006568 to 0.008983) | 0.008035 (0.006776 to 0.009284) |
| Chain 3 | |||
| Costs (£) | 2326 (1990 to 2662) | 1978 (1739 to 2216) | 2161 (1835 to 2487) |
| QALYs | 0.007602 (0.006330 to 0.008873) | 0.007783 (0.006565 to 0.009009) | 0.008039 (0.006793 to 0.009295) |
FIGURE 23.
Markov chain Monte Carlo history plots for mean costs and QALYs.

FIGURE 24.
Brooks–Gelman–Rubin plots for chains 1, 2 and 3, representing three different sets of starting/initial values.

FIGURE 25.
Markov chain Monte Carlo autocorrelation plots for mean costs and QALYs.

C. Alternative model (model B) – WinBUGS and R code
WinBUGS code
Model
Data
Starting values
R code
Matrices and (which are on the logit(QALY) scale) are obtained from model above using files and .
List of abbreviations
- 3WS
- Three Winters Study
- A&E
- accident and emergency
- AFC
- Agenda for Change
- ANOVA
- analysis of variance
- ARI
- acute respiratory infection
- AUC
- area under the curve
- BHQ
- black hole quencher
- BMI
- body mass index
- BTS
- British Thoracic Society
- CAP
- community-acquired pneumonia
- cDNA
- complementary deoxyribonucleic acid
- CEAC
- cost-effectiveness acceptability curve
- CI
- confidence interval
- CLIA
- Clinical Laboratory Improvement Amendments
- COPD
- chronic obstructive pulmonary disease
- CPAP
- continuous positive airway pressure
- CPE
- cytopathic effect
- CRF
- case report form
- CrI
- credible interval
- DH
- Department of Health
- DNAse
- deoxyribonuclease
- dNTP
- deoxynucleotide triphosphate
- EoU
- ease of use
- EQ-5D
- European Quality of Life-5 Dimensions
- EuroQoL
- European quality of life
- FDA
- US Food and Drug Administration
- GP
- general medical practitioner
- HA
- haemagglutinin
- HAd
- haemadsorption
- HAI
- haemagglutination inhibition
- hMPV
- human metapneumovirus
- HPA
- Health Protection Agency
- HR
- hazard ratio
- HRG
- Healthcare Resource Group
- HRQoL
- health-related quality of life
- HSV
- herpes simplex virus
- ICD-10
- International Classification of Diseases, Tenth Edition
- ICER
- incremental cost-effectiveness ratio
- IF
- immunofluorescence
- ILI
- influenza-like illness
- IQR
- interquartile range
- ITT
- intention to treat
- ITU
- intensive treatment unit
- i.v.
- intravenous
- MAU
- Medical Admissions Unit
- MCMC
- Markov chain Monte Carlo
- MCSD
- minimum clinically significant difference
- MeSH
- medical subject heading
- MGB
- minor groove binder
- MMLV
- Moloney murine leukaemia virus
- MRC-5
- Medical Research Council 5 cells
- MRSA
- methicillin-resistant Staphylococcus aureus
- NA
- not applicable
- NI
- neuraminidase inhibitor
- NICE
- National Institute for Health and Care Excellence
- NPT
- near-patient test
- NPV
- negative predictive value
- PCR
- polymerase chain reaction
- PLC/PRF5
- primary liver cells
- POC
- point of care
- POCT
- point-of-care test
- PPV
- positive predictive value
- PRISMA
- Preferred Reporting Items for Systematic Reviews and Meta-Analyses
- PSSRU
- Personal Social Services Research Unit
- QALY
- quality-adjusted life-year
- QoL
- quality of life
- QUADAS
- quality assessment tool for diagnostic accuracy studies
- RCT
- randomised controlled trial
- RETCIF
- rapid enhanced tissue culture immunofluorescence
- RFT
- rapid flu test
- RMK
- Rhesus monkey kidney
- RNA
- ribonucleic acid
- RNAse
- ribonuclease
- ROC
- receiver operating characteristic
- RSV
- respiratory syncytial virus
- RT
- reverse transcriptase
- RT-PCR
- reverse transcriptase-polymerase chain reaction
- SBCMV
- soil-borne cereal mosaic virus
- SD
- standard deviation
- SE
- standard error
- SROC
- summary receiver operating characteristic
- STARD
- Standards for Reporting Diagnostic Accuracy
- TS
- total score
- TtR
- time to reporting
- UHL
- University Hospitals of Leicester
- VSOP
- virology standard operating procedure
- VTM
- virus transport medium