Notes
Article history
The research reported in this issue of the journal was commissioned and funded by the HTA programme on behalf of NICE as project number 12/75/01. The protocol was agreed in January 2013. The assessment report began editorial review in July 2013 and was accepted for publication in November 2013. The authors have been wholly responsible for all data collection, analysis and interpretation, and for writing up their work. The HTA editors and publisher have tried to ensure the accuracy of the authors’ report and would like to thank the reviewers for their constructive comments on the draft document. However, they do not accept liability for damages or losses arising from material published in this report.
Declared competing interests of authors
none
Permissions
Copyright statement
© Queen’s Printer and Controller of HMSO 2014. This work was produced by Westwood et al. under the terms of a commissioning contract issued by the Secretary of State for Health. This issue may be freely reproduced for the purposes of private research and study and extracts (or indeed, the full report) may be included in professional journals provided that suitable acknowledgement is made and the reproduction is not associated with any form of advertising. Applications for commercial reproduction should be addressed to: NIHR Journals Library, National Institute for Health Research, Evaluation, Trials and Studies Coordinating Centre, Alpha House, University of Southampton Science Park, Southampton SO16 7NS, UK.
Chapter 1 Objective
The overall objective of this project was to summarise the evidence on the clinical effectiveness and cost-effectiveness of Kirsten rat sarcoma viral oncogene (KRAS) mutation tests (commercial or in-house) for the differentiation of adults with metastatic colorectal cancer (mCRC) whose metastases are confined to the liver and are unresectable and who may benefit from first-line treatment with cetuximab in combination with standard chemotherapy from those who should receive standard chemotherapy alone, as recommended in the National Institute for Health and Care Excellence (NICE) technology appraisal TA176. 1 To address clinical effectiveness, data on the clinical validity of the different KRAS mutation tests (sensitivity/specificity for detection of mutations known to be linked to insensitivity to cetuximab) are required. Because methods of testing KRAS mutation status differ in terms of both the mutations targeted and the limit of detection (the lowest proportion of tumour cells with a mutation that can be detected), the definitions of KRAS mutant and KRAS wild type vary according to which test is used. All testing methods are essentially reference standard methods for classifying mutation status, as defined by the specific test characteristics, and it is therefore not useful to select any particular test as the reference standard. In addition, the relationship between insensitivity to cetuximab and the presence of specific mutations or combinations of mutations, as well as the relationship between insensitivity to cetuximab and the level of mutation present, are uncertain. Therefore, the following research questions were formulated to address the review objectives:
-
What is the technical performance of the different KRAS mutation tests {e.g. proportion of tumour cells needed, limit of detection [minimum percentage mutation detectable against a background of wild-type deoxyribonucleic acid (DNA)], failures, costs, turnaround time}?
-
What is the accuracy (clinical validity) of KRAS mutation testing, using any test, for predicting response to treatment with cetuximab in combination with standard chemotherapy?
-
How do clinical outcomes from treatment with cetuximab in combination with standard chemotherapy and, when reported, from treatment with standard chemotherapy alone vary according to which test is used to select patients for treatment?
-
What is the cost-effectiveness of the use of the different KRAS mutation tests to decide between standard chemotherapy or cetuximab in combination with standard chemotherapy?
First-line chemotherapy of unresectable colorectal liver metastases seeks to achieve a tumour response such that the tumour is judged to be resectable. For this reason, resection rate is considered the ideal reference standard for question 2 and the optimal outcome measure for question 3.
Chapter 2 Background and definition of the decision problem
Population
The indication for this assessment is the detection of mutations in the KRAS oncogene in adults with mCRC, in whom metastases are confined to the liver and are unresectable. The presence or absence of KRAS mutations can affect the choice of first-line chemotherapy in these patients and mutation testing is used to direct the treatment pathway. 1
The 2010 cancer registration data from the Office for National Statistics2 showed that colorectal cancer (CRC) was the third most common cancer in both men and women, accounting for approximately 13% of all new cancer cases. The 2010 age-standardised incidence rate for CRC in England was 56.5 per 100,000 in men and 36.1 per 100,000 in women and this has remained constant for both sexes over the last 10 years. 2 In 2009 there were approximately 36,000 new cases of CRC recorded in England and Wales,3 and in 2010 there were 14,691 recorded deaths from CRC in England and Wales, accounting for around 10% of all cancer deaths. 4 Age-standardised 5-year survival rates for CRC in England (2005–9) were 54.2% for men and 55.6% for women. 5 Approximately two-thirds of CRC cases (64% in 2009) are cancers of the colon and one-third (36%) are rectal (including the anus). Most (60%) rectal cancer cases occur in men and colon cancer cases are evenly distributed between the sexes. 3 CRC incidence is strongly related to age, with incidence rates increasing from age 50 years and peaking in the over 80s; in the UK (2007–9), 72% of new cases were diagnosed in people aged > 65 years. 3 There is some evidence in UK men of an association between incidence of CRC and deprivation; 2000–4 data show incidence rates approximately 11% higher for men living in more deprived areas than for men living in the least deprived areas. 6 National Bowel Cancer Audit data for 2011 included 28,260 new cases in England and Wales of which 21,306 (75.4%) were surgically treated; 3425 (16.1%) of these had confirmed liver metastases. 7 Reported estimates of the prevalence of KRAS mutations in codons 12 and 13 in the tumours of patients with mCRC range from 35% to 42%8–10 and are similar (approximately 36%) when samples taken from metastases are considered separately. 8,9 The three most common mutations, G12D, G12V and G13D, account for approximately 75% of all KRAS mutations. 8 Because not all patients whose tumours are wild type for KRAS codons 12 and 13 respond to treatment with epidermal growth factor receptor (EGFR)-inhibiting monoclonal antibodies, the potential effects of mutations in codons 61 and 146 of KRAS have also been investigated. A US study,11 which found KRAS codon 12 or 13 mutations in 900/2121 (42.4%) CRC patients, conducted further analysis of the 513 wild-type samples and found 19 additional mutations in KRAS codon 61 and 17 in KRAS codon 146; these additional mutations represent < 2% of the total study population.
Intervention technologies
There are a variety of tests available for KRAS mutation testing (Table 1) in NHS reference laboratories currently providing testing [laboratories participating in the UK National External Quality Assurance Scheme (NEQAS)]. The tests used can be broadly divided into two subgroups: mutation screening and targeted mutation detection. Mutation screening tests screen samples for all KRAS mutations (known and novel) whereas targeted tests analyse samples for specific known mutations. Successful mutation analysis is dependent on adequate sample quality and a sufficient quantity of tumour tissue in the sample. The sample requirements vary between test methods, with some (e.g. Sanger sequencing) requiring up to 25% tumour cells. The limit of detection (the percentage of mutation detectable in a tumour sample against a background of wild-type DNA) may also vary between different test methods, with some studies reporting mutation detection at as little as 1% against a background of wild-type DNA (see Table 1). This is an important issue as it is unclear whether detecting diminishingly small proportions of mutation is clinically useful – should patients with very low proportions of mutation be treated as mutant or wild type? There is some evidence that the results of KRAS mutation testing in plasma samples correlate well with those obtained from tumour tissue. 13,14 However, tissue samples remain the gold standard. Clinical opinion, provided by specialist advisors during scoping, suggested that plasma testing is currently a ‘research only’ application that should not be included in this assessment.
| Sequencing method | Targeted (mutations targeted)/screening test | Limits of detection (% mutation) | Number of laboratories using the method | |
|---|---|---|---|---|
| NEQAS reporta | Laboratory contactb | |||
| Commercial tests | ||||
| Therascreen® KRAS RGQ PCR Kit (QIAGEN, Hilden, Germany) | Targeted (7 mutations: six codon 12 and one codon 13) | 0.77–6.43% | 3 | 1 |
| Therascreen® KRAS Pyro Kit (QIAGEN) | Targeted (12 mutations: six codon 12, one codon 13 and five codon 61) | 1.0–3.5% | 2 | |
| cobas® KRAS Mutation Test (Roche Molecular Systems, Branchburg, NJ, USA) | Targeted (19 mutations: six codon 12, six codon 13 and seven codon 61) | 1.6–6.3% depending on mutation | 4 | 4 |
| KRAS LightMix® kit (TIB MOLBIOL, Berlin, Germany) | Targeted (9 mutations: seven codon 12 and two codon 13) | Unclear | 0 | 0 |
| KRAS StripAssay® (ViennaLab, Vienna, Austria) | Targeted (13 mutations: eight codon 12, two codon 13 and three codon 61) | Unclear | 0 | 0 |
| In-house tests | ||||
| Sanger sequencing | All mutations within specific codons of the KRAS gene | Unclear | 6 | 1 |
| Pyrosequencing | All mutations within specific codons of the KRAS gene | 5–10%b | 15 | 8 |
| Real-time PCR | Targeted (details unclear) | Unclear | 2 | 0 |
| High-resolution melt analysis | All mutations within specific codons of the KRAS gene | ∼5%b | 2 | 2 |
| Next-generation sequencing | All mutations within specific codons of the KRAS gene | ∼5%b | 0 | 0 |
| MALDI-TOF mass spectrometry | All mutations within selected codons in the KRAS oncogene | ∼10% | 1 | 0 |
A Provisional Clinical Opinion (PCO) from the American Society of Clinical Oncology (ASCO), published in 2009,15 recommended universal KRAS mutation testing in patients with mCRC in whom treatment with EGFR inhibitors is being considered. The recommendation also stated that testing should be carried out in an accredited laboratory and that patients whose tumours have KRAS mutations in codons 12 or 13 should not be treated with EGFR inhibitors. At the time that this guidance was published there were no US Food and Drug Administration (FDA)-approved tests for KRAS mutations. The ASCO PCO specified that samples should be selected by a pathologist to include predominantly tumour cells without significant necrosis or inflammation; be freshly extracted or stored in an appropriate preservation solution or rapidly frozen; be neutral-buffered formalin fixed and paraffin embedded and the area of interest selected by the pathologist. 15 Acceptable assay types were listed as real-time polymerase chain reaction (PCR) using probes specific for the most common mutations in codons 12 and 13; direct sequencing of exon 1 in the KRAS gene; and the Therascreen commercial kit (at that time manufactured by DxS, Manchester, UK). 15 Subsequently, the QIAGEN Therascreen KRAS Rotar-Gene Q (RGQ) PCR Kit has been approved by the FDA when used with the QIAGEN QIAamp® DSP DNA FFPE (formalin fixed paraffin embedded) Tissue Kit and the QIAGEN Rotor-Gene® Q MDx (software version 2.1.0) and KRAS Assay Package. 16
Targeted mutation detection tests
All targeted tests are commercial kits and these look for different numbers of mutations within specific codons of the KRAS gene and have differing limits of detection. They may therefore differ in their ability to accurately differentiate patients who are likely to benefit from treatment with cetuximab in combination with standard chemotherapy from those who should receive standard chemotherapy alone.
The Therascreen KRAS RGQ PCR Kit is a CE-marked real-time PCR assay for the qualitative detection of seven mutations in codons 12 and 13 of the KRAS gene. It has been approved by the US FDA for the application covered by this assessment, that is, the selection of patients with mCRC for treatment with cetuximab. The Therascreen KRAS RGQ PCR Kit uses two technologies for the detection of mutations: ARMS (amplification-refractory mutation system) for mutation-specific DNA amplification and Scorpions for detection of amplified regions. Scorpions are bifunctional molecules containing a PCR primer covalently linked to a fluorescently labelled probe. A real-time PCR instrument [Rotor-Gene Q 5-Plex HRM (high-resolution melt) for consistency with CE marking] is used to perform the amplification and to measure fluorescence. 17 There is an earlier version of the Therascreen KRAS PCR Kit that also uses ARMS and Scorpions for the detection of KRAS mutations and is designed to detect the same KRAS mutations as the current, reformulated and revalidated version. Evidence for both versions will be included in this assessment.
The Therascreen KRAS Pyro Kit is a CE-marked test for the quantitative measurement of 12 mutations in codons 12, 13 and 61 of the KRAS gene. The kit is based on pyrosequencing technology and consists of two assays, one for detecting mutations in codons 12 and 13 and a second for detecting mutations in codon 61. The two regions are amplified separately by PCR and then amplified DNA is immobilised on streptavidin sepharose high-performance beads. Single-stranded DNA is prepared and sequencing primers added. The samples are then analysed using the PyroMark® Q24 System (QIAGEN). The KRAS Plug-in Report is recommended by the manufacturer for the analysis of the results; however, the analysis tool within the pyrosequencer can also be used. 18
The cobas KRAS Mutation Test from Roche Molecular Systems is a CE-marked TaqMelt™ real-time PCR assay intended for the detection of 19 mutations in codons 12, 13 and 61 of the KRAS gene. The assay uses DNA extracted from FFPE tissue and is validated for use with the cobas® 4800 System.
The KRAS LightMix Kit from TIB MOLBIOL is a CE-marked test designed for the detection and identification of mutations in codons 12 and 13 of the KRAS gene. The first part of the test involves PCR amplification of the KRAS gene. To reduce amplification of the wild-type KRAS gene and therefore enrich the mutant KRAS gene, a wild type-specific competitor molecule is added to the reaction mix. This is called clamped mutation analysis. The second part of the test procedure involves melting curve analysis with hybridisation probes. The melting temperature is dependent on the number of mismatches between the amplification product and the probe and allows the detection and identification of a mutation within the sample. The test is run on the LightCycler® instrument (Roche). 19
The KRAS StripAssay from ViennaLab is a CE-marked test for the detection of mutations in the KRAS gene. The test procedure involves three steps: the DNA is first isolated from the specimen; PCR amplification is then performed; and the amplification product is then hybridised to a test strip containing allele-specific probes immobilised as an array of parallel lines. Colour substrates are used to detect bound sequences, which can then be identified with the naked eye or by using a scanner and software. 20 There are two versions of the KRAS StripAssay: one is designed to detect 10 mutations in codons 12 and 13 of the KRAS gene and the other is designed to detect the same 10 mutations in codons 12 and 13 plus three mutations in codon 61.
Mutation screening tests
‘In-house’ laboratory-based tests are designed to detect all mutations within specific codons of the KRAS gene.
Pyrosequencing assays are the most commonly used method of KRAS mutation testing in UK laboratories (see Table 1). The process involves first extracting DNA from the sample and amplifying it using PCR. The PCR product is then cleaned up before the pyrosequencing reaction. The reaction involves the sequential addition of nucleotides to the mixture. A series of enzymes incorporate nucleotides into the complementary DNA strand, generate light proportional to the number of nucleotides added and degrade unincorporated nucleotides. The DNA sequence is determined from the resulting pyrogram trace. 21
Sanger sequencing is also a commonly used method (see Table 1); however, there is much variation in the detail of how the method is carried out. In general, after DNA is extracted from the sample it is amplified using PCR. The PCR product is then cleaned up and sequenced in both forward and reverse directions. The sequencing reaction uses dideoxynucleotides labelled with coloured dyes that randomly terminate DNA synthesis, creating DNA fragments of various lengths. The sequencing reaction product is then cleaned up and analysed using capillary electrophoresis. The raw data are analysed using analysis software to generate the DNA sequence. All steps are performed at least in duplicate to increase confidence that an identified mutation is real. It should be noted that sequencing works well only when viable tumour cells constitute at least 25% or more of the sample. 22
National Institute for Health and Care Excellence contact with laboratories (October/November 2012) suggested that several laboratories were planning to convert to next-generation sequencing in the coming year. As with Sanger sequencing, there is much variation in the methodology used to perform next-generation sequencing. The concept is similar to Sanger sequencing; however, the sample DNA is first fragmented into a library of small segments that can be sequenced in parallel reactions. 23
High-resolution melt analysis assays are also commonly used by laboratories participating in the UK NEQAS scheme (see Table 1). For this technique the DNA is first extracted from the sample and amplified using PCR. The HRM reaction is then performed. This involves a precise warming of the DNA during which the two strands of DNA ‘melt’ apart. Fluorescent dye that binds only to double-stranded DNA is used to monitor the process. A region of DNA with a mutation will ‘melt’ at a different temperature to the same region of DNA without a mutation. These changes are documented as melt curves and the presence or absence of a mutation can be reported. 24
Matrix-assisted laser desorption ionisation time-of-flight (MALDI-TOF) mass spectrometry is currently used by one laboratory participating in the UK NEQAS scheme. This technique involves extracting DNA and amplifying it using PCR. The PCR products are then cleaved and fragments separated based on mass by the MALDI-TOF mass spectrometer. This generates a ‘fingerprint’ of the DNA with each fragment represented as a peak with a certain mass. The ‘fingerprint’ of the test sample is compared with the ‘fingerprint’ of the wild-type DNA. A mutation would appear as a peak shift due to a change in the mass of a fragment caused by a base change. 25 MALDI-TOF mass spectrometry can be used to identify all mutations within selected codons in the KRAS oncogene and has a limit of detection of approximately 10% tumour DNA in a background of wild-type DNA. 26
Subgroup analyses of patients tested for KRAS mutation status from randomised controlled trials (RCTs) have shown that treatment with the EGFR-inhibiting monoclonal antibody cetuximab in combination with standard chemotherapy can increase progression-free survival (PFS) and tumour response in patients with KRAS wild-type tumours compared with standard chemotherapy alone. 27,28 In contrast, patients whose tumours were positive for KRAS mutations had reduced PFS and tumour response when treated with cetuximab in combination with standard chemotherapy compared with standard chemotherapy alone. 27,28 These two trials formed the basis of NICE technology appraisal 176,1 which recommends cetuximab in combination with standard chemotherapy for the first-line treatment of mCRC in patients whose tumours are KRAS wild type and whose metastases are confined to the liver and are unresectable. However, both of these trials used a pre-CE-marked version of the LightMix KRAS Kit (TIB MOLBIOL), which is not currently in use by any laboratory participating in the UK NEQAS scheme.
Care pathway
National Institute for Health and Care Excellence guidance on the diagnosis and management of CRC was updated in 2011. 29
Diagnosis of colorectal cancer
This guideline states that patients referred to secondary care for suspected CRC should be assessed using colonoscopy, flexible sigmoidoscopy followed by barium enema or computed tomography (CT), dependent on comorbidities and local expertise and test availability. When a lesion suspicious of cancer is detected a biopsy should be performed to confirm the diagnosis. 29
All patients with histologically confirmed CRC should be offered contrast-enhanced CT of the chest, abdomen and pelvis to estimate the stage of the disease. Further imaging [e.g. contrast-enhanced magnetic resonance imaging (MRI) or positron emission tomography/computed tomography (PET/CT)] may be considered if the CT scan shows metastatic disease only in the liver. 29 The aim of further imaging is to identify those patients who have resectable metastases, or metastases that may become resectable following response to chemotherapy. For the second group of patients, European Society for Medical Oncology (ESMO) 2010 clinical practice guidelines for the treatment of advanced CRC30 recommend establishing KRAS mutation status to determine the best treatment regimen. These guidelines do not stipulate which specific mutations should be analysed or which test method should be used. The KRAS status of a patient’s tumour is identified through analysis of a biopsy sample or, more frequently, a section of resected tumour tissue. The tissue is fixed in formalin and embedded in a block of paraffin for storage by the pathologist, who also examines the histology and evaluates the tumour content of the sample. Macro dissection may be performed before DNA is extracted and mutation analysis is carried out to determine the KRAS status of the tumour.
To minimise turnaround time, guidance from the Royal College of Pathologists31 recommends that mutation testing should be ordered by the pathologist reporting on the cellular make-up of the tumour. However, this is not currently universal practice and often the decision to perform a KRAS mutation test is often taken at the multidisciplinary team (MDT) meeting. If a sample is stored as a FFPE specimen for a long time this can lead to DNA degradation, which can result in a higher chance of failure when testing for KRAS mutations. The timing of the KRAS test varies between patients, with some clinicians preferring to test at diagnosis, potentially before the disease becomes metastatic, and other clinicians waiting until the cancer has progressed to metastatic disease. If the KRAS status is tested early, then the result is then referred to if metastatic disease develops. It has been suggested that analysing multiple resection or biopsy samples from the same patient increases the chances of identifying a KRAS mutation because of potential heterogeneity between tumour sites. The evidence on this is conflicting, with studies reporting that testing a single site only will potentially misclassify between 2% and 10% of tumours as KRAS wild type. 32,33
Treatment of colorectal cancer
In patients with unresectable liver metastases whose primary tumour has been resected or is potentially operable and who are fit enough to undergo liver surgery, the aim of chemotherapy is to induce tumour response such that resection becomes possible. The KRAS mutation status of a patient’s tumour is used to determine the optimal chemotherapy regimen for this purpose. Evidence suggests that patients with KRAS wild-type tumours are more likely to benefit from treatment with an EGFR-inhibiting monoclonal antibody (cetuximab) in combination with standard chemotherapy. However, patients whose tumours are positive for KRAS mutations are more likely to benefit from standard chemotherapy alone. In addition, the overall health and the preferences of the patient should be taken into consideration when selecting treatment.
The choice of standard chemotherapy is covered by NICE clinical guideline 131,29 which recommends that one of the following sequences of chemotherapy is considered:
-
oxaliplatin in combination with infusional fluorouracil plus folinic acid (FOLFOX) as first-line treatment and then single-agent irinotecan as second-line treatment
-
FOLFOX as first-line treatment and then irinotecan in combination with infusional fluorouracil plus folinic acid (FOLFIRI) as second-line treatment
-
oxaliplatin and capecitabine (XELOX) as first-line treatment and then FOLFIRI as second-line treatment.
The guideline further states that raltitrexed should be considered only for patients who are intolerant to fluorouracil and folinic acid or for patients for whom these drugs are not suitable. 29 NICE technology appraisal 6134 suggests that oral therapy with either capecitabine or tegafur with uracil (in combination with folinic acid) can also be considered as an option for the first-line treatment of mCRC.
With respect to the use of biological agents (EGFR inhibitors), NICE technology appraisal 1761 recommends cetuximab in combination with FOLFOX or FOLFIRI, within its licensed indication, for the first-line treatment of mCRC:
-
in patients in whom the primary colorectal tumour has been resected or is potentially operable
-
in patients in whom the metastatic disease is confined to the liver and is unresectable
-
when the patient is fit enough to undergo surgery to resect the primary colorectal tumour and to undergo liver surgery if the metastases become resectable after treatment with cetuximab.
The European Medicines Agency marketing authorisation for cetuximab states that it is ‘indicated for the treatment of patients with EGFR-expressing, KRAS wild-type metastatic colorectal cancer’. 35 Therefore, KRAS mutation testing is an important component of the care pathway. Cetuximab (monotherapy or combination therapy) and bevacizumab (in combination with non-oxaliplatin chemotherapy) for the treatment of mCRC after first-line chemotherapy are not recommended in NICE technology appraisal 242. 36 However, these treatments may be given to some patients through the Cancer Drugs Fund. If cetuximab is considered in the third-line setting, KRAS status is often not retested but a decision will be made based on the result of the KRAS test performed earlier in the care pathway. No other biological agents are currently recommended by NICE for the first-line treatment of patients with unresectable liver metastases from CRC.
National Institute for Health and Care Excellence clinical guideline 13129 stipulates that all patients with primary CRC undergoing treatment with curative intent should be followed up at a clinic visit 4–6 weeks after the potentially curative treatment. They should then have regular surveillance including:
-
a minimum of two CT scans of the chest, abdomen and pelvis in the first 3 years and
-
regular serum carcinoembryonic antigen tests (at least every 6 months in the first 3 years).
They should also have a surveillance colonoscopy at 1 year after initial treatment and, if the result is normal, further colonoscopic follow-up after 5 years and thereafter as determined by cancer networks.
Measuring response to treatment
In 1979 the World Health Organization (WHO) and the International Union Against Cancer introduced criteria for the classification of the response of solid tumours to treatment. 37 These criteria were an early attempt to standardise reporting of response outcomes and were widely adopted; however, some problems with their use have subsequently developed. There has been variation in the methods used for incorporating into response assessments the change in size of measurable lesions, as defined by WHO; the minimum lesion size and number of lesions to be recorded have also varied; the definitions of progressive disease (PD) have sometimes been related to change in a single lesion and sometimes to change in overall tumour load (sum of the measurements of all lesions); and there has been confusion around how to use three-dimensional measures from new technologies such as CT and MRI in the context of the WHO criteria. 38 The Response Evaluation Criteria in Solid Tumours (RECIST) Group is a collaborative initiative that was instigated to review the WHO criteria. The RECIST criteria use the same categories as the WHO criteria [complete response (CR), partial response (PR), stable disease (SD) and PD]. 38 RECIST guidance recommends CT and MRI for measuring target lesions in response assessment and that imaging-based evaluation is generally preferable to clinical examination. It is suggested that follow-up assessments every 6–8 weeks is a ‘reasonable norm’. 38 Taking into account the longest diameter only for all target lesions, the RECIST criteria, as they are applicable to this assessment, can be summarised as follows:38
-
CR – disappearance of all target lesions and no new lesions
-
PR – at least a 30% decrease in the sum of the longest diameters of target lesions, taking the sum of the baseline diameters as the reference, and no new lesions
-
PD – at least a 20% increase in the sum of the longest diameters of target lesions, taking the smallest sum of the longest diameters recorded since treatment started as the reference, or the appearance of one or more new lesions
-
SD – neither sufficient shrinkage to be classified as PR or sufficient increase to be classified as PD, taking the smallest sum of the longest diameters recorded since treatment started as the reference, and no new lesions.
Best overall response is defined as the best response recorded from the start of treatment to disease progression. 38
First-line chemotherapy of unresectable colorectal liver metastases seeks to achieve a tumour response such that the tumour is judged to be resectable. For this reason, resection rate is considered the ideal reference standard for research question 2 and the optimal outcome measure for research question 3 (see Chapter 1). Objective response rate (ORR), defined as best overall response = CR + PR, is also of interest as there is some evidence that ORR correlates well with resection rate. 39 Tumour status following treatment/resection is defined by the residual tumour (R) classification, in which R0 = no residual tumour, R1 = microscopic residual tumour and R2 = macroscopic residual tumour.
This assessment compares the performance and cost-effectiveness of KRAS mutation testing options, currently available in the UK NHS, for the differentiation of adults with mCRC whose metastases are confined to the liver and are unresectable and who may benefit from first-line treatment with cetuximab in combination with standard chemotherapy from those who should receive standard chemotherapy alone.
Chapter 3 Assessment of clinical effectiveness
A systematic review was conducted to summarise the evidence on the clinical effectiveness of the different KRAS mutation testing options, currently available in the UK NHS, for differentiating adults with mCRC whose metastases are confined to the liver and are unresectable and who may benefit from first-line treatment with cetuximab in combination with standard chemotherapy from those who should receive standard chemotherapy alone. Systematic review methods followed the principles outlined in the Centre for Reviews and Dissemination (CRD) guidance for undertaking reviews in health care40 and the NICE Diagnostic Assessment Programme manual. 41 In addition to the effectiveness review, additional data were obtained from an online survey of laboratories participating in the UK NEQAS pilot scheme for KRAS mutation testing.
Systematic review methods
Search strategy
Search strategies were based on target condition and intervention, as recommended in the CRD guidance for undertaking reviews in health care40 and the Cochrane Handbook for DTA Reviews. 42
Candidate search terms were identified from target references, browsing database thesauri [e.g. MEDLINE medical subject headings (MeSH) and EMBASE Emtree], existing reviews identified during the rapid appraisal process and initial scoping searches. These scoping searches were used to generate test sets of target references, which informed text mining analysis of high-frequency subject indexing terms using EndNote X5 reference management software (Thomson Reuters, CA, USA). Strategy development involved an iterative approach testing candidate text and indexing terms across a sample of bibliographic databases and aimed to reach a satisfactory balance of sensitivity and specificity.
The following databases were searched for relevant studies from 2000 to January 2013:
-
MEDLINE (OvidSP) (2000 to Week 2 January 2013)
-
MEDLINE In-Process and Other Non-Indexed Citations and Daily Update (OvidSP) (up to 21 January 2013)
-
EMBASE (OvidSP) (2000 to Week 3 2013)
-
Cochrane Database of Systematic Reviews (CDSR) (Wiley) (The Cochrane Library 2000 to Issue 12, 2012)
-
Cochrane Central Register of Controlled Trials (CENTRAL) (Wiley) (The Cochrane Library 2000 to Issue 12, 2012)
-
Database of Abstracts of Reviews of Effects (DARE) (Wiley) (The Cochrane Library 2000 to Issue 4, 2012)
-
Health Technology Assessment (HTA) database (Wiley) (The Cochrane Library 2000 to Issue 4, 2012)
-
Science Citation Index (SCI-EXPANDED) (Web of Knowledge) (2000 to 22 January 2013)
-
Conference Proceedings Citation Index (CPCI-S) (Web of Knowledge) (2000 to 22 January 2013)
-
Latin American and Caribbean Health Sciences Literature (LILACS) (Internet) (up to 24 January 2013), http://regional.bvsalud.org/php/index.php?lang=en
-
BIOSIS Previews (Web of Knowledge) (2000 to 22 January 2013)
-
National Institute for Health Research (NIHR) HTA programme (Internet) (up to 25 January 2013)
-
International Prospective Register of Systematic Reviews (PROSPERO) (Internet) (up to 25 January 2013), www.crd.york.ac.uk/prospero/.
Completed and ongoing trials were identified by searches of the following resources:
-
National Institutes of Health ClinicalTrials.gov (Internet) (2000 to 23 January 2013), www.clinicaltrials.gov/
-
Current Controlled Trials (Internet) (2000 to 29 January 2013), www.controlled-trials.com/
-
WHO International Clinical Trials Registry Platform (ICTRP) (Internet) (2000 to 25 January 2013), www.who.int/ictrp/en/.
Searches were undertaken to identify studies of KRAS testing for mCRC. The main EMBASE strategy for each set of searches was independently peer reviewed by a second information specialist using the Peer Review of Electronic Search Strategies Evidence-Based Checklist (PRESS-EBC). 43 Search strategies were developed specifically for each database and the keywords associated with CRC were adapted according to the configuration of each database. Searches took into account generic and other product names for the intervention. No restrictions on language or publication status were applied. Full search strategies are reported in Appendix 1.
Electronic searches were undertaken for the following conference abstracts:
-
ASCO conference proceedings (Internet) (2007–13), www.asco.org/ASCOv2/Meetings/Abstracts
-
ESMO conference proceedings (Internet) (2007–13), www.esmo.org/education-research/abstracts-virtual-meetings-and-meeting-reports.html
-
American Association for Cancer Research conference proceedings (Internet) (2007–13), www.aacrmeetingabstracts.org/search.dtl
-
Association for Molecular Pathology conference proceedings (Internet) (2007–13), www.amp.org/meetings/past_meetings.cfm.
Identified references were downloaded into EndNote X4 software for further assessment and handling.
References in retrieved articles were checked for additional studies. The final list of included papers was also checked on PubMed for retractions, errata and related citations. 44–46
Inclusion and exclusion criteria
Separate inclusion criteria were developed for each of the three clinical effectiveness questions; these are summarised in Table 2.
| What is the technical performance of the different KRAS mutation tests? | What is the accuracy of KRAS mutation testing, using any test, for predicting response to treatment with cetuximab in combination with standard chemotherapy? | How do outcomes from treatment with cetuximab in combination with standard chemotherapy and, when reported, from treatment with standard chemotherapy vary according to which test is used to select patients for treatment? | |
|---|---|---|---|
| Participants | Adult patients (≥ 18 years) with mCRC and a resected or resectable primary tumour whose metastases are confined to the liver and are unresectable but may become resectable after response to chemotherapy | Adult patients (≥ 18 years) with mCRC and a resected or resectable primary tumour whose metastases are confined to the liver and are unresectable but may become resectable after response to chemotherapy | Adult patients (≥ 18 years) with mCRC and a resected or resectable primary tumour whose metastases are confined to the liver and are unresectable but may become resectable after response to chemotherapy Patients who have been tested for KRAS mutation status |
| Setting | Secondary or tertiary care | ||
| Interventions (index test) | Any commercial or in-house KRAS mutation test listed in Table 1 | Any commercial or in-house KRAS mutation test listed in Table 1 | First-line chemotherapy with cetuximab in combination with standard chemotherapya |
| Comparators | Not applicable | Not applicable | Standard chemotherapya |
| Reference standard | Not applicable | Response to treatment with cetuximab in combination with standard chemotherapy (e.g. PFS, ORR, disease control rate) | Not applicable |
| Outcomes | Proportion of tumour cells needed, failures, limit of detection, turnaround time, costs, expertise/logistics of test | Overall survival or PFS in patients whose tumours are KRAS mutant vs. overall survival or PFS in patients whose tumours are KRAS wild type. Test accuracy – the numbers of true positives, false negatives, false positives and true negatives | PFS, overall survival, ORR, disease control rate |
| Study design | To be addressed by survey (see Survey of laboratories providing KRAS mutation testing); publications from UK laboratories | RCTs (CCTs and cohort studies will be considered if no RCTs are identified) | RCTs (CCTs will be considered if no RCTs are identified) |
Inclusion screening and data extraction
Two reviewers (MW and PW) independently screened the titles and abstracts of all reports identified by searches and any discrepancies were discussed and resolved by consensus. Full copies of all studies deemed potentially relevant were obtained and the same two reviewers independently assessed these for inclusion; any disagreements were resolved by consensus. Details of studies excluded at the full paper screening stage are presented in Appendix 5.
Studies cited in materials provided by the manufacturers of the Therascreen KRAS RGQ PCR Kit and Therascreen KRAS Pyro Kit (QIAGEN), the cobas KRAS Mutation Test Kit (Roche Molecular Systems), the KRAS LightMix Kit (TIB MOLBIOL) and the KRAS StripAssay (ViennaLab) were first checked against the project reference database, in EndNote X4; any studies not already identified by our searches were screened for inclusion following the process described above.
Data were extracted on the following: study design/details, participant details (e.g. inclusion/exclusion criteria, age, liver metastases details, criteria for unresectability, performance status, previous treatments), KRAS mutation test(s) and mutations targeted, intervention details, clinical outcomes, test performance outcome measures (against treatment response as reference standard), details of specific mutations identified by outcome measure (when reported), test failure rates and limits of detection. Data were extracted by one reviewer, using a piloted standard data extraction form, and checked by a second (MW and PW); any disagreements were resolved by consensus. Full data extraction tables are provided in Appendix 2.
Quality assessment
The risk of bias in included RCTs was assessed using the Cochrane Collaboration’s tool for assessing risk of bias in randomised trials. 47 Studies used to derive accuracy data, for the ability of KRAS mutation tests to predict treatment response, were assessed using the QUADAS-2 tool. 48 Risk of bias assessments were undertaken by one reviewer and checked by a second reviewer (MW and PW) and any disagreements were resolved by consensus.
The results of the risk of bias assessments were summarised and are presented in tables and graphs in the results section of this chapter and in full, by study, in Appendix 3.
Survey of laboratories providing KRAS mutation testing
We conducted a web-based survey to gather data on the technical performance characteristics of KRAS mutation tests. We sent an e-mail invitation via NEQAS to laboratories participating in the UK NEQAS pilot scheme for KRAS mutation testing. We used SurveyMonkey online software to run the survey. We structured the survey into sections on:
-
laboratory details
-
KRAS testing methods
-
logistics
-
technical methods
-
costs.
When possible we used multiple choice options with tick boxes to make the survey quick and easy to complete. A copy of the survey is provided in Appendix 4.
Methods of analysis/synthesis
The results of studies included in this review were summarised by research question (see Chapter 1), that is, studies providing technical information on KRAS mutation testing in NHS laboratories in the UK (see What are the technical performance characteristics of the different KRAS mutation tests?), studies providing information on the accuracy of KRAS mutation tests for predicting response to treatment (see What is the accuracy of KRAS mutation testing for predicting response to treatment with cetuximab plus standard chemotherapy and subsequent resection rates?) and studies reporting information on how clinical outcomes may vary according to which test is used to select patients for treatment (see How do outcomes from treatment with cetuximab plus standard chemotherapy vary according to which test is used to select patients for treatment?). We planned to use a bivariate/hierarchical summary receiver operating characteristic (HSROC) random-effects model to generate summary estimates and a summary receiver operating characteristic (SROC) curve for test accuracy data49–51 and a DerSimonian and Laird random-effects model to generate summary estimates of treatment effects. However, because the review identified a small number of studies with between-study variation in participant characteristics, methods used to test for KRAS mutations and mutations targeted, we did not consider meta-analysis to be appropriate and have provided a structured narrative synthesis.
For all studies that provided data on accuracy for the prediction of response to treatment with cetuximab in combination with standard chemotherapy, the absolute numbers of true-positive (TP), false-negative (FN), false-positive (FP) and true-negative (TN) test results, as well as sensitivity and specificity values with 95% confidence intervals (CIs), are presented in results tables, for each reference standard response (e.g. ORR or resection rate) reported. When reported, data on the numbers of failed KRAS mutation tests and reasons for failure were also included in the results tables. The results of individual studies were plotted in the receiver operating characteristic (ROC) plane to illustrate the trade-off between sensitivity and specificity and for ease of comparison between test methods; separate plots were provided for each reference standard response. For RCTs providing information on how clinical outcomes may vary according to which test is used to select patients for treatment with cetuximab in combination with standard chemotherapy, hazard ratios (HRs) with 95% CIs were provided for PFS and odds ratios (ORs) with 95% CIs were reported for tumour response outcomes (ORR and resection rate). The results of individual studies were illustrated in forest plots. Between-study clinical heterogeneity was assessed qualitatively. There were insufficient studies to assess heterogeneity statistically using the chi-squared test or I2 statistic.
Results of the assessment of clinical effectiveness
The literature searches of bibliographic databases identified 7903 references. After initial screening of titles and abstracts, 100 were considered to be potentially relevant and were ordered for full paper screening. No additional papers were ordered based on screening of papers provided by test manufacturers; all studies cited in documents supplied by the test manufacturers had already been identified by the bibliographic database searches. No additional studies were identified from searches of clinical trials registries or from hand searching of conference abstracts. Figure 1 shows the flow of studies through the review process and Appendix 5 provides details, with reasons for exclusions, of all publications excluded at the full paper screening stage.
FIGURE 1.
Flow of studies through the review process.
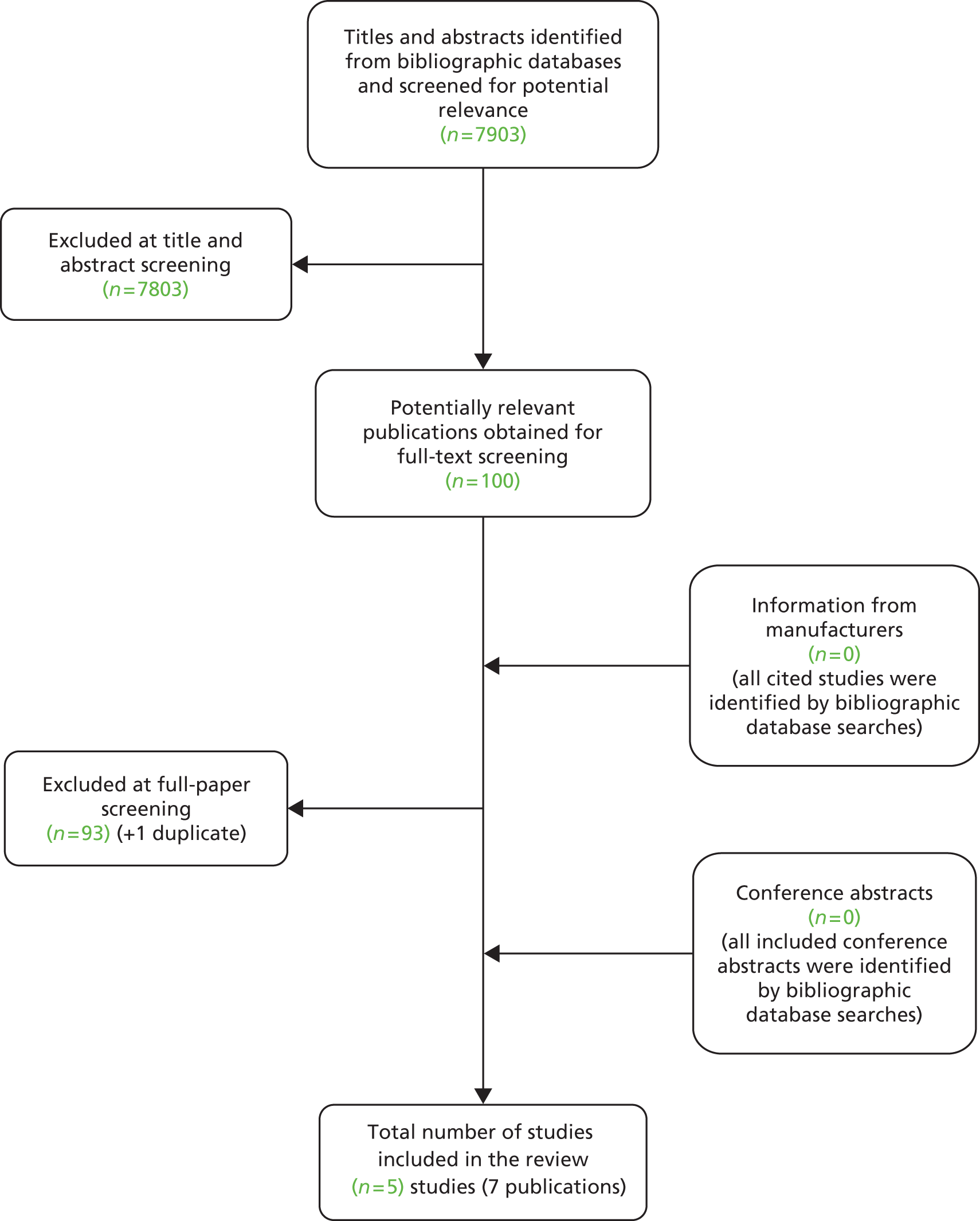
Based on the searches and inclusion screening described earlier, seven publications of five studies were included in the review. 27,28,52–56 Because data for participants with colorectal metastases and no extrahepatic metastases were frequently reported as subgroup analyses of larger trials, the authors of two additional potentially relevant trials in patients with mCRC were contacted to request subgroup data. The author of the Central European Cooperative Oncology Group (CECOG) trial57 reported that only 23 (15%) participants had metastases that were limited to the liver and that no subgroup data were available for these participants. The authors of the NORDIC-VII trial58 did not respond to our request.
No studies conducted in UK NHS laboratories were identified that reported information on the technical performance characteristics of KRAS mutation tests. One study52 reported data on tumour response following treatment with cetuximab plus FOLFOX6 or cetuximab plus FOLFIRI in a group of patients with unresectable colorectal liver metastases who were tested for tumour KRAS mutation status. This study provided information on the accuracy of the Therascreen KRAS PCR test for the prediction of response to treatment. Additional data, supplied by the COIN trial investigators, allowed calculation of accuracy for prediction of resection of liver metastases following treatment with cetuximab plus FOLFOX or XELOX, in which a combination of pyrosequencing and MALDI-TOF mass spectrometry was used to asses KRAS mutation status [D Fisher, Medical Research Council (MRC) Clinical Trials Unit, London, 19 April 2013, personal communication]. Four RCTs, reported in six publications,27,28,53–56 compared the effectiveness of cetuximab plus standard chemotherapy with that of standard chemotherapy alone in patients whose tumours were KRAS wild type. Because the method used to determine mutation status varied between trials, these RCTs provide some information on how clinical outcomes may vary according to which test is used to select patients for treatment.
All included studies were published in 2009 or later. The study providing information on test accuracy was a multicentre European study funded by Merck Serono, Sanofi Aventis and Pfizer. 52 Two of the four RCTs were multicentre European studies funded by Merck Serono,27,28,53,56 one was a multicentre study conducted in the UK and the Republic of Ireland and funded by the UK MRC54 and one was a single-centre study conducted in China and published as an abstract only (no funding details reported). 55
Full details of the characteristics of the study participants, study inclusion and exclusion criteria, KRAS mutation test used and mutations targeted, and treatment groups are reported in the data extraction tables presented in Appendix 2. For studies providing test accuracy data, full details of the KRAS mutation testing process are reported as part of the QUADAS-2 risk of bias assessment in Appendix 3.
What are the technical performance characteristics of the different KRAS mutation tests?
Literature review
No studies reporting the technical performance of KRAS mutation tests on clinical samples in UK laboratories were identified. Data on the technical performance characteristics of KRAS mutation tests, as experienced by UK laboratories, were therefore derived solely from the results of the online survey.
Laboratory survey results
A total of 31 laboratories participated in the 2012–13 UK NEQAS pilot scheme for KRAS mutation testing. The survey was completed by 21 laboratories; however, five of these were based outside the UK (Norway, Belgium, Switzerland, Italy and Ireland) and one was excluded as KRAS mutation testing was carried out for haematological malignancies only. Therefore, survey results were analysed for 15 laboratories (response rate 50%).
KRAS mutation test methods (Figure 2)
Fifteen laboratories stated that they used one method of KRAS mutation testing; one of these stated that they sometimes use a single KRAS mutation testing method and sometimes multiple methods (e.g. to confirm mutations).
FIGURE 2.
KRAS mutation tests used in NHS laboratories in the UK participating in the UK NEQAS pilot scheme for KRAS mutation testing.
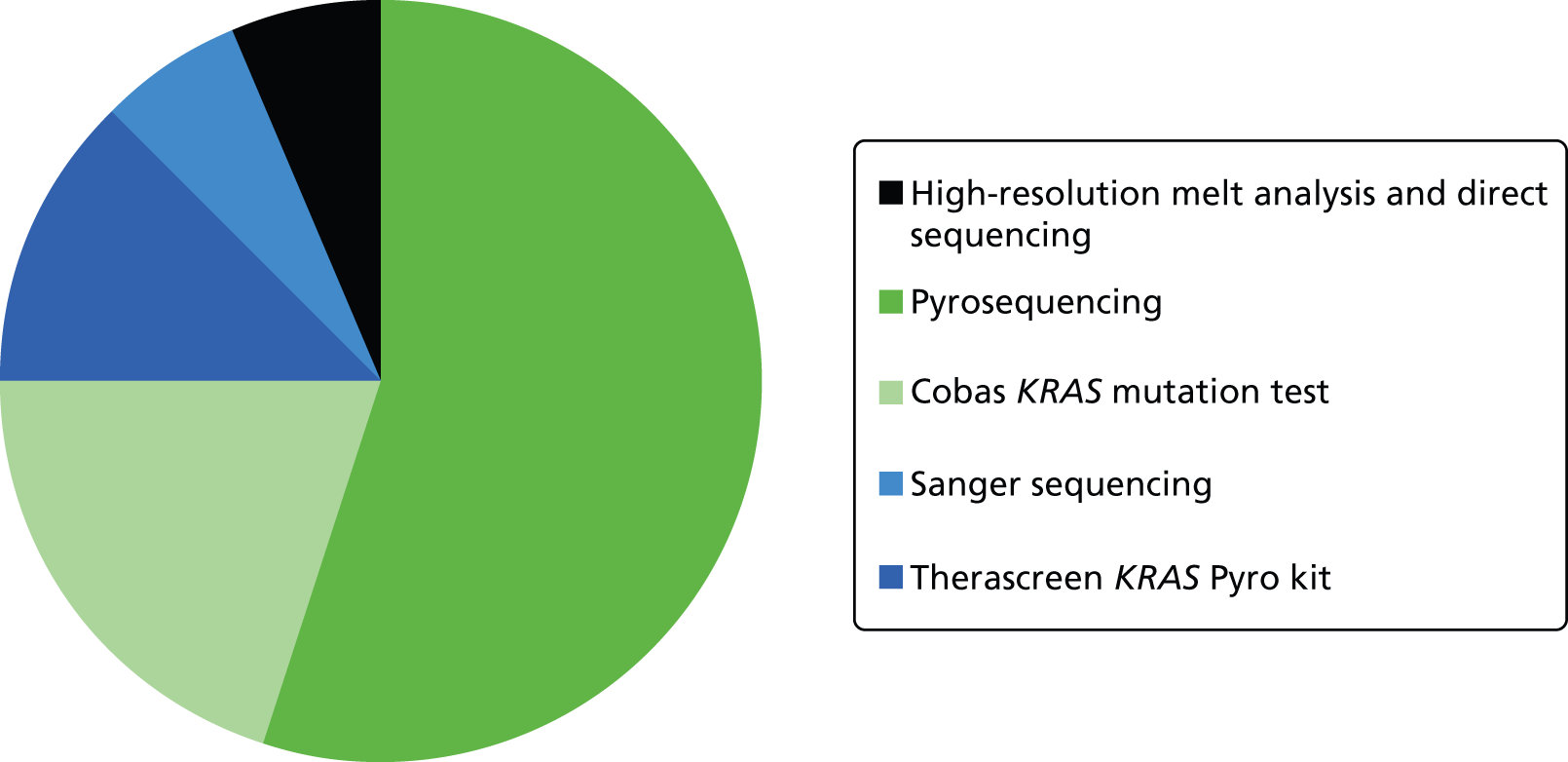
Pyrosequencing, using in-house methods, was the most commonly used KRAS mutation test with nine laboratories using this approach, although one of the laboratories using pyrosequencing stated that it was in the process of switching to HRM analysis because of its quicker turnaround time. The cobas KRAS Mutation Test was used by three laboratories, Sanger sequencing was used by two laboratories and only a single laboratory used the Therascreen KRAS Pyro Kit. The final laboratory, which had initially reported using only a single method of KRAS mutation testing, stated that it used HRM analysis and direct sequencing. The laboratory that reported sometimes using multiple methods reported details for only one testing method (Sanger sequencing); however, this laboratory did state that it used Sanger sequencing in the event of an unusual pyrosequencing result. It also stated that its reason for using more than one testing method was the ability to fully characterise detected mutations. This suggests that its first choice method was in fact pyrosequencing not Sanger sequencing. This laboratory is therefore included in both methods in Figure 2 and the numbers above. All other laboratories stated that they used the reported KRAS mutation testing method for 100% of the samples.
Nine laboratories reported that samples were referred to their laboratory for testing on demand (one specified that this was MDT meetings, via pathologist, or oncologist), two reported mixed referral (some centres on demand, some all CRC samples), one laboratory reported that all resected primary CRC samples were sent for testing, one reported that samples were sent through clinical trials, one reported that all resected primary CRC plus metastatic samples were sent for testing and one laboratory did not answer this question.
The main reasons cited for choice of KRAS mutation testing method were mutation coverage (n = 13, 87%), ease of use (n = 12, 80%) and cost (n = 11, 73%). Nine laboratories (60%) also selected sensitivity (proportion of tumour cells required) and seven (47%) selected turnaround time. One laboratory did not answer this question. There was no apparent association between test method and reason for choice.
Of the eight laboratories that completed the questionnaire for pyrosequencing, all reported that they targeted mutations in codons 12, 13 and 61. Two of these laboratories reported that all mutations were targeted, one using commercial primers and one using self-designed primers. Two laboratories reported that they targeted specific mutations using self-designed primers. The others all used self-designed primers but did not state whether they targeted all or specific mutations; one laboratory stated that it also targeted mutations in codon 146. Two laboratories used Sanger sequencing. One stated that it targeted specific mutations but did not provide any further details. The other stated that it targeted mutations in codons 12, 13 and 61 using self-designed primers. One laboratory stated that it used a single testing method only, HRM analysis, and subsequently stated that it used HRM analysis and direct sequencing; mutations in codons 12, 13 and 61 and all mutations in exons 2 and 3 were targeted. Details on primers were not reported. The other four laboratories used commercial KRAS mutation testing kits.
KRAS mutation test logistics (Table 3 and Figure 3)
The number of samples screened for KRAS mutations in a typical week varied by laboratory, from less than five (three laboratories) to > 20 (four laboratories). The batch size ranged from less than one to two to 15–20 samples. Only laboratories with five or less samples screened per week ran batches of three or less. Only one laboratory had a batch size of > 15 (reported as 15–20 samples per week) and this laboratory screened > 20 samples per week; most other laboratories had batch sizes between five and 10. The two laboratories using Sanger sequencing both reported screening five or less samples per week and reported batch sizes of one or two. Of the four laboratories using commercial kits, one did not report on number of samples screened or batch size, two reported screening > 20 samples per week with batch sizes of 10 and 15, and one reported screening 6–10 samples per week with a similar batch size. Only one laboratory reported that it waited until it had a certain number of samples before running the KRAS mutation test; this laboratory waited until it had 10 samples before running the test.
| KRAS mutation test | Samples per week | Batch size | Frequency of test | Wait for batch size? | Time from receiving sample to returning result to clinician |
|---|---|---|---|---|---|
| cobas KRAS Mutation Test Kit | 6–10 | 6–10 | Weekly | No | 6–7 days |
| > 20 | 10 | Two to three times per week | Yes, 10 | 3–5 days | |
| NR | NR | NR | NR | NR | |
| HRM analysis | 6–10 | 4 | Two to three times per week | No | 3–5 days |
| Pyrosequencing | ≤ 5 | 3 | On demand | No | 3–5 days |
| 6–10 | 6–10 | Weekly | No | 3–5 days | |
| > 20 | 15–20 | Two to three times per week | No | 3–5 days | |
| 6–10 | 8 | Weekly | No | 6–7 days | |
| 11–15 | 6–10 | Two to three times per week | No | 24–48 hours | |
| 16–20 | 10 | Two to three times per week | No | 3–5 days | |
| 6–10 | 5–10 | Two to three times per week | No | 3–5 days | |
| > 20 | 12 | Daily | No | 3–5 days | |
| Sanger sequencing | ≤ 5 | ≥ 1 | Daily | No | 3–5 days |
| ≤ 5 | 1–2 patients | Variable | No | 3–5 days | |
| Therascreen KRAS Pyro Kit | > 20 | 15 | Two to three times per week | No | 6–7 days |
FIGURE 3.
Summary of logistic information.
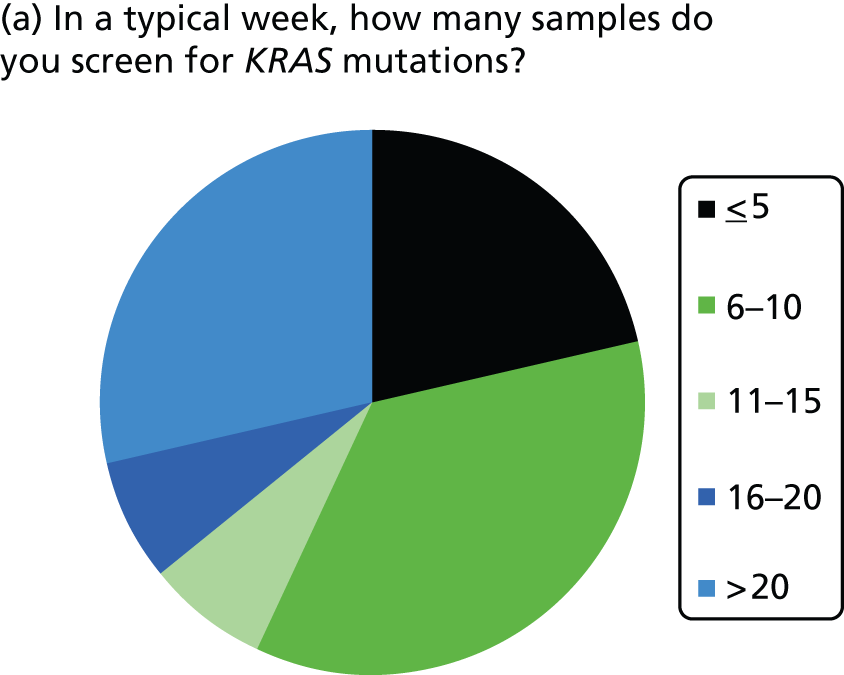
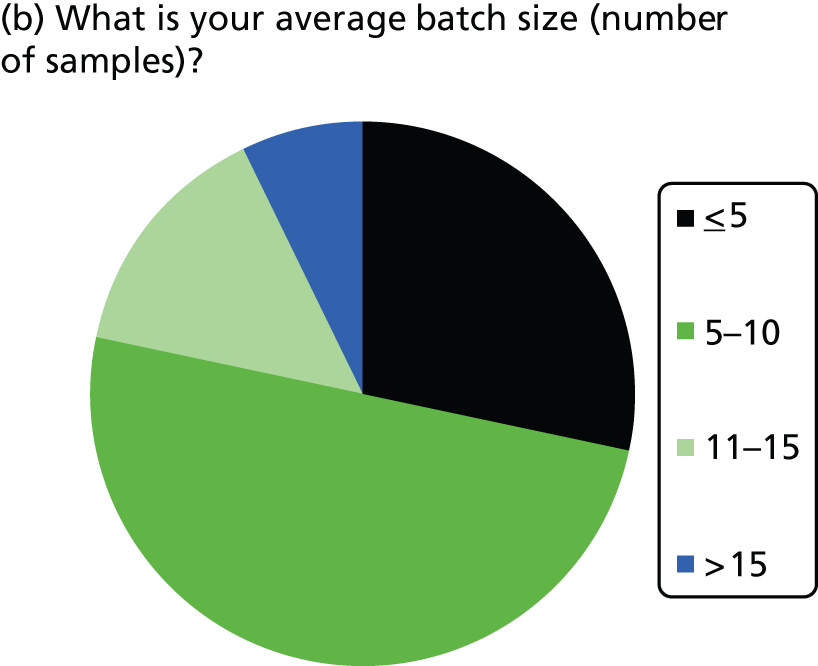
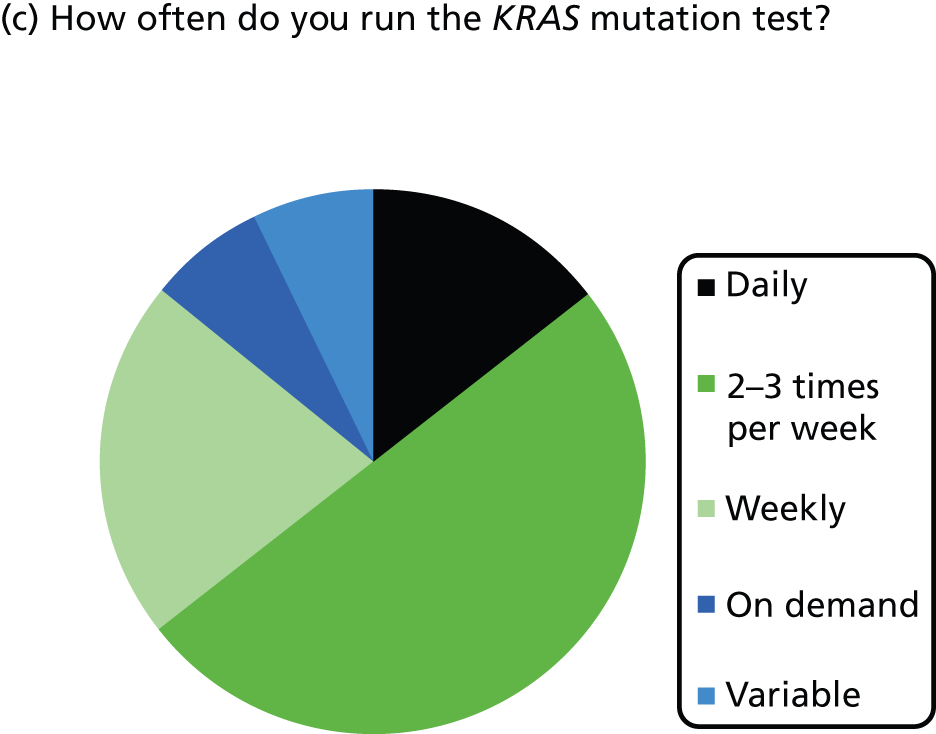
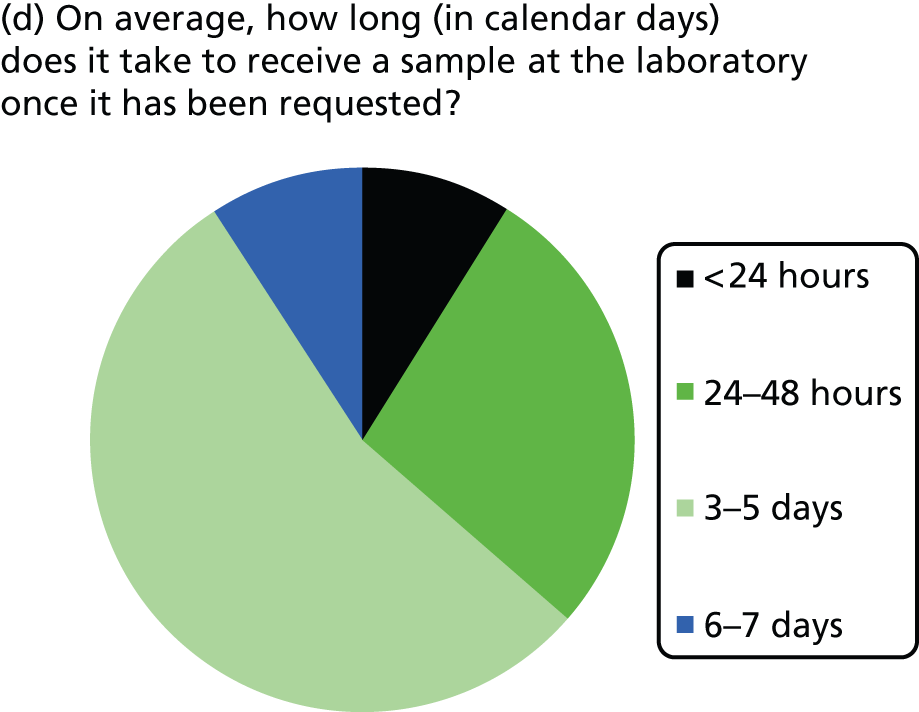
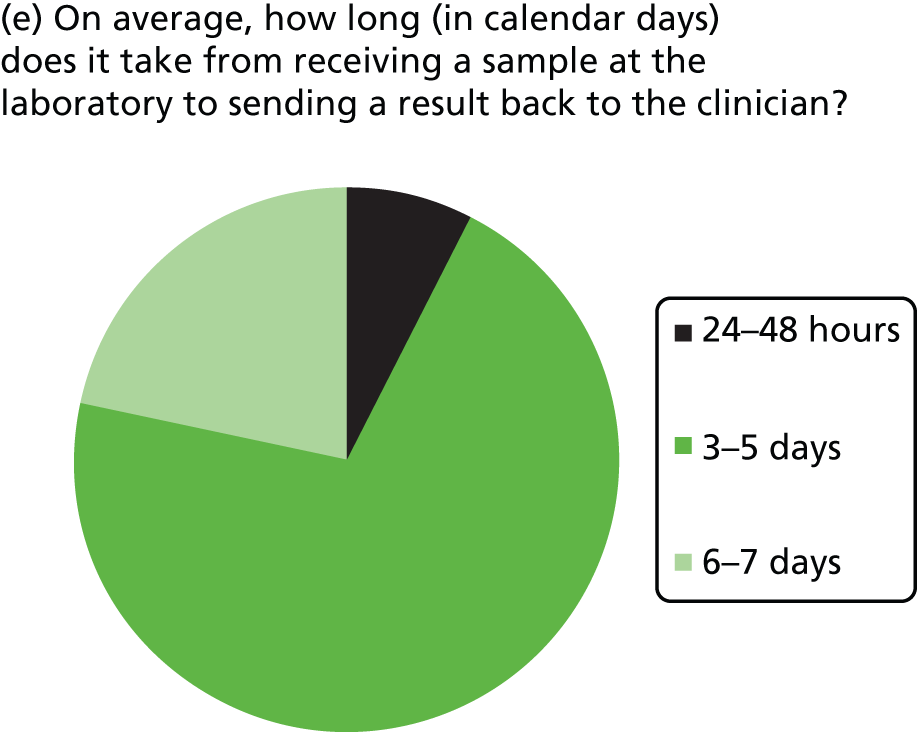
Most laboratories reported an average waiting time from requesting the sample for KRAS testing to receiving the sample in the laboratory of 24–48 hours (three laboratories) or 3–5 days (six laboratories), although one laboratory reported a waiting time of < 24 hours and one reported a waiting time of 6–7 days. The range in waiting times was reported by four laboratories and was 1–10 days in two and 2–30 days in one, with the fourth stating that occasionally request dates are included in referral and so the range is 1–3 weeks. Four laboratories did not report data on time to receive samples at the laboratory once the sample had been requested. The majority of laboratories had a turnaround time from receiving the sample to reporting the result to the clinician of 3–5 or 6–7 days, with only one laboratory reporting a time of 24–48 hours. The laboratory with the shortest turnaround time was one that used pyrosequencing and tested 11–15 samples per week.
KRAS mutation test technical performance (Table 4)
The minimum reported percentage of tumour cells required varied between laboratories (range < 1% to > 30%), even for those using the same KRAS mutation test. All laboratories using the cobas KRAS Mutation Test reported that the limit of detection was 1–5%; the limit of detection was reported to be either 1–5% (three laboratories) or 6–10% (five laboratories) for pyrosequencing, > 10% for Sanger sequencing and 1–5% for HRM. A variety of methods were used to determine the limit of detection. Ten laboratories reported using microdissection, with two stating that they always used this technique and others using microdissection at thresholds of < 10–50%. The laboratory that used the Therascreen KRAS Pyro Kit did not provide any data on technical performance.
| KRAS mutation test | Minimum tumour cells required (%) | Limit of detection (%) | How was limit of detection determined? | Use of microdissection? | Threshold below which microdissection used (%) |
|---|---|---|---|---|---|
| cobas KRAS Mutation Test Kit | 1–5 | 1–5 | Manufacturer guidance | Yes | 10 |
| NR | 1–5 | In-house validation | No | NA | |
| 6–10 | 1–5 | Artificial blends of tumour DNA in normal DNA | Yes | 10–15 | |
| HRM analysis | 11–20 | 1–5 | Serial dilutions of control samples | No | NA |
| Pyrosequencing | >30 | 6–10 | Horizon Diagnostics reference standards | Yes | 50 |
| 11–20 | 6–10 | Spiking of wild-type DNA with mutant DNA | Yes | 20 | |
| 11–20 | 6–10 | Dilution series of known mutations at known percentage | Yes | Always | |
| 11–20 | 6–10 | Dilution series of DNA from three cell lines each with a different KRAS mutation | No | NA | |
| 6–10 | 1–5 | Cell lines with known mutations | Yes | 20; all samples that contain adenoma or when dissection would greatly improve the tumour percentage | |
| 21–30 | 1–5 | CE-marked kit, in-house validation conducted | Yes | 20 | |
| 21–30 | 6–10 | Internal quality control | Yes | Always | |
| 6–10 | 1–5 | Cell lines with set percentage tumour burden | Yes | 50 | |
| Sanger sequencing | > 30 | > 10 | Cell line with known mutation | Yes | 30 |
| ≤ 1 | > 10 | Cell line control | No | NA | |
| Therascreen KRAS Pyro Kit | NR | NR | NR | NR | NR |
KRAS mutation test failure rates (Table 5)
The proportion of samples rejected before analysis was < 2% for all 13 laboratories that provided data on rejection rates. Reasons for rejection included insufficient tumour cells/tissue, sample type unsuitable for analysis and insufficient patient identifiers. The proportion of failed tests ranged from 3% to 6% for the cobas KRAS Mutation Test Kit and from 0.2% to 10% for pyrosequencing. The one laboratory using HRM analysis reported no failed tests and of the two laboratories using Sanger sequencing one reported no failed tests and the other did not provide information on the number of failed tests. Reasons for test failure included insufficient DNA, amplification failure, DNA degradation/quality, insufficient tumour cells and poor fixation. The laboratory that used the Therascreen KRAS Pyro Kit did not provide any data on failure rates.
| KRAS mutation test | No. of samples per yeara | |||||
|---|---|---|---|---|---|---|
| Submitted, n | Rejected, n (%) | Reason for rejection | Analysed, n | Failed, n (%) | Reason for failure | |
| cobas KRAS Mutation Test Kit | NR | NR | NR | 1000 | 29 (3) | Various, most commonly DNA yield too low |
| 1358 | 7 (0.5) | Sample type unsuitable for analysis, insufficient identifiers | 1351 | 86 (6) | Insufficient extracted DNA (8%), amplification failure (92%) | |
| 1058 | 5–10 (0.7) | Insufficient tumour cells | 1058 | 28 (3) | Insufficient tumour cells, DNA degradation | |
| HRM analysis | 1000 | 0 | NA | 1000 | 0 | NA |
| Pyrosequencing | 9 | 0 | NA | 9 | 0 | NA |
| 1000 | < 10 (< 1) | Insufficient tissue left in the block | 1000 | 100 (10) | NR | |
| 1500 | 15–20 (1.5) | Insufficient tumour cells | NR | NR (1) | Assumed to be because of fixation and DNA degradation | |
| 415 | 3 (0.7) | Insufficient tumour cells | 412 | 1 (0.2) | DNA quality | |
| 374 | 0 | NA; samples preselected by laboratory | 374 | 10–20 (4) | Poor fixation | |
| 1000 | 0 | NA | 1000 | 4 (0.4) | Unknown | |
| 1000 | 10 (1) | Insufficient tumour cells, unsuitable sample type | 1000 | 50 (5) | Insufficient tumour cells, DNA degradation | |
| 1736 | ∼20 (1) | Insufficient tumour cells, insufficient tissue | 1736 | ∼30 (2) | Insufficient tumour cells, DNA degradation | |
| Sanger sequencing | 65 | 0 | NA | 65 | 0 | NA |
| 1000 | 0 | NA | 1000 | NA | Insufficient tumour cells, DNA degradation | |
| Therascreen KRAS Pyro Kit | NR | NR | NR | NR | NR | NR |
KRAS mutation test costs (Table 6)
Only seven laboratories provided data on the cost of the KRAS mutation test. Two of these provided data on the costs of the reagents only, which were reported as £22 for Sanger sequencing and approximately £50 for pyrosequencing. A further laboratory also reported that the cost of pyrosequencing was £50. Three other laboratories reported costs for pyrosequencing, with one reporting a cost of £150, one reporting a cost of approximately £120 and the other reporting a cost of approximately £273 for a single sample, which reduced to approximately £110 per sample if running a batch of 10. The final laboratory to report cost data reported that the cost of the cobas KRAS Mutation Test Kit was £100–125. With the exception of two laboratories, all laboratories received some funding from Merck Serono. One laboratory did not provide any details on test costs or funding. The price charged to both the NHS and Merck Serono ranged from £99 to £150 per sample.
| KRAS mutation test | Cost to laboratory (£) | Funding | NHS price (£) | Merck Serono price (£) |
|---|---|---|---|---|
| cobas KRAS Mutation Test Kit | NR | Merck Serono | NA | NR |
| NR | Merck Serono, private | NA | NR | |
| 100–125 | Merck Serono, private | NA | Unable to disclose | |
| HRM analysis | NR | Merck Serono | NA | NR |
| Pyrosequencing | 150 | NHS, privately funded from abroad | 150 | NA |
| NR | Merck Serono, NHS | 140 | 100 | |
| ∼120 | Merck Serono, Cancer Research UK Stratified Medicine programme | 99 | 99 | |
| NR | Merck Serono | NA | 100 | |
| ∼273 for a single sample, ∼110 if running a batch of 10 | Merck Serono | NA | 150 | |
| ∼50 (reagents only) | Merck Serono | NA | 100 | |
| 50 | Trials unit | NA | NA | |
| NR | Merck Serono, NHS, private | 120 | 120 | |
| Sanger sequencing | 22 (reagents only) | Merck Serono | NA | 100 |
| NR | Merck Serono | NR | NR | |
| Therascreen KRAS Pyro Kit | NR | NR | NR | NR |
What is the accuracy of KRAS mutation testing for predicting response to treatment with cetuximab plus standard chemotherapy and subsequent resection rates?
One study, the CELIM trial,52 reported sufficient data to allow calculation of the accuracy of KRAS mutation testing for predicting response to treatment in patients with colorectal liver metastases who are treated with cetuximab plus FOLFOX6 or cetuximab plus FOLFIRI. This study is potentially useful in that it could provide full information on the extent to which KRAS mutation tests are able to discriminate between patients who will have benefit from the addition of cetuximab to standard chemotherapy regimens and those who will not. The utility of the study for this assessment is limited because reporting of outcome data by mutation status was limited to objective response. Thus, we defined TPs as those patients with KRAS wild-type tumours who have a positive response to treatment with cetuximab plus FOLFOX6 or cetuximab plus FOLFIRI (best observed response = CR or PR). FPs were defined as those patients with KRAS wild-type tumours who did not have a positive response to treatment with cetuximab plus FOLFOX6 or cetuximab plus FOLFIRI (SD or PD). FNs were defined as those with KRAS mutant tumours who had a positive response to treatment with cetuximab plus FOLFOX6 or cetuximab plus FOLFIRI. TNs were defined as those with KRAS mutant tumours who did not have a positive response to treatment with cetuximab plus FOLFOX6 or cetuximab plus FOLFIRI. Full definitions of CR, PR, SD and PD are provided in Chapter 2 (see Measuring response to treatment). The publication of the results of the CELIM trial included resection rates for liver metastases; however, these data were reported for all participants and by treatment group (cetuximab plus FOLFOX6 or cetuximab plus FOLFIRI) only and not by tumour KRAS mutation status. 52 For all study participants, the R0 resection rate was 36/106 (34%, 95% CI 25% to 44%) and the R0/R1 resection and/or radiofrequency ablation rate was 49/106 (46%, 95% CI 36% to 56%). 52 (Commercial-in-confidence information has been removed.)59 All participants in the CELIM trial received treatment with cetuximab in addition to standard chemotherapy; therefore, this trial could not contribute data to question 3 on how outcomes from treatment with cetuximab plus standard chemotherapy vary according to which test is used to select patients for treatment.
Additional data, supplied by the COIN trial investigators,54 allowed calculation of the accuracy of KRAS mutation testing, using a combination of pyrosequencing and MALDI-TOF and targeting mutations in codons 12, 13 and 61, for predicting response to treatment in patients with colorectal liver metastases who are treated with cetuximab plus FOLFOX or XELOX. These data could be viewed as being of limited applicability to this assessment because the standard chemotherapy regimen used in the COIN trial does not exactly match that in our inclusion criteria (some participants received XELOX rather than FOLFOX or FOLFIRI). However, the additional data supplied did allow the calculation of accuracy with respect to prediction of the more clinically relevant outcome of potentially curative resection. In this case we defined TPs as those patients with KRAS wild-type tumours who had a potentially curative resection following treatment with cetuximab plus FOLFOX or cetuximab plus XELOX. FPs were defined as those patients with KRAS wild-type tumours who did not have a potentially curative resection following treatment with cetuximab plus FOLFOX or cetuximab plus XELOX. FNs were defined as those with KRAS mutant tumours who had a potentially curative resection following treatment with cetuximab plus FOLFOX or cetuximab plus XELOX. TNs were defined as those with KRAS mutant tumours who did not have a potentially curative resection following treatment with cetuximab plus FOLFOX or cetuximab plus XELOX.
Study details
Participants in the CELIM trial52 all had unresectable colorectal liver metastases with no extrahepatic metastases. Non-resectability was defined as five or more liver metastases or metastases that were viewed as technically non-resectable by a liver surgeon and a radiologist on the basis of inadequate future remnant, infiltration of all hepatic liver veins, infiltration of both hepatic arteries or infiltration of both portal veins. Study participants had a median age of 64 years [interquartile range (IQR) 56 to 71] and 64% were male. The primary tumour site was the colon in 61 (55%) participants and the rectum in 49 (44%) participants; the primary site was unknown in one participant. Most patients (83%) had a primary tumour of stage T3/4. The primary reason for non-resectability of liver metastases was given as ‘technically unresectable’ for 61 (55%) participants and ‘five or more metastases’ for 50 (45%) participants. Full details of the study participants are reported in Appendix 2.
KRAS mutation testing used an older version of the Therascreen KRAS RGQ PCR Kit, which is identical to the current Therascreen KRAS RGQ PCR kit in terms of mutations targeted. Both versions of the kit detected seven mutations in codons 12 and 13 (Table 7) and the two versions will be treated as equivalent for the purpose of this assessment.
| Codon | Coding DNA | Protein/amino acid, three-letter code | Protein/amino acid, one-letter code |
|---|---|---|---|
| 12 | c.34G > A | p.Gly12Ser | p.G12S |
| c.34G > C | p.Gly12Arg | p.G12R | |
| c.34G > T | p.Gly12Cys | p.G12C | |
| c.35G > A | p.Gly12Asp | p.G12D | |
| c.35G > C | p.Gly12Ala | p.G12A | |
| c.35G > T | p.Gly12Val | p.G12V | |
| 13 | c.38G > A | p.Gly13Asp | p.G13D |
Tumour response was assessed according to the RECIST criteria38 to evaluate response to EGFR inhibitor treatment; response was defined as the best response to treatment with cetuximab plus FOLFOX6 or cetuximab plus FOLFIRI observed during treatment and was assessed every four cycles (8 weeks). Post-treatment surgical review, to assess resectability, was undertaken after eight cycles of chemotherapy by senior surgeons with experience in hepatobiliary surgery; CT and MRI scans were presented by a radiologist and the surgeons were blinded to when the scans were carried out and the participants’ clinical outcome data. 52
Details of the COIN trial54 are provided in ‘How do outcomes from treatment with cetuximab plus standard chemotherapy vary according to which test is used to select patients for treatment?’
KRAS mutation test accuracy
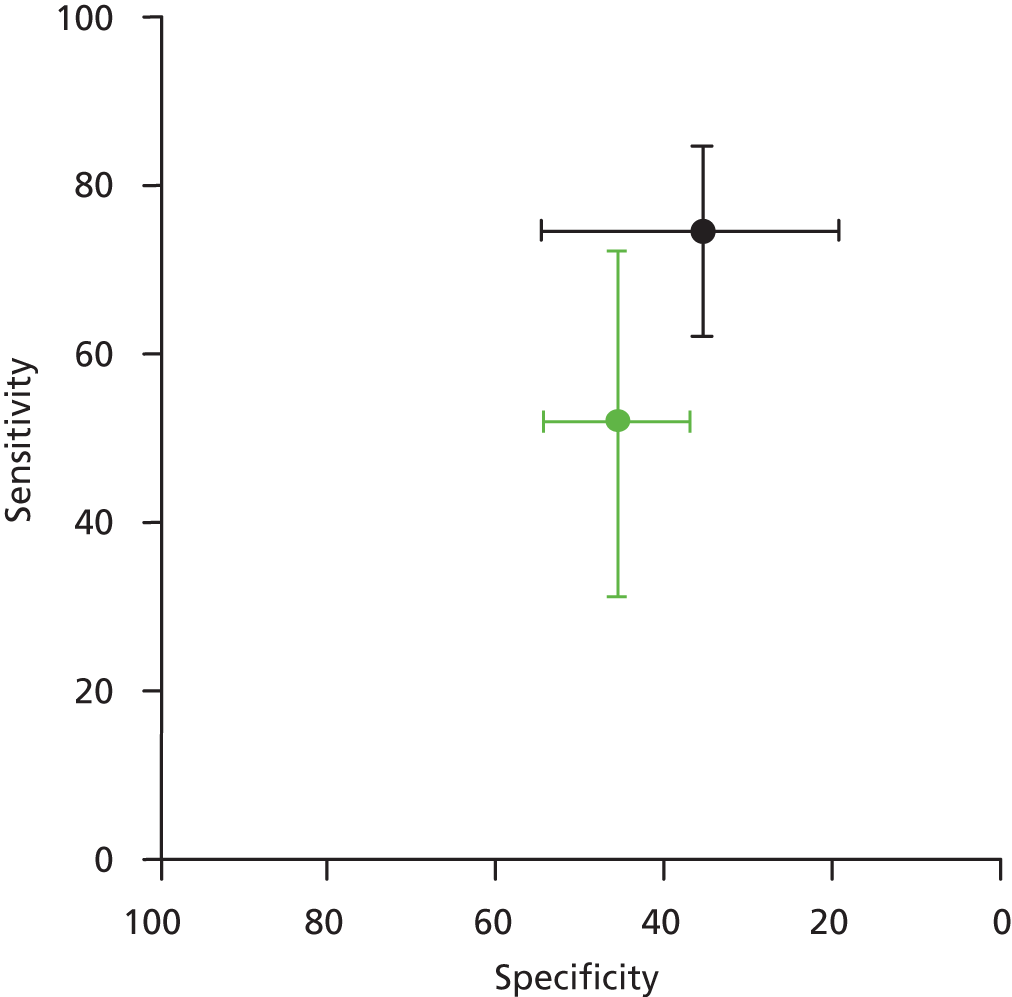
Data from the CELIM trial52 provided estimates for the accuracy of the Therascreen KRAS RGQ PCR Kit for discriminating between patients who are likely to benefit from addition of cetuximab to standard chemotherapy regimens and those who are not. Sensitivity for the prediction of objective response was moderate (74.6%, 95% CI 62.1% to 84.7%) and specificity was poor (35.5%, 95% CI 19.2% to 54.6%) (Table 8 and Figure 4). Additional data supplied by the COIN trial investigators allowed the calculation of estimates for the accuracy of pyrosequencing and MALDI-TOF, targeting mutations in codons 12, 13 and 61, for predicting potentially curative resection following treatment with cetuximab plus FOLFOX or XELOX (D Fisher, personal communication). Sensitivity and specificity were both poor at 52.0% (95% CI 31.3% to 72.2%) and 45.6% (95% CI 37.0% to 54.3%) respectively (see Table 8 and Figure 4).
| Study | KRAS mutation test method and mutations targeted | Non-evaluable samples | Reference standard | TP | FP | FN | TN | Sensitivity (95% CI) (%) | Specificity (95% CI) (%) | Prevalence (%) | PPV (95% CI) (%) | NPV (95% CI) (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Folprecht 201052 (CELIM) | Therascreen KRAS RGQ PCR Kit | 12/111 unknown mutation status (KRAS mutation testing not carried out successfully, no reasons reported) | Objective response | 47 | 20 | 16 | 11 | 74.6 (62.1 to 84.7) | 35.5 (19.2 to 54.6) | 67 | 70.1 (58.3 to 79.8) | 40.7 (24.5 to 59.3) |
| COINa | Pyrosequencing and MALDI-TOF (codons 12, 13 and 61) | Resection rate | 13 | 74 | 12 | 62 | 52.0 (31.3 to 72.2) | 45.6 (37.0 to 54.3) | 16 | 14.9 (8.9 to 23.9) | 83.9 (73.8 to 90.5) |
FIGURE 4.
Summary receiver operating characteristic plot showing estimates of sensitivity and specificity together with 95% CIs from the CELIM trial52 (black lines) and the COIN trial (green lines) (D FIsher, personal communication).
Quality Assessment of Diagnostic Accuracy Studies assessment
A summary of the results of the QUADAS-2 assessments for the CELIM and COIN trails are presented in Table 9 and the full assessments are reported in Appendix 3. The rating of high concern regarding the applicability of the reference standard reflects the absence of data on the ability of the test (KRAS mutation status) to predict resection of liver metastases following treatment with cetuximab plus FOLFOX6 or cetuximab plus FOLFIRI for the CELIM trial, and the use of standard chemotherapy in the COIN trial that did not fully match that in the inclusion criteria for this assessment. In addition, participants in the CELIM trial were described as having technically non-resectable or five or more liver metastases from CRC and it was therefore unclear whether some participants may have had potentially resectable metastases at baseline. 52 Both studies were rated as being at high risk of bias with respect to flow and timing because approximately 15% of participants were excluded from the analyses, in most cases because they were not evaluable for response.
How do outcomes from treatment with cetuximab plus standard chemotherapy vary according to which test is used to select patients for treatment?
Four RCTs (six publications)27,28,53–56 provided data on the clinical effectiveness of cetuximab plus standard chemotherapy compared with standard chemotherapy alone in patients with colorectal liver metastases and no extrahepatic metastases whose tumours were KRAS wild type. The trials compared cetuximab in combination with standard chemotherapy (FOLFOX or FOLFIRI) with standard chemotherapy alone (see Table 11); in one trial54 standard chemotherapy could be either FOLFOX or XELOX. One trial55 included only participants with unresectable colorectal liver metastases and no extrahepatic metastases whose tumours were KRAS wild type. This trial was reported as a conference abstract only and some additional information on this trial was derived from the trial registry entry. 60 The remaining three trials, CRYSTAL,27,53 OPUS28,53,56 and COIN54 (five publications), included participants with mCRC, conducted tumour KRAS mutation testing in a subgroup of these participants and reported data for a smaller subgroup of participants whose metastases were confined to the liver; in all cases outcome data on participants whose metastases were confined to the liver were reported only for those with KRAS wild-type tumours.
Study details
Participant characteristics varied across studies. The three studies that reported subgroup data for patients with colorectal metastases confined to the liver were multicentre studies conducted in continental Europe27,28,53,56 or the UK and the Republic of Ireland. 54 The subgroup data taken from these studies represented between 11% and 14% of the total study population (see Table 11). None of the studies reported separate participant characteristics for the relevant subgroup and none reported the criteria used to define unresectable liver metastases. For the larger KRAS wild-type subgroup, study participants were similar across the three studies. The median age of study participants was 61–62 years and 54–68% of participants were male. More than 90% of participants in all three studies had an Eastern Cooperative Oncology Group (ECOG) or a WHO performance status of 0 or 1 and two27,53,54 out of three studies included only participants with histologically confirmed adenocarcinoma. The trial that included only participants with unresectable colorectal liver metastases and no extrahepatic metastases whose tumours were KRAS wild type was reported as an abstract only and did not provide any further details of participant characteristics. 55 The trial registry entry specified histologically confirmed adenocarcinoma and ECOG performance status of 0 or 1 as inclusion criteria. 60 Full details of study participants are reported in Appendix 2.
The included trials used various methods to assess KRAS mutation status. The CRYSTAL27 and OPUS28 trials both used the LightMix k-ras Gly12 assay (TIB MOLBIOL). PCR reactions were performed on a LightCycler 2.0 system using a KRAS mutation detection-specific program. The LightMix k-ras Gly12 assay detects nine mutations in codons 12 and 13 (Table 10).
| Codon | Coding DNA | Protein/amino acid, three-letter code | Protein/amino acid, one-letter code |
|---|---|---|---|
| 12 | c.34G > A | p.Gly12Ser | p.G12S |
| c.34G > C | p.Gly12Arg | p.G12R | |
| c.34G > T | p.Gly12Cys | p.G12C | |
| c.35G > A | p.Gly12Asp | p.G12D | |
| c.35G > C | p.Gly12Ala | p.G12A | |
| c.35G > T | p.Gly12Val | p.G12V | |
| c.[34G > A; 35G > C] | p.Gly12Thr | p.G12T | |
| 13 | c.37G > T | p.Gly12Cys | p.G13C |
| c.38G > A | p.Gly13Asp | p.G13D |
The COIN trial54 used pyrosequencing of KRAS codons 12, 13 (amplification primers 5′-GGCCTGCTGAAAATGACTGA-3′ and 5′-AGAATGGTCCTGCACCAGTAATA-3′ and extension primers 5′-TGTGGTAGTTGGAGCTG-3′, 5′-TGTGGTAGTTGGAGCT-3′ and 5′-TGGTAGTTGGAGCTGGT-3′) and 61 (amplification primers 5′-CTTTGGAGCAGGAACAATGTC-3′ and 5′-CTCATGTACTGGTCCCTCATTG-3′ and extension primer 5′-ATTCTCGACACAGCAGGT-3′) together with MALDI-TOF mass spectrometry. The MALDI-TOF genotyping assay was designed using the Sequenom MassARRAY Assay Design 3.1 software (Sequenom Inc., San Diego, CA, USA) and 200 base pairs of sequence upstream and downstream of each known mutation (known mutations taken from the Catalogue of Somatic Mutations in Cancer (COSMIC) database; see www.sanger.ac.uk/genetics/CGP/cosmic). For discordant results (< 1%), Sanger sequencing of KRAS codons 12, 13 (primers 5′-AAAAGGTACTGGTGGAGTATTTGA-3′ and 5′-CATGAAAATGGTCAGAGAAACC-3′) and 61 (primers 5′-CTTTGGAGCAGGAACAATGTC-3′ and 5′-CTCATGTACTGGTCCCTCATTG-3) was undertaken. The final trial55 used pyrosequencing to identify mutations in KRAS codons 12 and 13 (information supplied in personal communication from the study author).
All four trials reported data on R0 resection rates in patients with colorectal metastases limited to the liver and KRAS wild-type tumours; for the CRYSTAL and OPUS trials these data were reported only in a conference abstract. 53 Three of the four trials also reported ORR. 28,53,55 Two trials, CRYSTAL27 and OPUS,56 used modified WHO criteria to assess tumour response, the COIN trial54 used RECIST criteria38 and the final trial55 did not specify criteria for assessing tumour response. None of the three trials that reported subgroup data for participants whose metastases were limited to the liver reported data on how resectability of liver metastases was assessed post treatment. 27,28,54 The trial registry entry for the study that included only patients with unresectable liver metastases whose metastases were confined to the liver stated that post-treatment resectability would be assessed after 4–12 cycles by a MDT of more than three liver surgeons and one radiologist, using CT and MRI images. 60 Two studies reported PFS in the relevant patient group28,54 and some limited data were also reported for overall survival (OS).
Clinical outcomes in patients with colorectal metastases limited to the liver and KRAS wild-type tumours who were treated with cetuximab plus standard chemotherapy compared with clinical outcomes in those treated with standard chemotherapy
All studies in this section reported that the addition of cetuximab to standard chemotherapy was associated with an increase in the rate of R0 resections (Table 11); however, this increase reached statistical significance only in the trial by Xu et al. 55 (OR 4.57, 95% CI 1.56 to 13.34). All three studies that assessed ORR reported a statistically significant higher response rate for participants treated with cetuximab plus standard chemotherapy than for those treated with standard chemotherapy alone; ORs ranged from 3.00 (95% CI 1.49 to 6.03)53 to 4.93 (95% CI 1.42 to 17.06). 28 Only the COIN trial54 reported an improvement in PFS associated with the addition of cetuximab to standard chemotherapy (HR 0.68, 95% CI 0.48 to 0.97). The study by Xu et al. 55 reported a significant improvement in 3-year survival rates for participants treated with cetuximab plus standard chemotherapy compared with those treated with standard chemotherapy alone (OR 2.76, 95% CI 1.12 to 6.26). There were no clear differences in treatment effect regardless of which KRAS mutation test was used to identify participants whose tumours were KRAS wild type (Figures 5–7). The median PFS for participants with KRAS wild-type tumours who were treated with cetuximab plus standard chemotherapy was 11.8 months in the CRYSTAL trial and 11.9 months in the OPUS trial; the corresponding PFS values in the standard chemotherapy groups were 9.2 months and 7.9 months. 53 The median OS for participants with KRAS wild-type tumours who were treated with cetuximab plus standard chemotherapy was 27.8 months in the CRYSTAL trial and 26.3 months in the OPUS trial; the corresponding OS values in the standard chemotherapy groups were 27.7 months and 23.9 months. 53
| Study | KRAS test (mutations targeted) | Participant details | Intervention | Comparator | Outcome | Effect estimate (95% CI) |
|---|---|---|---|---|---|---|
| Bokemeyer 201128,53,56 (OPUS) | LightMix k-ras Gly12 assay (KRAS codons 12 and 13 missense mutations) | n = 337 KRAS wild type 179; KRAS wild type with liver-limited metastases 48; KRAS mutation status unknown 22/337 (no reason reported) |
Cetuximab + FOLFOX-4 (n = 25) | FOLFOX-4 (n = 23) | PFS | HR 0.64 (0.23 to 1.79) |
| ORR | OR 4.93 (1.42 to 17.06)a | |||||
| R0R | OR 4.19 (0.43 to 40.62)a | |||||
| Van Cutsem 200927,53 (CRYSTAL) | LightMix k-ras Gly12 assay (KRAS codons 12 and 13 missense mutations) | n = 1198 KRAS wild type 348; KRAS wild type with liver-limited metastases 140; KRAS mutation status could not be evaluated 658/1198 (no reason reported) |
Cetuximab + FOLFIRI (n = 68) | FOLFIRI (n = 72) | ORR | OR 3.00 (1.49 to 6.03)a |
| R0R | OR 2.59 (0.76 to 8.86)a | |||||
| Maughan 201154 (COIN) | Pyrosequencing and MALDI-TOF mass array with Sanger sequencing for discordant samples (< 1%) (KRAS mutations in codons 12, 13 and 61) | n = 1630 KRAS wild type 729; KRAS wild type with liver-limited metastases 178; KRAS mutation status unknown 336/1630 (141 tumour blocks not available, 173 blocks contained insufficent tumour material for processing, 22 not successfully genotyped) |
Cetuximab + FOLFOX or XELOX (n = 87) | FOLFOX or XELOX (n = 91) | PFS | HR 0.68 (0.48 to 0.97) |
| R0R | OR 1.16 (0.50 to 2.70)a | |||||
| Xu 201255 | Pyrosequencing (KRAS mutations in codons 12 and 13) | n = 116 KRAS wild type 116; KRAS wild type with liver-limited metastases 116 |
Cetuximab + FOLFIRI or FOLFOX6 (n = 59) | FOLFIRI or FOLFOX6 (n = 57) | OSR | OR 2.76 (1.12 to 6.26)a |
| ORR | OR 3.90 (1.80 to 8.43)a | |||||
| R0R | OR 4.57 (1.56 to 13.34)a |
FIGURE 5.
Progression-free survival in patients with colorectal metastases limited to the liver and KRAS wild-type tumours who were treated with cetuximab plus standard chemotherapy compared with those treated with standard chemotherapy.
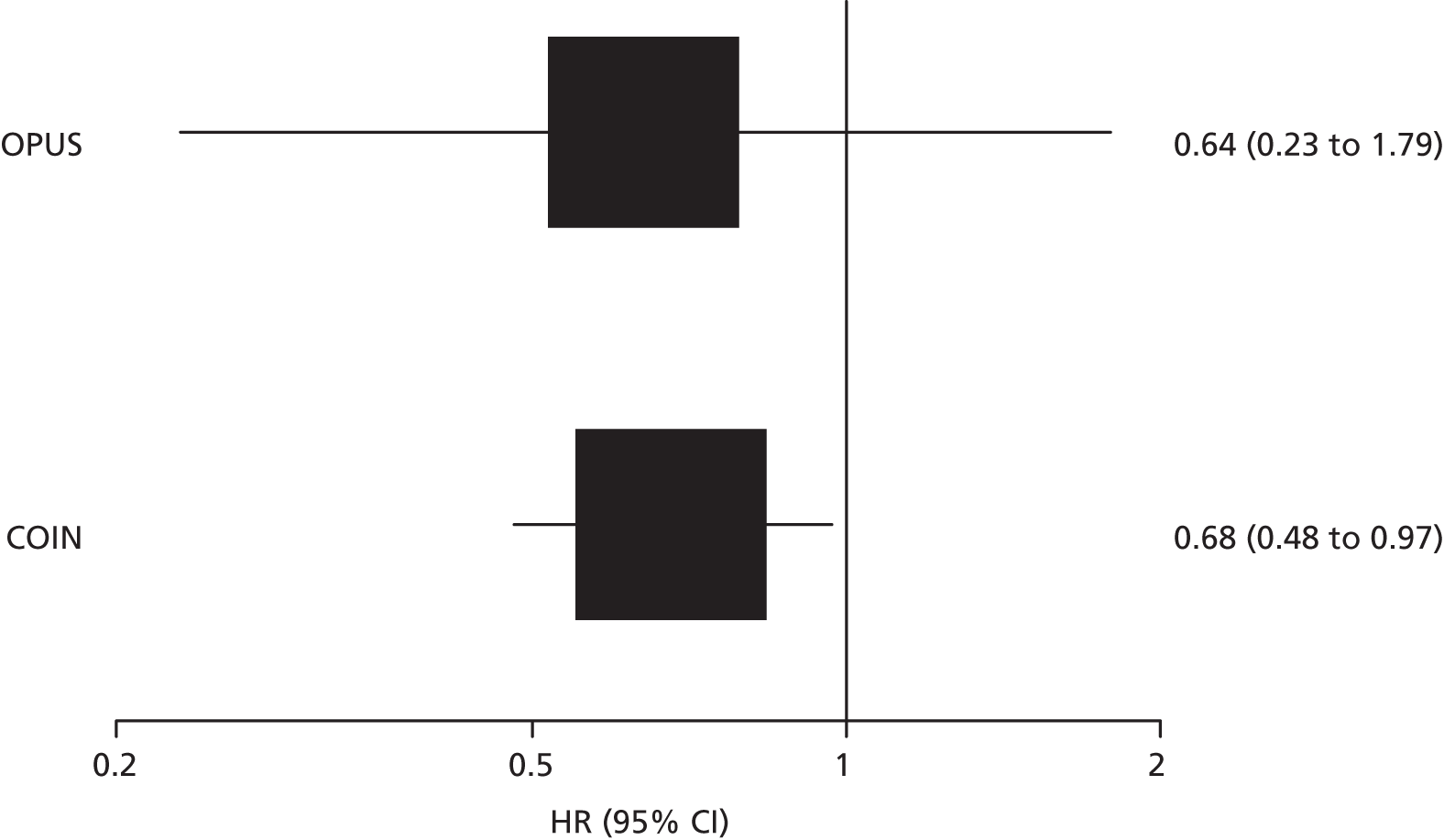
FIGURE 6.
Objective response in patients with colorectal metastases limited to the liver and KRAS wild-type tumours who were treated with cetuximab plus standard chemotherapy compared with those treated with standard chemotherapy.
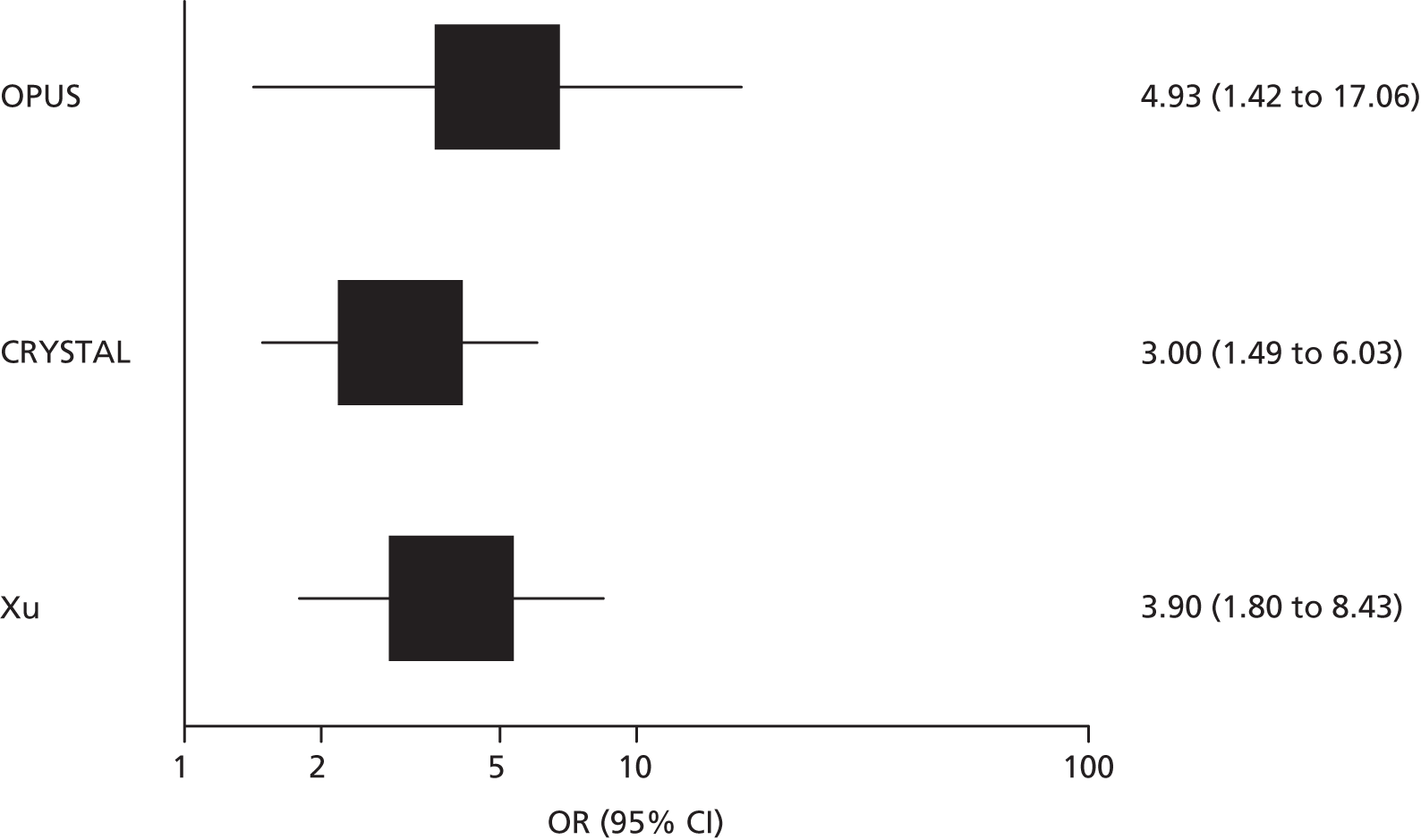
FIGURE 7.
R0 resection rate in patients with colorectal metastases limited to the liver and KRAS wild-type tumours who were treated with cetuximab plus standard chemotherapy compared with those treated with standard chemotherapy.
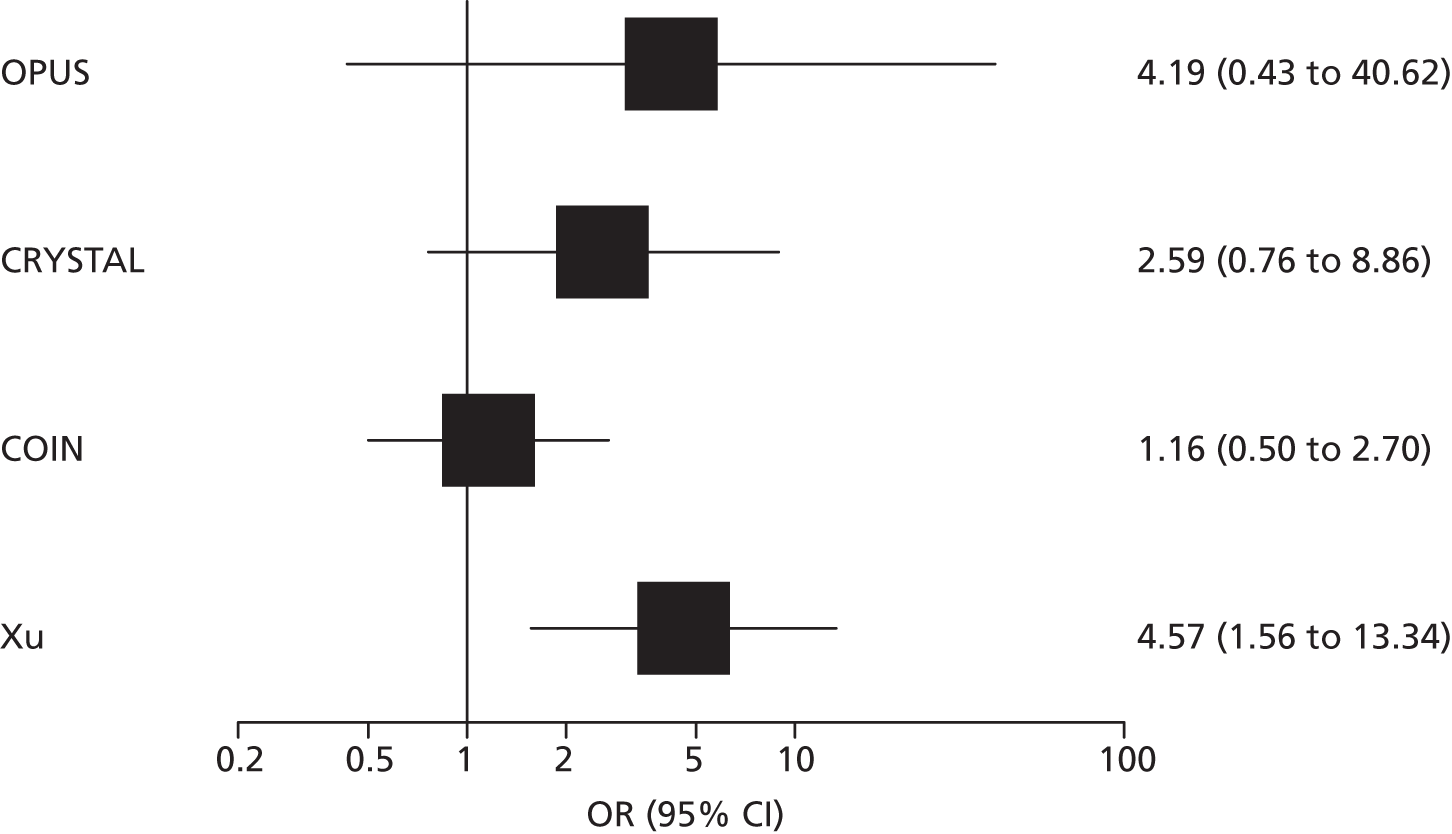
Clinical outcome for studies that provided data for patients according to KRAS mutation test status
Data from the COIN trial54 indicated that there was a slight increase in PFS for patients with initially unresectable liver metastases whose tumours were KRAS wild type who received cetuximab plus FOLFOX or XELOX compared with those who received FOLFOX or XELOX alone (HR 0.48, 95% CI 0.48 to 0.97). Additional data supplied by the COIN trial investigators indicated that, for patients with initially unresectable liver metastases whose tumours were KRAS mutant, there was no significant difference in PFS between the two treatment groups (HR 1.19, 95% CI 0.80 to 1.77). The reported rate of potentially curative resection in patients whose tumours were KRAS wild type was 15% (13/87) for the cetuximab plus FOLFOX or XELOX group and 13% (12/91) for the FOLFOX or XELOX only group. 54 The COIN trial investigators provided additional data for patients whose tumours were KRAS mutant; in these patients the potentially curative resection rate was 16% (12/74) for the cetuximab plus FOLFOX or XELOX group and 14% (6/44) for the FOLFOX or XELOX only group.
Risk of bias
All studies in this section were rated as having a low or unclear risk of bias for randomisation, incomplete outcome data and selective outcome reporting. All three of the trials that were reported as full papers stated that effectiveness analyses were conducted on an intention-to-treat (ITT) basis. 27,28,54 Details of allocation concealment were generally not reported, with the exception of the COIN trial,54 which stated that treatment allocation was not masked. All studies were rated as having a high risk of bias for blinding of study participants and personnel; all were open-label studies. However, two studies55,56 were rated as having a low risk of bias for blinding of outcome assessors as they reported some independent/blinded assessment of outcomes. The results of the risk of bias assessments are summarised in Table 12 and a full risk of bias assessment for each study is provided in Appendix 3.
| Study | Risk of bias | |||||
|---|---|---|---|---|---|---|
| Randomisation | Allocation concealment | Participant and personnel blinding | Outcome assessor blinding | Incomplete outcome data | Selective outcome reporting | |
| Bokemeyer 201128,56 (OPUS) | ? | ? | ☹ | ☺ | ☺ | ☺ |
| Van Cutsem 200927,53 (CRYSTAL) | ? | ? | ☹ | ? | ☺ | ☺ |
| Maughan 201154 (COIN) | ☺ | ☹ | ☹ | ? | ☺ | ☺ |
| Xu 201255,60 | ? | ? | ☹ | ? | ? | ? |
Chapter 4 Assessment of cost-effectiveness
This chapter explores the cost-effectiveness of the use of different KRAS mutation tests to decide between standard chemotherapy and cetuximab in combination with standard chemotherapy in adults with mCRC in whom metastases are confined to the liver and are unresectable.
Review of economic analyses of KRAS mutation testing
Search strategy
Searches were undertaken to identify cost-effectiveness studies of KRAS mutation testing in mCRC. As with the clinical effectiveness searching, the main EMBASE strategy for each set of searches was independently peer reviewed by a second information specialist using the PRESS-EBC. 43 Search strategies were developed specifically for each database and searches took into account generic and other product names for the intervention. All search strategies are reported in Appendix 1.
The following databases were searched for relevant studies from 2000 to January 2013:
-
MEDLINE (OvidSP) (2000 to Week 3 January 2013)
-
MEDLINE In-Process and Other Non-Indexed Citations and Daily Update (OvidSP) (up to 28 January 2013)
-
EMBASE (OvidSP) (2000 to Week 4 2013)
-
NHS Economic Evaluation Database (NHS EED) (Wiley) (The Cochrane Library 2000 to Issue 4, 2012)
-
Health Economic Evaluations Database (HEED) (Wiley) (up to 30 January 2013), http://onlinelibrary.wiley.com/book/10.1002/9780470510933
-
EconLit (EBSCO) (2000 to 30 January 2013)
-
Science Citation Index (SCI-EXPANDED) (Web of Science) (2000 to 25 January 2013).
Inclusion criteria
Studies reporting a full economic analysis that related explicitly to the test–treat combination of KRAS mutation testing and treatment with cetuximab were eligible for inclusion. Specifically, one of the comparators included KRAS mutation testing and for this comparator the treatment decision was guided by the test result; patients whose tumour was KRAS mutant were also included in the treatment pathway.
Results
The search retrieved 445 references. Following title and abstract screening, 416 references were excluded. Of the remaining 29 titles, the full papers were screened, which led us to exclude another 24, leaving five references: one HTA report (from Ontario)61 and four papers. 62–65 A summary of the included studies is provided in Table 13 with a quality checklist based on Drummond et al. 66 provided in Table 14.
| Study details | Health Quality Ontario61 |
|---|---|
| Population | Patients diagnosed with (stage IV) mCRC for whom cetuximab or panitumumab monotherapies or cetuximab and irinotecan combination therapy were indicated as third-line treatment |
| Time horizon | Lifetime |
| Objective | To determine the cost-effectiveness of KRAS mutation testing for the third-line treatment of mCRC in Ontario |
| Source of effectiveness information | Survival, utility weights and adverse events taken from various studies |
| Comparators | (0) BSC; (1a) cetuximab (perform KRAS mutation test); (1b) cetuximab (no KRAS mutation test); (2a) panitumumab (perform KRAS mutation test); (2b) panitumumab (no KRAS mutation test); (3a) cetuximab + irinotecan (perform KRAS mutation test); (3b) cetuximab + irinotecan (no KRAS mutation test) |
| Unit costs | Taken from the literature and the 2009 Ontario Health Insurance Plan and Ontario Case Costing Initiative administrative databases |
| Measure of benefit | QALYs |
| Study type | Cost–utility analysis: Markov model |
| Model assumptions | None mentioned |
| Perspective | Ontario Ministry of Health and Long-Term Care |
| Discount rate | 5% for effects and costs |
| Uncertainty around cost-effectiveness ratio expressed | Only in text and only for the strategies for which PSA was performed. CEACs, etc., not shown as the report is very concise |
| Sensitivity analysis | PSA for strategies 0, 1a, 2a and 3a (i.e. does not include the ‘no KRAS mutation testing’ strategies). The PSA varied parameters only on a highly aggregated level: PFS, OS, total costs and utilities. It seems as if a uniform distribution was used for all of them? |
| Outcomes (cost and LYs/QALYs) per comparator | 0: C$1414, 0.7455; 2a: C$12,236, 0.9719; 1a C$18,305, 1.0537; 2b C$20,424, 0.9985; 3a C$23,373, 1.2596; 1b C$29,399, 1.0447; 3b C$44,798, 1.3907 |
| Summary of incremental analysis | For all strategies involving KRAS mutation testing, cetuximab with irinotecan combination therapy was the cost-effective option for increasing values of WTP. For lower WTP values, the probabilities of specific KRAS mutation testing strategies being cost-effective varied. At a WTP of C$50K, the probability of cetuximab monotherapy, panitumumab monotherapy and cetuximab with irinotecan combination therapy being cost-effective was approx. 14%, 44% and 42% respectively. The BSC strategy was not cost-effective (0% probability) for WTP values < C$45K |
| Study details | Vijayaraghavan et al.62 |
| Population | Patients with mCRC in whom previous chemotherapy had failed |
| Time horizon | Lifetime |
| Objective | To assess the cost-effectiveness of testing for KRAS mutations before administering EGFR inhibitors such as cetuximab and panitumumab in the USA and Germany |
| Source of effectiveness information | Three recently published studies on the efficacy of EGFR inhibitors in patients with and without KRAS mutations |
| Comparators | (1) Combination therapy (cetuximab + irinotecan/FOLFIRI) with KRAS mutation testing; (2) combination therapy (cetuximab + irinotecan/FOLFIRI) without KRAS mutation testing; (3) cetuximab alone with KRAS mutation testing; (4) cetuximab alone without KRAS mutation testing; (5) panitumumab alone with KRAS mutation testing; (6) panitumumab alone without KRAS mutation testing |
| Unit costs | For the US model, costs were taken from the Medicare fee schedule. For the German model, costs were taken from published literature and expert opinion |
| Measure of benefit | LYs |
| Study type | Cost-effectiveness analysis: Markov model |
| Model assumptions | (1) Patients with KRAS mutant tumours received no benefit from EGFR inhibitors; (2) patients with KRAS mutant tumours received some benefit from combination therapy containing FOLFIRI or irinotecan; (3) KRAS mutation testing has a sensitivity of 95% and a specificity of 100% |
| Perspective | Health-care payer perspective |
| Discount rate | None mentioned |
| Uncertainty around cost-effectiveness ratio expressed | No, only uncertainty around cost-savings presented by means of one-way sensitivity analyses |
| Sensitivity analysis | One-way sensitivity analyses varying percentage of patients with KRAS wild-type tumours, cost of cetuximab, cost of BSC and cost of KRAS mutation test |
| Outcomes (cost and LYs/QALYs) per comparator | Panitumumab with KRAS mutation testing: LYs 18.26, €13,787, US$19,656; panitumumab without KRAS mutation testing: LYs 18.26, €18,399, US$27,202; cetuximab with KRAS mutation testing: LYs 19.78, €13,588, US$22,893; cetuximab without KRAS testing: LYs 19.78, €17,444, US$30,933; combination therapy 1 with KRAS mutation testing: LYs 24.26, €26,292, US$35,075; combination therapy 2 with KRAS mutation testing: LYs 25.83, €–, US$36,148; combination therapy without KRAS mutation testing: LYs 25.83, €35,852, US$48,576 |
| Summary of incremental analysis | KRAS mutation testing to select patients eligible for EGFR inhibitors is cost-saving at equivalent clinical outcome |
| Study details | Behl et al.63 |
| Population | Patients with mCRC who are chemorefractory |
| Time horizon | 10 years |
| Objective | To assess the cost-effectiveness of screening for KRAS and BRAF mutations before EGFR inhibitor treatment |
| Source of effectiveness information | RCTs |
| Comparators | (1) No anti-EGFR therapy (BSC); (2) anti-EGFR therapy without screening; (3) screening for KRAS mutations only (before providing anti-EGFR therapy); (4) screening for KRAS and BRAF mutations (before providing anti-EGFR therapy) |
| Unit costs | For cost of chemotherapy, average selling price plus median Medicare average payment for physician services for administration of the chemotherapies. Total costs also include cost of liver resection(s) |
| Measure of benefit | LYs |
| Study type | Cost-effectiveness analysis: Markov model |
| Model assumptions | Many small assumptions |
| Perspective | Not specified: probably US third-party payer |
| Discount rate | 3% for costs and effects |
| Uncertainty around cost-effectiveness ratio expressed | Yes, with scatter plots, acceptability curves and frontier |
| Sensitivity analysis | (1) One way sensitivity analyses: conversion probability for chemotherapy is 30%; conversion probability for bevacizumab is +10%, cetuximab is +20%; cost of surgery is +50%; cost of screening is (a) +50% and (b) –50%; prognostic decrease in OS with BRAF mutation (regardless of treatment). (2) Cohort simulation: a cohort of 50,000 patients is analysed 10,000 times. No PSA mentioned |
| Outcomes (cost and LYs/QALYs) per comparator | No anti-EGFR therapy: US$34,291, LYs 0.6686; KRAS and BRAF mutation screening with anti-EGFR therapy: US$56,324, LYs 0.7025; KRAS screening with anti-EGFR therapy: US$57,348, LYs 0.7029; anti-EGFR therapy without screening: US$64,841, LY 0.7055 |
| Summary of incremental analysis | ICER (cost/LY) for KRAS and BRAF mutation screening compared with no anti-EGFR therapy was US$648,396. Other ICERS (KRAS mutation screening compared with KRAS and BRAF mutation screening, no screening compared with KRAS mutation screening) were > US$2M/LY gained |
| Study details | Shiroiwa et al.64 |
| Population | Japanese patients with mCRC in whom previous chemotherapy (including fluoropyrimidine, irinotecan and oxaliplatin) had failed or who had contraindications to these drugs |
| Time horizon | 2.5 years |
| Objective | To determine the cost-effectiveness of cetuximab treatment after KRAS mutation testing compared with BSC |
| Source of effectiveness information | PFS and OS were taken from the National Cancer Institute of Canada Clinical Trials Group CO.17 (NCIC CO.17) trial67 |
| Comparators | (1) KRAS testing strategy: patients with KRAS wild-type tumours received cetuximab and those with KRAS mutations received BSC; (2) no KRAS mutation testing strategy: all patients received cetuximab; (3) no cetuximab: all patients received BSC |
| Unit costs | Costs were calculated according to the social insurance reimbursement schedule and the drug tariff based on Japanese ‘fee for service’ |
| Measure of benefit | LYs and QALYs |
| Study type | Cost-effectiveness and cost–utility analysis: Markov model |
| Model assumptions | (1) Utility of PFS was assumed to be 0.7 for all treatments (cetuximab and BSC); (2) 40% of patients were assumed to have KRAS mutant tumours |
| Perspective | Health-care payer’s perspective |
| Discount rate | 3% for both costs and effects |
| Uncertainty around cost-effectiveness ratio expressed | CEACs |
| Sensitivity analysis | One-way sensitivity analyses were performed for discount rates, body surface area, percentage of patients with KRAS mutant tumours, BSC costs, HR of cetuximab for wild-type patients and costs of KRAS mutation testing. A PSA was also performed |
| Outcomes (cost and LYs/QALYs) per comparator | KRAS mutation testing strategy: US$29,000, LYs 0.70, QALYs 0.49; no KRAS mutation testing strategy: US$35,000, LYs 0.69, QALYs 0.48; no cetuximab strategy: US$6800, LYs 0.52, QALYs 0.36 |
| Summary of incremental analysis | KRAS testing vs. no KRAS mutation testing: KRAS mutation testing dominant; KRAS mutation testing vs. no cetuximab: US$180,000 per QALY gained; no KRAS mutation testing vs. no cetuximab: US$230,000 per QALY gained |
| Study details | Blank et al.65 |
| Population | Patients with mCRC who are chemorefractory |
| Time horizon | Lifetime |
| Objective | To assess the cost-effectiveness of testing for KRAS and BRAF mutations before cetuximab treatment |
| Source of effectiveness information | NCIC CO.17 trial67 |
| Comparators | (1) KRAS mutation testing; (2) KRAS mutation testing with subsequent BRAF testing of KRAS wild-type patients (KRAS/BRAF); (3) cetuximab without testing. Comparison was against a reference strategy of no cetuximab treatment. In the testing strategies, cetuximab was administered if no mutations were detected |
| Unit costs | Unit costs were drawn from the official Swiss tariff list (Tarmed). Drug costs were based on official Swiss pharmacy prices |
| Measure of benefit | QALYs |
| Study type | Cost–utility analysis: Markov cohort simulation model |
| Model assumptions | 70% of patients were assumed to have KRAS wild-type tumours |
| Perspective | Swiss health-care system |
| Discount rate | 3% for both costs and effects |
| Uncertainty around cost-effectiveness ratio expressed | CEACs and cost-effectiveness acceptability frontier |
| Sensitivity analysis | One-way sensitivity analyses were performed for different values of utilities, sensitivity and specificity of mutation analyses, prevalence of KRAS and BRAF mutations and OS and PFS. A PSA was also performed |
| Outcomes (cost and LYs/QALYs) per comparator | Reference treatment (no cetuximab): €3983, QALYs 0.4430; KRAS and BRAF mutation testing: €34,771, QALYs 0.934; KRAS mutation testing: €35,361, QALYs 0.936; no testing: €38,662, QALYs 0.947 |
| Summary of incremental analysis | KRAS and BRAF mutation testing compared with reference strategy: €62,653 per QALY; KRAS mutation testing compared with KRAS and BRAF mutation testing: €313,537 per QALY; no testing compared with KRAS mutation testing: €314,588 per QALY |
| Health Quality Ontario61 | Vijayaraghavan et al.62 | Behl et al.63 | Shiroiwa et al.64 | Blank et al.65 | |
|---|---|---|---|---|---|
| Study design | |||||
| The research question is stated | ✓ | ✓ | ✓ | ✓ | ✓ |
| The economic importance of the research question is stated | ✗ | ✓ | ✓ | ✓ | ✓ |
| The viewpoint(s) of the analysis are clearly stated and justified | ✓ | ✓ | ✗ | ✓ | ✓ |
| The rationale for choosing alternative programmes or interventions compared is stated | ✓ | ✓ | ✓ | ✓ | ✓ |
| The alternatives being compared are clearly described | ✓ | ✓ | ✓ | ✓ | ✓ |
| The form of economic evaluation used is stated | ✓ | ✓ | ✓ | ✓ | ✓ |
| The choice of form of economic evaluation is justified in relation to the questions addressed | ✓ | ✓ | ✓ | ✓ | ✓ |
| Data collection | |||||
| The source(s) of effectiveness estimates used are stated | ✓ | ✓ | ✓ | ✓ | ✓ |
| Details of the design and results of the effectiveness study are given (if based on a single study) | ✓ | ✓ | ✓ | ✗ | ✓ |
| Details of the methods of synthesis or meta-analysis of estimates are given (if based on a synthesis of a number of effectiveness studies) | NA | NA | NA | NA | NA |
| The primary outcome measure(s) for the economic evaluation are clearly stated | ✓ | ✓ | ✓ | ✓ | ✓ |
| Methods to value benefits are stated | ✓ | ✓ | ✓ | ✓ | ✓ |
| Details of the subjects from whom valuations were obtained are given | ✗ | NA | NA | ✗ | ✗ |
| Productivity changes (if included) are reported separately | NA | NA | NA | NA | NA |
| The relevance of productivity changes to the study question is discussed | ✗ | ✗ | ✗ | ✗ | ✗ |
| Quantities of resource use are reported separately from their unit costs | ✓ | ✗ | ✗ | ✓ | ✗ |
| Methods for the estimation of quantities and unit costs are described | ✓ | ✓ | ✓ | ✓ | ✓ |
| Currency and price data are recorded | ✓ | ✓ | ✓ | ✓ | ✓ |
| Details of currency of price adjustments for inflation or currency conversion are given | ✓ | ✓ | ✓ | ✓ | ✓ |
| Details of any model used are given | ✓ | ✓ | ✓ | ✓ | ✓ |
| The choice of model used and the key parameters on which it is based are justified | ✓ | ✓ | ✓ | ✓ | ✓ |
| Analysis and interpretation of results | |||||
| Time horizon of costs and benefits is stated | ✓ | ✓ | ✓ | ✓ | ✓ |
| The discount rate(s) is stated | ✓ | ✗ | ✓ | ✓ | ✓ |
| The choice of discount rate(s) is justified | ✓ | ✗ | ✓ | ✓ | ✓ |
| An explanation is given if costs and benefits are not discounted | NA | ✗ | NA | NA | NA |
| Details of statistical tests and CIs are given for stochastic data | ✗ | ✗ | ✗ | ✗ | ✗ |
| The approach to sensitivity analysis is given | ✓ | ✓ | ✓ | ✓ | ✓ |
| The choice of variables for sensitivity analysis is justified | ✗ | ✓ | ✓ | ✗ | ✓ |
| The ranges over which the variables are varied are justified | ✗ | ✗ | ✗ | ✗ | ✓ |
| Relevant alternatives are compared | ✓ | ✓ | ✓ | ✓ | ✓ |
| Incremental analysis is reported | ✓ | ✓ | ✓ | ✓ | ✓ |
| Major outcomes are presented in a disaggregated as well as aggregated form | ✓ | ✓ | ✓ | ✓ | ✓ |
| The answer to the study question is given | ✓ | ✓ | ✓ | ✓ | ✓ |
| Conclusions follow from the data reported | ✓ | ✓ | ✓ | ✓ | ✓ |
| Conclusions are accompanied by the appropriate caveats | ✗ | ✓ | ✓ | ✓ | ✓ |
The Ontario HTA report61 aimed to determine the cost-effectiveness of KRAS mutation testing for the third-line treatment of (stage IV) mCRC in Ontario. For this purpose, seven strategies were compared:
-
0 best supportive care (BSC)
-
1(a) cetuximab with KRAS mutation testing
-
1(b) cetuximab without KRAS mutation testing
-
2(a) panitumumab with KRAS mutation testing
-
2(b) panitumumab without KRAS mutation testing
-
3(a) cetuximab plus irinotecan with KRAS mutation testing
-
3(b) cetuximab plus irinotecan without KRAS mutation testing.
In the strategies with KRAS mutation testing, only patients with wild-type KRAS tumours receive the therapy in question. In the strategies without KRAS mutation testing, all patients receive the therapy. A cost–utility analysis was performed by means of a Markov model with a lifetime time horizon. Inputs for PFS, OS, utility weights and adverse events were obtained from various clinical studies. Although a probabilistic sensitivity analysis (PSA) was performed for the BSC strategy and all KRAS mutation testing strategies, the information on uncertainty around the incremental cost-effectiveness ratio (ICER) presented in the report was limited to some percentages taken from the cost-effectiveness acceptability curve (CEAC) (which was not shown). Also, it appears that the parameters that were varied within the PSA were highly aggregated (PFS, OS, utility scores and total costs) and the distribution used was not mentioned (possibly uniform, as only a range was given). The deterministic results showed that, compared with BSC, all monotherapy strategies are either dominated or extendedly dominated. The ICER for cetuximab plus irinotecan with KRAS mutation testing compared with BSC is C$42,710. The ICER for cetuximab plus irinotecan without KRAS mutation testing compared with cetuximab plus irinotecan with KRAS mutation testing is C$163,396. The authors concluded that, although KRAS mutation testing is cost-effective for all strategies considered, it is not equally cost-effective for all treatment options.
Vijayaraghavan et al. 62 developed a Markov model to compare six hypothetical strategies for the second-line treatment of patients with mCRC who have failed previous chemotherapy:
-
combination therapy (cetuximab plus irinotecan/FOLFIRI) with KRAS mutation testing
-
combination therapy (cetuximab plus irinotecan/FOLFIRI) without KRAS mutation testing
-
cetuximab alone with KRAS mutation testing
-
cetuximab alone without KRAS mutation testing
-
panitumumab alone with KRAS mutation testing
-
panitumumab alone without KRAS mutation testing.
In treatment strategies without KRAS mutation testing, all patients received EGFR inhibitor-based chemotherapy, as did the patients with KRAS wild-type tumours in the treatment strategies with KRAS mutation testing. Patients with KRAS mutant tumours received chemotherapy without EGFR inhibitors or BSC. The model results were calculated for a situation in the USA as well as in Germany, with country-specific chemotherapy regimens and associated costs. Clinical effects were assumed to be the same for the USA and Germany and were based on published studies. In the results section the treatment strategies were compared as KRAS mutation testing compared with no KRAS mutation testing within a certain treatment regimen. For all of these comparisons KRAS mutation testing saved costs at equivalent clinical outcomes.
The cost-effectiveness of both KRAS and v-raf murine sarcoma viral oncogene homolog B (BRAF) mutation screening in patients with mCRC who are chemorefractory was the subject of a paper by Behl et al. 63 A decision-analytic model was developed comparing the following strategies:
-
KRAS plus BRAF mutation screening (before providing anti-EGFR therapy)
-
KRAS mutation screening (before providing anti-EGFR therapy)
-
anti-EGFR therapy (no screening)
-
no anti-EGRF therapy (no screening).
Inputs for the model were estimated using observations from RCTs. The model followed each patient for a maximum of 10 years. The no cetuximab strategy was least costly (US$34,291) but also least effective [0.6686 (LYs)], followed by the KRAS plus BRAF screening therapy, which offered more LYs (0.7025) but at a higher cost (US$56.324). Screening for KRAS mutations again added more LYs (0.7029) at a higher cost (US$57,348) but at an unfavourable ratio compared with KRAS plus BRAF mutation screening (ICER > US$2M). Finally, the anti-EGFR strategy – providing cetuximab to all patients without screening – was the most effective (0.7055 LYs) and the most expensive (US$64,841) strategy, which only proved cost-effective at a willingness to pay of > US$3M. Therefore, KRAS plus BRAF mutation screening appeared to be the most cost-effective option compared with no anti-EGFR therapy, but still had an ICER of US$648,396 per life-year gained.
Shiroiwa et al. 64 performed a cost-effectiveness analysis of KRAS testing in a Japanese population of patients with mCRC in whom previous chemotherapy had failed. The included strategies were KRAS mutation testing, no KRAS mutation testing (all patients receive cetuximab) and no cetuximab (all patients receive BSC). In the KRAS mutation testing strategy, patients with KRAS wild-type tumours received cetuximab whereas patients with KRAS mutant tumours received BSC. The analysis involved a three-state Markov model to estimate and extrapolate survival curves and treatment costs. Transition probabilities for disease progression and survival after disease progression were derived from the NCIC CO.17 trial. 67 The time horizon of the analysis was 2.5 years. As expected, the no cetuximab strategy was least costly (US$6800) and least effective [0.36 quality-adjusted life-years (QALYs)]. The no KRAS mutation testing strategy (all patients receive cetuximab) cost US$35,000 and resulted in 0.48 QALYs whereas the KRAS mutation testing strategy cost US$29,000 and resulted in 0.49 QALYs. Therefore, KRAS mutation testing is considered dominant compared with no KRAS mutation testing. The ICER for KRAS mutation testing compared with no cetuximab was US$180,000 per QALY. The authors concluded that, although KRAS mutation testing compared with no KRAS mutation testing could be considered dominant, the ICER for cetuximab treatment is too high, even if treatment is limited to patients with KRAS wild-type tumours.
Blank et al. 65 constructed a model comparing the cost-effectiveness of four strategies for chemorefractory patients with mCRC: KRAS mutation testing, KRAS mutation testing with subsequent BRAF mutation testing of KRAS wild-type tumours, cetuximab treatment without testing and the reference strategy of no cetuximab. In the testing strategies, cetuximab treatment was initiated if no mutations were detected. BSC was given to all patients. Survival times and utilities were derived from published RCTs. Costs were assessed from the perspective of the Swiss health system. Adding cetuximab to BSC increased costs considerably, but the increase in costs in the testing strategies was distinctly lower than that in the no testing strategy. The costs of mutation testing were overcompensated for by savings associated with the restriction of cetuximab administration. The least costly and least effective strategy was the reference strategy (no cetuximab). Testing for KRAS and BRAF mutations led to an ICER of €62,653 per QALY compared with the reference strategy. Testing for KRAS mutations only compared with testing for KRAS and BRAF mutations, as well as the no testing strategy compared with KRAS mutation testing, both had ICERs well above €300,000 per QALY. The authors concluded that testing for KRAS and BRAF mutations before cetuximab treatment of chemorefractory mCRC patients is clinically appropriate and economically favourable, despite high costs for predictive testing [i.e. given that cetuximab should be the next step in the treatment pathway, it is worthwhile to test for mutations first; it appears that the reference strategy (no cetuximab) was not included in this recommendation].
Based on all of these publications it can be said that, in general, although KRAS testing is obviously a more cost-effective option than administering cetuximab to all patients, the ICER of KRAS testing and treating with cetuximab only those patients with KRAS wild-type tumour status compared with treating all patients with standard chemotherapy alone seems rather high.
Model structure and methodology
KRAS mutation tests considered in the model
The health economic analysis will determine the cost-effectiveness of different methods for KRAS mutation testing to decide between standard chemotherapy or standard chemotherapy plus cetuximab in adults with mCRC and a resected or resectable primary tumour, whose metastases are confined to the liver and are unresectable but may become resectable after response to chemotherapy. Standard chemotherapy regimens considered include FOLFOX and FOLFIRI. A range of methods for KRAS mutation testing are currently used in NHS laboratories in England and Wales. Ideally, the performance of these tests would be assessed against an objective measure of the true presence/absence of a clinically relevant KRAS mutation (the ‘reference standard’). The comparative effectiveness of treatment (cetuximab plus chemotherapy vs. chemotherapy alone) conditional on the true or false presence/absence of the KRAS mutation could then be determined. However, each testing method targets a different range of mutations and has different limits of detection (the lowest proportion of mutation detectable in tumour cells) and the exact combination of mutation type and level that will provide optimal treatment selection remains unclear. For this reason, assessment of test performance based on comparison with a conventional ‘reference standard’ is currently not possible. In this situation, an alternative way to determine the relative value of diagnostic methods for KRAS mutation testing is to use studies that report on the comparative treatment effect in patients with different KRAS mutation status (positive, negative or unknown), as defined using different KRAS mutation tests. As outlined in the previous chapter, information on the accuracy of tests (either based on ORR or tumour resection rate) for distinguishing between patients with KRAS wild-type tumours and patients with KRAS mutant tumours with metastases confined to the liver was available only for the Therascreen KRAS RGQ PCR Kit52 and pyrosequencing and MALDI-TOF mass spectometry. 54 A major assumption underlying the use of these accuracy data in the health economic modelling is that differences in response rates and resection rates between the two trials are solely due to the use of different KRAS mutation tests.
In the COIN trial54 patients were tested using both pyrosequencing and the MALDI-TOF mass array, with a reported concordance of > 99%. It was therefore assumed that, for the economic evaluation, MALDI-TOF and pyrosequencing are equal, that is, all results reported for pyrosequencing also apply to MALDI-TOF. However, survey data were available only for pyrosequencing and therefore pyrosequencing data are reported in the results tables.
For all other KRAS mutation tests listed in the scope, no accuracy data were available. As a result, for the remaining tests it was only possible to make a comparison based on differences in technical performance and test costs retrieved from the online survey of NHS laboratories in England and Wales (see Chapter 3, What are the technical performance characteristics of the different KRAS mutation tests?), whilst assuming a prognostic value equal to pyrosequencing across all tests. The latter assumption was not based on evidence of equality but rather on the absence of any reliable evidence to model a difference in prognostic value for these tests.
Based on the information available, two analyses were performed:
-
‘Linked evidence’ analysis – for all tests for which information on accuracy was available. In this analysis the Therascreen KRAS RGQ PCR Kit was compared with pyrosequencing. For the Therascreen KRAS RGQ PCR Kit, accuracy based on ORRs was taken from the CELIM trial. 52 Resection rates for patients with a KRAS wild-type test result treated with cetuximab plus chemotherapy were also based on CELIM trial data (data not reported in the published study but provided as part of the submissions that informed NICE guidance TA1761) whereas resection rates for patients with KRAS mutant and unknown test results who were treated with chemotherapy alone were taken from the Groupe Coopérateur Multidisciplinaire en Oncologie (GERCOR) study68 as the CELIM trial did not contain a chemotherapy-only strategy. For pyrosequencing, both accuracy data (based on resection rates) and resection rates for cetuximab plus chemotherapy as well as chemotherapy alone were taken from the COIN trial54 and from additional data supplied by the COIN triallists. PFS and OS after successful resection were assumed to be conditional on resection and treatment independent.
-
‘Assumption of equal prognostic value’ analysis – for all tests for which information on technical performance was available from the online survey. In this analysis we assessed whether the tests were likely to be cost-effective given an assumption of equal prognostic value and test-specific information on failure rate only. The equal prognostic value assigned was based on data for the pyrosequencing test (as this was the only test for which accuracy data were available on resection rates following treatment with chemotherapy, with and without cetuximab, for patients with initially inoperable liver metastases and both KRAS mutant and KRAS wild-type tumours). The following tests were included in this analysis:
-
cobas KRAS Mutation Test Kit (Roche Molecular Systems)
-
Therascreen KRAS RGQ PCR Kit (QIAGEN)
-
Therascreen KRAS Pyro Kit (QIAGEN)
-
KRAS LightMix Kit (TIB MOLBIOL)
-
KRAS StripAssay (ViennaLab)
-
HRM analysis
-
pyrosequencing
-
MALDI-TOF mass spectrometry
-
next-generation sequencing
-
Sanger sequencing.
-
Consistency with related assessments
This assessment does not update the appraisal of cetuximab for the first-line treatment of mCRC. 1 To ensure consistency between the modelling approach used in technology appraisal TA1761 and the assessment of the cost-effectiveness of different methods for KRAS mutation testing in this report, the assessment group received the electronic health economic model submitted by Merck Serono for technology appraisal TA176. This model calculates the expected cost-effectiveness of chemotherapy plus cetuximab compared with chemotherapy for the first-line treatment of mCRC patients whose metastases are confined to the liver and are unresectable and whose tumours are KRAS wild type as tested with a pre-CE-marked version of the KRAS LightMix Kit.
This model, together with the amendments suggested and made by the Evidence Review Group and NICE, was used to inform the development of a de novo model in which the long-term consequences of using different KRAS mutation tests were assessed not only in patients with KRAS wild-type tumours but also in patients with KRAS mutant tumours or an unknown test result.
Model structure
In the health economic model the mean expected costs and QALYs were calculated for each alternative. As specified in the protocol, this economic evaluation takes a ‘no comparator’ approach, which implies that the cost-effectiveness of each strategy will be presented compared with the next most cost-effective strategy.
The health economic analysis considers the long-term consequences of the technical performance and accuracy of the different tests followed by treatment with cetuximab plus standard chemotherapy or standard chemotherapy alone in patients (average starting age 60 years) with mCRC whose metastases are confined to the liver and are unresectable. For this purpose a decision tree and a Markov model were developed. The decision tree was used to model the test result (KRAS wild type, KRAS mutant or unknown) and the accompanying treatment decision. In the model, patients with a KRAS wild-type tumour receive cetuximab plus standard chemotherapy. It is assumed that patients with a KRAS mutant tumour will receive standard chemotherapy (i.e. FOLFOX). Patients with an unknown KRAS status are also assumed to receive standard chemotherapy as cetuximab is indicated only for patients with KRAS wild-type tumour status. 1 The decision tree is shown in Figure 8.
FIGURE 8.
Decision tree structure. Ctx, chemotherapy.
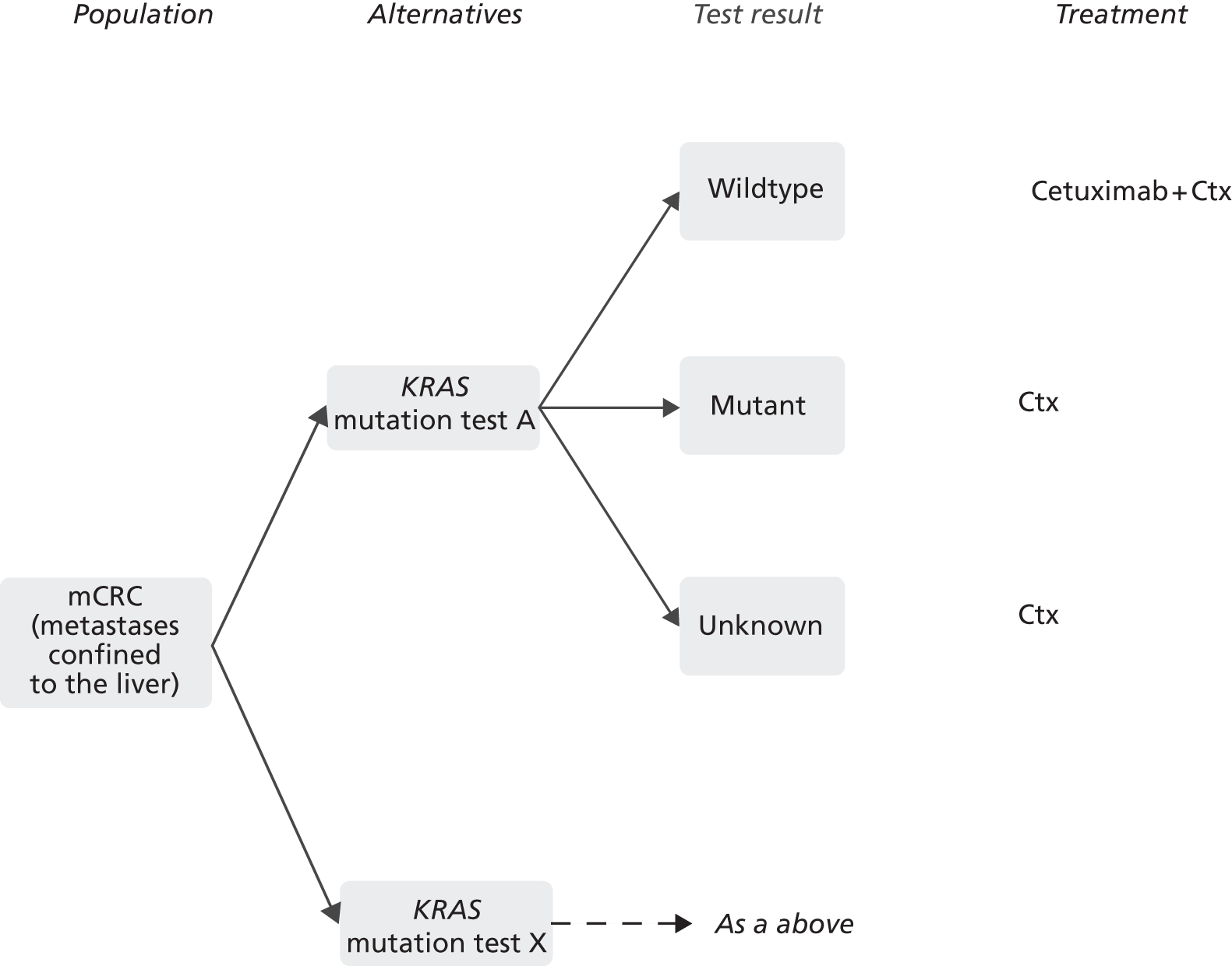
The long-term consequences in terms of costs and QALYs were estimated using a Markov model with a cycle time of 1 week and a lifetime time horizon (23 years were modelled using 1200 cycles). Health states in the Markov model are numbered according to NICE technology appraisal TA176:1
-
progression-free first line – never operated
-
PD second line – never operated
-
PD second line – unsuccessful resection
-
survival after curative resection
-
progression-free first line – unsuccessful resection
-
PD third line – never operated
-
PD third line – unsuccessful resection
-
dead.
The Markov model structure is shown in Figure 9. The model is described in more detail in NICE technology appraisal TA176. 1
FIGURE 9.
Markov model structure.
Model parameters
Estimates for model input parameters were retrieved from NICE technology appraisal TA1761 and the manufacturer’s submission for TA176,59,69 the assessment of the clinical effectiveness of different KRAS mutation tests (see Chapter 3, What is the accuracy of KRAS mutation testing for predicting response to treatment with cetuximab plus standard chemotherapy and subsequent resection rates?) and an online survey of NHS laboratories in England and Wales (see Chapter 3, What are the technical performance characteristics of the different KRAS mutation tests?).
Test result
The proportions of test failures in the laboratory for the KRAS mutation tests were based on the online survey of NHS laboratories in England and Wales. The proportions of KRAS wild-type and KRAS mutant test results were based on the estimated proportions of patients with KRAS wild-type tumours in the population (65.2% with standard error 0.8%),70 the test accuracy (sensitivity and specificity with objective response to cetuximab or resection rate as reference standard; see Table 8) and the proportion of patients with an unknown test result. The proportion of patients with an unknown test result was based on the proportions of patients with unknown tumour mutation status relative to the number of patients for whom a tissue sample was available in the clinical trials. The proportion of patients with an unknown test result may be an overestimate as the clinical trials are unlikely to be representative of the true situation in current clinical practice in the trails, sample were not typically taken for the purpose of KRAS testing. By contrast, the results of the online survey of laboratories in England and Wales are likely to provide an underestimation of the total proportion of patients with an unknown test result as the laboratories may not have insight into the total proportion of pretest failures (samples considered inadequate by the pathologist and therefore not sent to the laboratory). In the linked evidence analysis, the proportion of patients with a unknown tumour mutation status was taken from the clinical trials. For the equal prognostic value analysis, the proportion of patients with an unknown tumour mutation status for all tests was assumed to be equal to that for the pyrosequencing test, as the COIN trial54 (using pyrosequencing) was the only study reporting on resection rates following treatment with chemotherapy, with and without cetuximab, in patients with initially inoperable liver metastases and both KRAS mutant and KRAS wild-type tumours.
The proportions of TP (wild-type), TN (mutant), FN (mutant) and FP (wild-type) test results were calculated by:
-
TP = proportion of wild-types × sensitivity × (1 – proportion of unknown tests)
-
TN = (1 – proportion of wild-types) × specificity × (1 – proportion of unknown tests)
-
FN = proportion of wild-types × (1 – sensitivity) × (1 – proportion of unknown tests)
-
FP = (1 – proportion of wild-types) × (1 – specificity) × (1 – proportion of unknown tests).
Subsequently, the proportions of patients with a wild-type (TP + FP) and mutant (TN + FN) test result were calculated. The results are listed in Tables 15 and 16.
| Input parameter | Estimated value (SE) | Distribution | Source |
|---|---|---|---|
| Proportion of patients with KRAS mutation-positive tumours in England and Wales (%) | 65.2 (0.8) | Beta | Andreyev et al.70 |
| Test accuracy | |||
| Therascreen KRAS RGQ PCR Kit | Sensitivity: 74.6 (5.4); specificity: 35.5 (8.5) | Beta | CELIM52 |
| Pyrosequencing | Sensitivity: 52.0 (9.8); specificity: 45.6 (4.3) | Beta | COIN54 |
| Probability of unknown test result (%) | |||
| Therascreen KRAS RGQ PCR Kit | 10.8 (2.9) | Beta | CELIM52 |
| Pyrosequencing | 1.7 (0.4) | Beta | COIN54 |
Resection rate
Patients who are in the ‘progression-free first line – never operated’ state can move to ‘survival after curative resection’, ‘progression-free first line – unsuccessful resection’, ‘progressive/disease second line – never operated’ or ‘death’, based on tumour resection rates, rate for failure of resection and postoperative mortality. For patients with KRAS wild-type tumours, the resection rate after treatment with cetuximab and chemotherapy (Table 17) was used; for the remaining patients (KRAS mutant or unknown test results), the resection rate after treatment with chemotherapy alone was used. As the CELIM trial52 did not contain a chemotherapy-only strategy, it was not possible to use the resection rates from this trial for patients with KRAS mutant and unknown tumour status. Therefore, the resection rates for the Therascreen KRAS RGQ PCR Kit for these groups were taken from the GERCOR trial,68 which was in line with NICE technology appraisal TA176. 1 However, the GERCOR trial also included patients with metastases outside the liver, which is not in line with the scope for this assessment. The resection rates reported and used in technology appraisal TA1761 for the chemotherapy-only strategy were calculated based on all patients (thus including patients with metastases not confined to the liver) and are therefore probably an underestimation of the true resection rate in the population with metastases confined to the liver. In the assumption of equal prognostic value analysis, however, the resection rate used was based on the COIN trial,54 which included a population with liver-only metastases.
| Mutation test | Resection rate (SE)a,b | Source | ||
|---|---|---|---|---|
| Wild type | Unknown | Mutant | ||
| Therascreen KRAS RGQ PCR Kit | 0.433 (0.060) | 0.092 (0.028) | 0.092 (0.028) | CELIM,52 GERCOR68 |
| Pyrosequencing | 0.149 (0.038) | 0.132 (0.035) | 0.132 (0.035) | COIN54 |
The resection failure rate was set at 5%71 and the probability of postoperative mortality was 2.8% (standard error based on PSA: 1.2%; beta PERT distribution),72 both consistent with NICE technology appraisal TA176. 1
Progression-free and overall survival
To ensure consistency with NICE technology appraisal TA176,1 parametric survival models were obtained from this technology appraisal for patients without resection or patients with unsuccessful resection to estimate cycle-dependent PFS in the first and second line and OS in the first and third line. For patients with successful resection, parametric survival models were obtained from the technology appraisal to calculate cycle-dependent PFS and OS probabilities (Table 18).
| Probability of | Model distribution | Parameter | Estimated value | Standard error | Distribution | Sourcea |
|---|---|---|---|---|---|---|
| First line (Figure 10) | ||||||
| Progression to second line | Log-normal | Cetuximabb | (Commercial-in-confidence information has been removed) | (Commercial-in-confidence information has been removed) | Multivariate normalc | OPUS, Merck Serono59,69 |
| Constantb | (Commercial-in-confidence information has been removed) | (Commercial-in-confidence information has been removed) | Multivariate normalc | OPUS, Merck Serono59,69 | ||
| LN(Sigma) | (Commercial-in-confidence information has been removed) | (Commercial-in-confidence information has been removed) | Multivariate normalc | OPUS, Merck Serono59,69 | ||
| Survival | Log-normal | Cetuximabb | (Commercial-in-confidence information has been removed) | (Commercial-in-confidence information has been removed) | Multivariate normalc | CRYSTAL, Merck Serono59,69 |
| Constantb | (Commercial-in-confidence information has been removed) | (Commercial-in-confidence information has been removed) | Multivariate normalc | CRYSTAL, Merck Serono59,69 | ||
| LN(Sigma) | (Commercial-in-confidence information has been removed) | (Commercial-in-confidence information has been removed) | Multivariate normalc | CRYSTAL, Merck Serono59,69 | ||
| Second line (Figure 11) | ||||||
| Progression to third line | Weibull | Constantb | (Commercial-in-confidence information has been removed) | (Commercial-in-confidence information has been removed) | Multivariate normalc | GERCOR,68 Merck Serono59,69 |
| LN(Sigma) | (Commercial-in-confidence information has been removed) | (Commercial-in-confidence information has been removed) | Multivariate normalc | GERCOR,68 Merck Serono59,69 | ||
| Survival | Based on age-dependent background mortality | Fixed | TA176,1 Merck Serono59,69 | |||
| Third line (Figure 12) | ||||||
| Survival | Weibull | Constantb | (Commercial-in-confidence information has been removed) | (Commercial-in-confidence information has been removed) | Multivariate normalc | Jonker et al.,73 Mittmann et al.,74 Merck Serono59,69 |
| LN(Gamma) | (Commercial-in-confidence information has been removed) | (Commercial-in-confidence information has been removed) | Multivariate normalc | Jonker et al.,73 Mittmann et al.,74 Merck Serono59,69 | ||
| After successful resection (Figure 13) | ||||||
| Progression | Log-logistic | Constantb | (Commercial-in-confidence information has been removed) | (Commercial-in-confidence information has been removed) | Multivariate normalc | Adam et al.,75 Merck Serono59,69 |
| LN(Gamma) | (Commercial-in-confidence information has been removed) | (Commercial-in-confidence information has been removed) | Multivariate normalc | Adam et al.,75 Merck Serono59,69 | ||
| Survival | Log-logistic | Constantb | (Commercial-in-confidence information has been removed) | (Commercial-in-confidence information has been removed) | Multivariate normalc | Adam et al.,75 Merck Serono59,69 |
| LN(Gamma) | (Commercial-in-confidence information has been removed) | (Commercial-in-confidence information has been removed) | Multivariate normalc | Adam et al.,75 Merck Serono59,69 | ||
FIGURE 10.
Progression-free survival and OS in the first line (Commercial-in-confidence information has been removed).
FIGURE 11.
Progression-free survival in the second line (Commercial-in-confidence information has been removed).
FIGURE 12.
Overall survival in the third line (Commercial-in-confidence information has been removed).
FIGURE 13.
Progression-free survival and OS after successful resection (Commercial-in-confidence information has been removed).
Progression-free survival and OS in the first line for standard chemotherapy were based on data from the OPUS and CRYSTAL trials respectively, (Commercial-in-confidence information has been removed) and were estimated separately for patients treated with or without cetuximab. All PFS and OS probabilities for the first line are presented in Figure 10.
Time-dependent probabilities for PFS in the second line and OS in the third line were equal for patients who were treated with or without cetuximab and were converted to constant probabilities based on the median PFS and OS (see Figures 11 and 12 respectively). These constant probabilities were used to prevent an unfeasible amount of tunnel states of 7200 per comparator (1200 cycles × two health states × three possible test results).
Although PFS after successful resection was not incorporated as a separate health state, the probability of progression was estimated to incorporate the utility loss and increased costs associated with progression after successful curative resection. Estimated PFS and OS are equal for patients who were treated with cetuximab and patients who were treated without cetuximab (see Figure 13).
Adverse events
The occurrence of adverse events was assumed to be dependent on treatment and independent of tumour KRAS mutation status, that is, the occurrence of adverse events for patients with KRAS wild-type, KRAS unknown and KRAS mutant tumours was assumed to be equal among different test strategies. Consistent with NICE technology appraisal TA176,1 the occurrence of adverse events was included in the model only by incorporating the additional costs related to the adverse events based on the CRYSTAL27 and OPUS28 trials. These costs are discussed in Resource use and costs.
Health state utilities
Utility scores were retrieved from NICE technology appraisal TA1761 and are presented in Table 19.
| Health state | Utility score | SE | Distribution | Source |
|---|---|---|---|---|
| Progression free (first line) | 0.777 | (Commercial-in-confidence information has been removed) | Beta | TA1761 |
| PD (second line) | 0.730 | (Commercial-in-confidence information has been removed) | Beta | TA1761 |
| PD (third line) | 0.680 | (Commercial-in-confidence information has been removed) | Beta | TA1761 |
| Progression free (after successful resection) | (Commercial-in-confidence information has been removed) | (Commercial-in-confidence information has been removed) | Beta | TA1761 |
| PD (after successful resection) | (Commercial-in-confidence information has been removed) | (Commercial-in-confidence information has been removed) | Beta | TA1761 |
If the mutation tests were to differ substantially in turnaround time, there could be a difference in process disutility associated with waiting for a test result, or even in health outcome because of the delay to the start of treatment. To investigate this, an item on turnaround time was included in the online survey. The results showed that the tests were very similar in terms of turnaround time (see Chapter 3, What are the technical performance characteristics of the different KRAS mutation tests?). In most laboratories the turnaround time was generally between 3 and 7 days. One laboratory (out of eight reporting on the use of pyrosequencing) had a turnaround time of 1–2 days. There was no clear association, however, between the specific test used and the turnaround time reported. Turnaround times are probably impacted most by the number of received samples and batch size. Therefore, it was assumed in the health economic analysis that the turnaround times were not test driven and that the tests did not differ with respect to process disutility or health outcomes associated with waiting for test results.
Resource use and costs
Resource use and costs were taken from NICE technology appraisal TA176,1 with the exception of the KRAS test costs. These costs were based on the online survey of NHS laboratories in England and Wales. Total test costs are calculated based on Table 20; other costs are reported in Table 21.
| Test | Total proportion (SE) of patients with an unknown test result (%) | Distribution | Source | Proportion of technical failures in the laboratories (%) | No. of reporting laboratories | Proportion (SE) of technical failures of patients with an unknown test result (%)a | Distribution |
|---|---|---|---|---|---|---|---|
| Analysis 1b | |||||||
| Therascreen KRAS RGQ PCR Kit | 10.8 (2.9) | Beta | CELIM52 | 1.9c | 0 | 15.9 (8.3) | Beta |
| Pyrosequencing | 1.7 (0.4) | Beta | COIN54 | 3.1 | 7 | 100.0 (12.0)d | Beta |
| Analysis 2b | |||||||
| Pyrosequencing | 1.7 (0.4) | Beta | COIN54 | 3.1 | 7 | 100.0 (13.8)d | Beta |
| Therascreen KRAS RGQ PCR Kit | As for pyrosequencing | 1.9b | 0 | 100.0 (16.6)d | Beta | ||
| cobas KRAS Mutation Test | As for pyrosequencing | 4.5 | 2 | 100.0 (5.6)d | Beta | ||
| HRM analysis | As for pyrosequencing | 0.0 | 1 | 0.0 (0.0) | Beta | ||
| Sanger sequencing | As for pyrosequencing | 0.0 | 1 | 0.0 (0.0) | Beta | ||
| Therascreen KRAS Pyro Kitb | NA | NR | 0 | NA | Beta | ||
| KRAS LightMix kitb | NA | NR | 0 | NA | Beta | ||
| KRAS StripAssayb | NA | NR | 0 | NA | Beta | ||
| MALDI-TOF mass spectrometryb | NA | NR | 0 | NA | Beta | ||
| Next-generation sequencingb | NA | NR | 0 | NA | Beta | ||
| Type of costs | Cost (£) | Distribution | Source |
|---|---|---|---|
| Erbitux® (cetuximab, Merck Serono) (1 mg) | 1.37 | Fixed | TA1761 |
| Irinotecan (1 mg) | 1.30 | Fixed | TA1761 |
| Folinic acid (1 mg) | 0.39 | Fixed | TA1761 |
| 5-Fluorouracil (1 mg) | 0.01 | Fixed | TA1761 |
| Oxaliplatin (1 mg) | 3.30 | Fixed | TA1761 |
| Oncology outpatient attendance | 123.00 | Beta PERTb | TA1761 |
| Outpatient attendance for grade 3/4 adverse event (CRYSTAL27) | 161.51 | Beta PERTb | TA1761 |
| Outpatient attendance for grade 3/4 adverse event (OPUS28) | (Commercial-in-confidence information has been removed) | Beta PERTb | TA1761 |
| Outpatient attendance for serious adverse event (CRYSTAL27) | 165.91 | Beta PERTb | TA1761 |
| Outpatient attendance for serious adverse event (OPUS28) | (Commercial-in-confidence information has been removed) | Beta PERTb | TA1761 |
| Adverse event in second line (outpatient visit) (CRYSTAL27) | 191.27 | Beta PERTb | TA1761 |
| Adverse event in second line (outpatient visit) (OPUS28) | (Commercial-in-confidence information has been removed) | Beta PERTb | TA1761 |
| Serious adverse event requiring hospitalisation (CRYSTAL27) | 1170.83 | Beta PERTb | TA1761 |
| Serious adverse event requiring hospitalisation (OPUS28) | (Commercial-in-confidence information has been removed) | Beta PERTb | TA1761 |
| Hospitalisation for non-serious adverse event | 1050.70 | Beta PERTb | TA1761 |
| Abdomen CT scan | 214.00 | Beta PERTb | TA1761 |
| Chest CT scan | 350.00 | Beta PERTb | TA1761 |
| Hepatic ultrasound | 95.00 | Beta PERTb | TA1761 |
Test costs
For patients with a KRAS wild-type or KRAS mutant test result, the full test costs were accounted for. For this purpose the NHS prices from the online survey of NHS laboratories in England and Wales (see Table 6) were used. As a price was reported for only one test in the online survey, it was decided to assume an equal test cost across all tests of £127.25, which was the average of the four available NHS prices from the survey for this one test (pyrosequencing). To calculate test costs for patients with an unknown tumour mutation status, it is necessary to differentiate between patients for whom the sample was considered inadequate by the pathologist before sending the specimen to the laboratory (pre-laboratory clinical failure) and patients for whom the sample was considered adequate by the pathologist but which resulted in failure once inside the laboratory (technical failures within the laboratory). In the case of an unknown mutation status because of a pre-laboratory clinical failure, no test costs were taken into account. In the case of an unknown mutation status because of a technical failure within the laboratory, full test costs were taken into account. This proportion was calculated based on the proportion of patients with an unknown mutation status taken from the literature and the total proportion of technical failures in the laboratories reported in the online survey (see Table 5), using the following formula:
Proportion of technical failures within the laboratories of all patients with an unknown test result
The results of the calculations are presented in Table 20.
Model analyses
Expected mean costs, LYs and QALYs were estimated for all KRAS mutation testing methods. Long-term costs, LYs and QALYs were discounted using the UK discount rates of 3.5%. Based on the estimated outcomes (probabilistic), the ICER was calculated by dividing the incremental costs by the incremental QALYs. The ICER represents the cost of an additional QALY gained and was used to estimate the cost-effectiveness of a strategy compared with the next best alternative, as in the absence of a comparator strategy it was not possible to calculate ICERs relative to the comparator. All outcomes are based on PSA with 5000 simulations using parameter distributions as presented in this section.
Overview of the main model assumptions
The main assumptions in the health economic analysis were:
-
The differences between ORRs and resection rates for cetuximab plus chemotherapy compared with chemotherapy alone reported in the CELIM trial52 combined with the GERCOR trial68 and those reported in the COIN trial54 are solely the result of the different tests used (Therascreen KRAS RGQ PCR Kit and pyrosequencing respectively) to distinguish between patients whose tumours are KRAS wild type (and who receive cetuximab) and patients whose tumours are KRAS mutant (and who receive chemotherapy) (linked evidence analysis).
-
To calculate the sensitivity and specificity of the tests, required to calculate the proportion of KRAS wild-type and KRAS mutant test results (see Table 8), patients tested as KRAS wild type were categorised as FP if no objective response was observed (for the Therascreen KRAS RGQ PCR Kit) or no liver resection was performed (for pyrosequencing) after treatment with cetuximab whereas patients were categorised as TP if an objective response was observed (Therascreen KRAS RGQ PCR Kit) or a liver resection was performed (pyrosequencing). Similarly, patients tested as tumour KRAS mutant were categorised as FN if an objective response was observed (for the Therascreen KRAS RGQ PCR Kit) or a liver resection was performed after treatment with cetuximab (for pyrosequencing) whereas patients were categorised as TN if no objective response was observed (Therascreen KRAS RGQ PCR Kit) or no liver resection was performed (pyrosequencing).
-
Test accuracy based on objective response can be compared with accuracy based on resection rates. 39
-
The number of patients with unknown mutation status relative to the number of patients for whom a tissue sample was available in the trials52,54 provides a realistic approximation of the proportion of patients with an unknown test result in clinical practice (both analyses).
-
As the COIN trial54 tests for KRAS mutations using both pyrosequencing and MALDI-TOF, with a reported concordance of > 99%, it was assumed that the accuracy derived from this trial and also the resection rates reported apply to both pyrosequencing and MALDI-TOF, that is, all pyrosequencing results in this report also apply to MALDI-TOF.
-
The standard chemotherapy used in the COIN trial54 (FOLFOX or XELOX) is comparable to FOLFOX6 as used in the CELIM trial. 52
Sensitivity analyses
For both the linked evidence and the assumption of equal prognostic value analysis, the following sensitivity analyses were performed:
-
mortality in the second line was based on the average of the first- and third-line mortality instead of background mortality as in NICE technology appraisal TA176. 1
-
the proportion of patients with unknown mutation status was based on the results of the online survey instead of the literature (see Table 5).
Results of the cost-effectiveness analyses
This section reports the results of the linked evidence analysis and the assumption of equal prognostic value analysis. As this economic evaluation takes a ‘no comparator’ approach, ICERs for each strategy are calculated compared with the next most cost-effective strategy.
Linked evidence analysis
The linked evidence analysis includes two tests, that is, only those tests for which evidence on test accuracy based on either resection rate or ORR was available. Table 22 shows the probabilistic results of this analysis. It should be noted that this analysis was based on a number of substantial assumptions, which are outlined in the previous section. In short, we have only the COIN54 and CELIM52 trials to rely on, of which the COIN trial used pyrosequencing to test for KRAS mutations and the CELIM trial used the Therascreen KRAS RGQ PCR Kit. We assumed that the differences between the outcomes of these trials are exclusively caused by the different tests used (assumption 1). Table 23 provides a summary of the comparability of the study populations across the COIN,54 CELIM52 and GERCOR68 trials used in the linked evidence analysis. In addition, we assumed that all KRAS wild-type patients would respond perfectly to cetuximab – or would all have a liver resection after cetuximab – and that all KRAS mutant patients would not (assumption 2), and that test accuracy based on objective response can be compared with accuracy based on resection rates (assumption 3).
| Strategy | Cost (£) | QALYs | Δ Costs (£) | Δ QALYs | ICER (£) |
|---|---|---|---|---|---|
| Base case | |||||
| Pyrosequencinga | 30,870 | 1.49 | |||
| Therascreen KRAS RGQ PCR Kit | 33,995 | 1.67 | 3125 | 0.18 | 17,019 |
| Sensitivity analysis: mortality second line based on average of first- and third-line mortality | |||||
| Pyrosequencinga | 29,704 | 1.28 | |||
| Therascreen KRAS RGQ PCR Kit | 33,132 | 1.51 | 3428 | 0.23 | 14,860 |
| Sensitivity analysis: proportion with unknown mutation status based on survey | |||||
| Pyrosequencinga | 30,714 | 1.48 | |||
| Therascreen KRAS RGQ PCR Kit | 34,799 | 1.69 | 4085 | 0.20 | 20,528 |
| Study details | Participant selection | Population characteristics |
|---|---|---|
| Folprecht 201052 (CELIM) Country: Germany and Austria Study design: RCT No. randomised: 111 No. of KRAS wild-type patients randomised: 70 No. of patients with liver-limited metastases randomised: 111 Intervention: Cetuximab + FOLFOX vs. cetuximab + FOLFIRI |
Inclusion criteria: Unresectable, histologically confirmed colorectal liver metastases; no extrahepatic metastases (patients with synchronous liver metastases were eligible if the primary tumour had been resected before chemotherapy); Karnofsky perfomance score ≥ 80%; adeuqate hepatic, renal and bone marrow function Exclusion criteria: Previous chemotherapy (except adjuvant chemotherapy with an interval of ≥ 6 months); previous EGFR-targeted therapy; concurrent antitumour therapy; clinically relevant coronary artery disease; inflammatory bowel disease; previous malignancy; age < 18 years |
Age (years), median (range): 63 (56–71) Male, n: 71 Liver metastases, n: < 5: 30; 5–10: 58; > 10: 19; NR: 4 No. with previous liver resection: 14 Criteria for unresectability: Five or more liver metastases or metastases that were viewed as technically non-resectable by the local liver surgeon and radiologist on the basis of inadequate future liver remnant or one of the following critera: infiltration of all hepatic liver veins, infiltration of both hepatic arteries or both protal vein branches Previous treatments: 9 patients had adjuvant radiotherapy, 18 had adjuvant chemotherapy |
| Maughan 201154 (COIN) Country: UK and Republic of Ireland Study design: RCT No. randomised: 1630 No. of KRAS wild-type patients randomised: 729 No. of patients with liver-limited metastases randomised: 178 Intervention: Cetuximab + FOLFOX or XELOX vs. FOLFOX or XELOX |
Inclusion criteria: Adults aged ≥ 18 years; histologically confirmed adenocarcinoma of the colon or rectum; inoperable metastatic or locoregional disease; no previous chemotherapy for metastatic disease; WHO performance status 0–2; adequate hepatic, renal and haematological function; no adjuvant chemotherapy or rectal chemoradiotherapy within 1 month of the start of the trial Exclusion criteria: Unfit for chemotherapy; severe, uncontrolled medical illness; psychiatric illness inhibitig informed consent; partial or complete bowel obstruction; pre-existing neuropathy greater than grade 1; requirement for treatment with contraindicated medication; another previous or current malignant disease that may affect treatment response; known hypersensitivity to any study treatment; brain metastases |
Age (years), median (range): 64 (56–70) Male, n: 498 Liver metastases: Resection rates reported separately for patients with liver-only metastases Criteria for unresectability: NR Previous treatments: NR |
| Tournigand 200468 (GERCOR) Country: France Study design: RCT No. randomised: 226 (of whom 6 not eligible) No. of KRAS wild-type patients randomised: NA No. of patients with liver-limited metastases randomised: NR Intervention: FOLFIRI + FOLFOX (arm A) vs. FOLFOX + FOLFIRI (arm B) |
Inclusion criteria: Adults aged 18–75 years; adenocarcinoma of the colon or rectum; unresectable metastases; at least one bidimensionally measurable lesion of ≥ 2 cm or a residual non-measurable lesion; adequate bone marrow, liver and renal function; WHO performance status 0–2. Previous adjuvant chemotherapy, if given, must have been completed at least 6 months before inclusion Exclusion criteria: Central nervous system metastases; second malignancies; bowel obstruction; current diarrhoea of grade 2 or higher; symptomatic angina pectoris; disease confined to previous radiation fields |
Age (years), median (range): Arm A: 61 (29–75); arm B: 65 (40–75) Male, n: 142 (of 220) Metastases: Liver: 184 (84%); lung: 67 (30%); other: 98 (45%) Resection rates not reported separately for patients with liver-only metastases No. of sites of metastases: 1: 130 (59%); ≥ 2: 90 (41%) Criteria for unresectability: NR Previous treatments: 17% and 21% of arm A and arm B, respectively, had adjuvant chemotherapy |
As is apparent from Table 22, pyrosequencing has the lowest total cost. The Therascreen KRAS RGQ PCR Kit is the more expensive but also more effective strategy, with an ICER of £17,019 per QALY gained. The CEAC in Figure 14 shows that, for lower values of the threshold, pyrosequencing is the preferred strategy and that at thresholds of ≥ £17,000 the Therascreen KRAS RGQ PCR Kit is the most cost-effective option. The results of the sensitivity analyses (see Table 22) do not differ substantially from the base-case results in the sense that the Therascreen KRAS RGQ PCR Kit is consistently more expensive and more effective than pyrosequencing, with ICERs ranging from £14,860 to £20,528 per QALY gained. CEACs for the sensitivity analyses are presented in Appendix 6.
FIGURE 14.
Cost-effectiveness acceptability curve for the linked evidence analysis: base case.
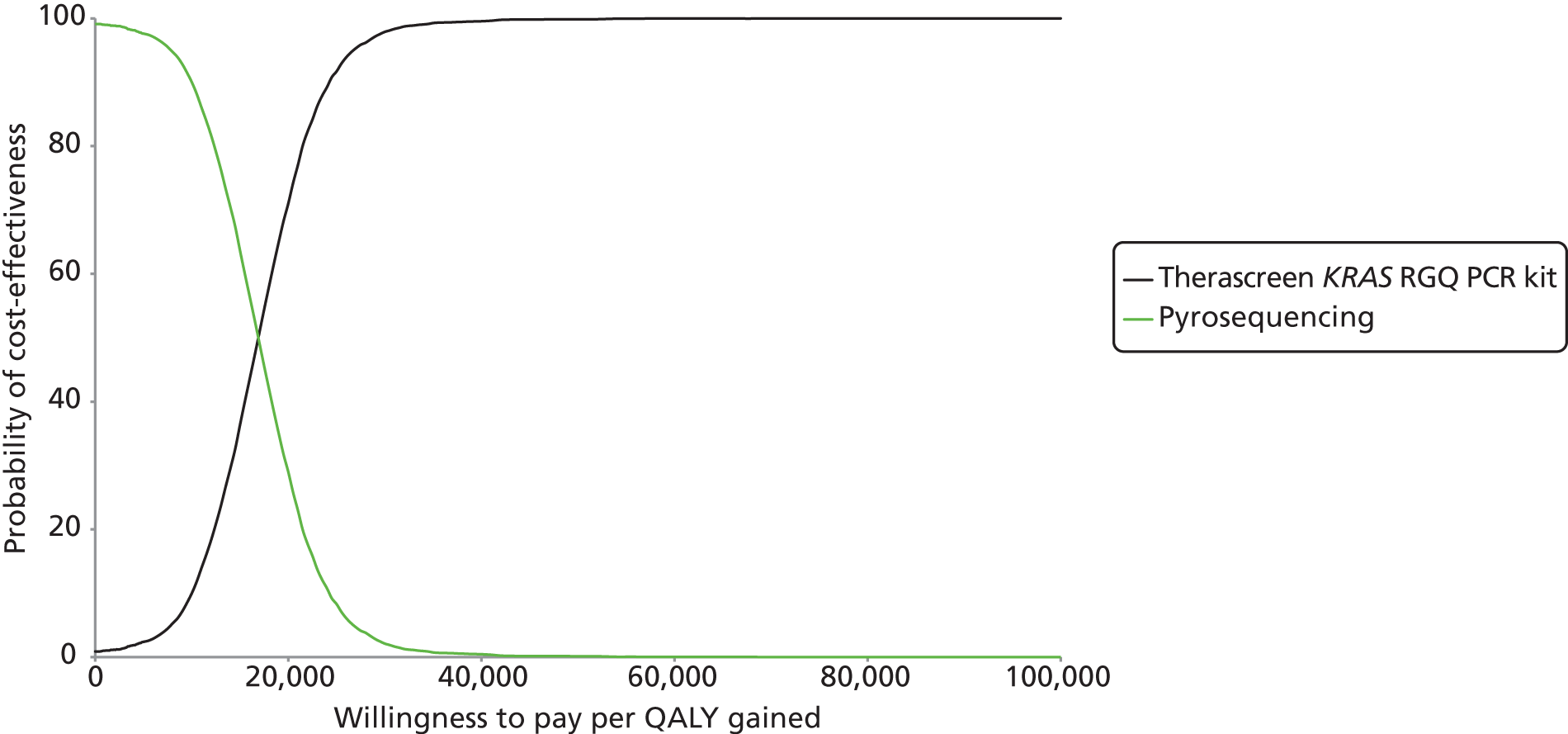
Assumption of equal prognostic value analysis
The assumption of equal prognostic value analysis includes all tests for which information on technical performance was available from the online survey of NHS laboratories in England and Wales. This includes the tests for which accuracy data, based on either response rates or resection rates, were not available. Therefore, this analysis assessed whether the tests were likely to be cost-effective given an assumption of equal prognostic value based on the prognostic value of testing with pyrosequencing (as this was the only test for which full data were available on resection rates following treatment with chemotherapy, with and without cetuximab, for patients with initially inoperable liver metastases and both KRAS mutant and KRAS wild-type tumours) and test-specific information on technical failures within the laboratory only (Table 24). In the base case and the first sensitivity analysis, the total technical failure rate (pre-laboratory plus within-laboratory technical failures) is assumed to be equal for all tests. As a result, the strategies in these analyses differ only with respect to costs (because of differences in within-laboratory technical failures). In the base case the average QALYs for all comparators were 1.48 (95% CI 1.33 to 1.64). The total costs associated with the various testing strategies (see Table 24) are highly similar. The same applies to the first sensitivity analysis (Table 25): costs are similar across strategies and average QALYs are equal by assumption at 1.28 (95% CI 1.12 to 1.44).
| Test | Cost (95% CI) (£) | Δ Costa (95% CI) (£) |
|---|---|---|
| HRM analysis | 30,857.09 (27,079.58 to 34,736.14) | |
| Sanger sequencing | 30,857.09 (27,079.58 to 34,736.14) | 0.00 (0.00 to 0.00)b |
| Therascreen KRAS RGQ PCR Kit | 30,857.46 (27,079.91 to 34,736.60) | 0.37 (0.12 to 0.88) |
| Pyrosequencingc | 30,857.70 (27,080.27 to 34,737.03) | 0.61 (0.14 to 1.64) |
| cobas KRAS Mutation Test | 30,857.99 (27,080.25 to 34,737.14) | 0.91 (0.23 to 2.28) |
| Test | Cost (95% CI) (£) | Δ Costa (95% CI) (£) |
|---|---|---|
| HRM analysis | 29,661.10 (25,991.06 to 33,401.42) | |
| Sanger sequencing | 29,661.10 (25,991.06 to 33,401.42) | 0.00 (0.00 to 0.00)b |
| Therascreen KRAS RGQ PCR Kit | 29,661.47 (25,991.81 to 33,401.80) | 0.37 (0.12 to 0.85) |
| Pyrosequencingc | 29,661.71 (25,992.12 to 33,401.81) | 0.61 (0.14 to 1.59) |
| cobas KRAS Mutation Test | 29,662.00 (25,993.07 to 33,402.58) | 0.90 (0.23 to 2.18) |
In the second sensitivity analysis the total technical failure rate is also test specific, which impacts on the proportion of patients with unknown (and therefore also wild-type and mutant) tumour KRAS status. Therefore, in this sensitivity analysis the strategies differ with respect to both effects and costs. All other input parameters, such as test costs and test accuracy, are still considered equal. The probabilistic results in Table 26 show that the cobas KRAS Mutation Test is the least costly and least effective strategy. HRM analysis and Sanger sequencing have equal costs and effects and their ICER compared with the cobas KRAS Mutation Test is £69,815 per QALY gained. Pyrosequencing and the Therascreen KRAS RGQ PCR Kit are ruled out by extended dominance in this analysis. From the CEAC (Figure 15) it is apparent that the cobas KRAS Mutation Test is the preferred strategy for all threshold values of < £60,000.
| Test | Cost (£) | QALYs | Comparator | Δ Cost (£) | Δ QALYs | ICER (£) |
|---|---|---|---|---|---|---|
| cobas KRAS Mutation Test | 30,663 | 1.48 | ||||
| Pyrosequencinga | 30,796 | 1.48 | cobas KRAS Mutation Test | 133.66 | 0.002 | Extended dominance |
| Therascreen KRAS RGQ PCR Kit | 30,876 | 1.48 | Pyrosequencing | 80.06 | 0.001 | Extended dominance |
| HRM analysis | 31,006 | 1.49 | cobas KRAS Mutation Test | 343.64 | 0.005 | 69,815b |
| Sanger sequencing | 31,006 | 1.49 | cobas KRAS Mutation Test | 343.64 | 0.005 | 69,815b |
FIGURE 15.
Cost-effectiveness acceptability curve for assumption of equal prognostic value sensitivity analysis: proportion of patients with unknown mutation status based on the survey.
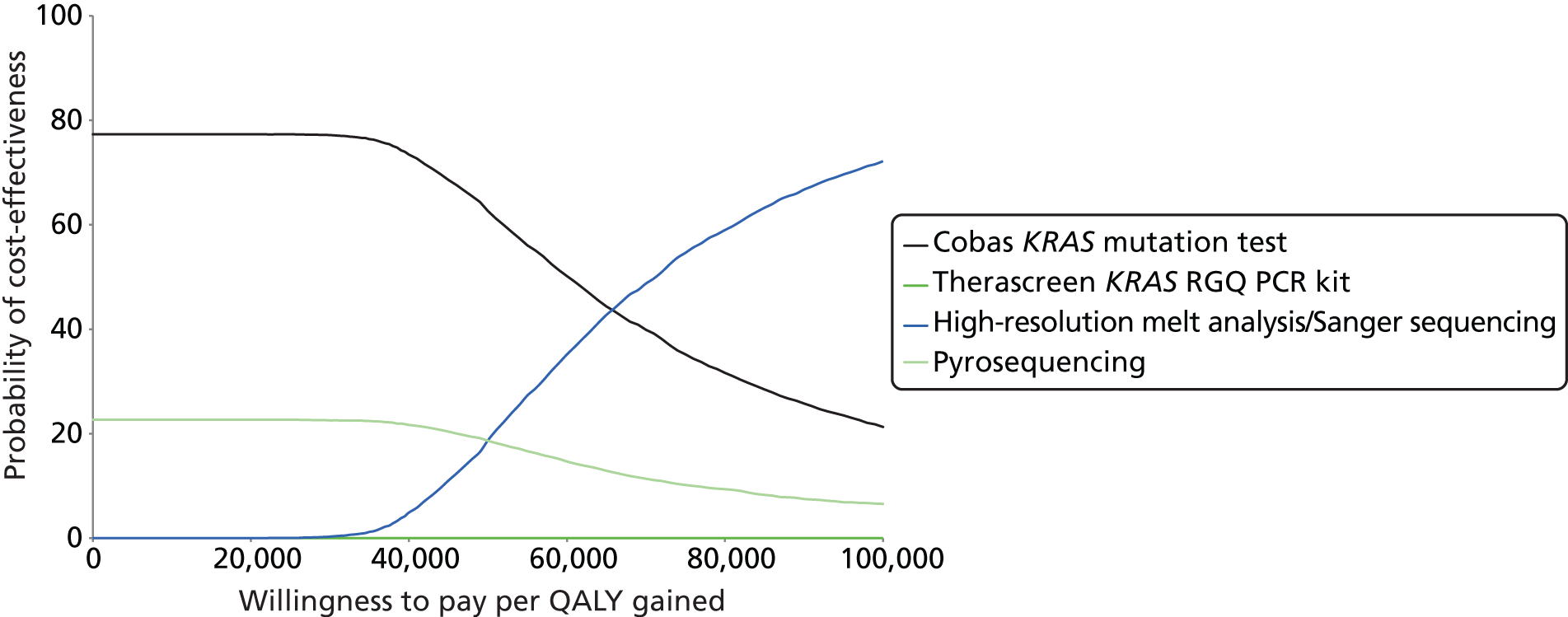
Chapter 5 Discussion
Statement of principal findings
Clinical effectiveness
There was no clear evidence to suggest any differences between the KRAS mutation testing techniques for any of the measures assessed (technical performance, accuracy for predicting response to treatment with cetuximab in combination with standard chemotherapy, or variation in clinical outcomes following treatment with cetuximab in combination with standard chemotherapy depending on which method is used to classify patients as having KRAS wild-type tumours).
The survey of laboratories providing KRAS mutation testing indicated that in-house pyrosequencing methods, targeting KRAS mutations in codons 12, 13 and 61 and using self-designed primers, were the most commonly used approaches (9/15 respondents); reasons cited by respondents for their choice of this technique were the proportion of tumour cells required, ease of use, cost, mutations covered, turnaround time and experience of pyrosequencing techniques available in the laboratory. There was no apparent association between test method and reason for choice. Commercial kits used were the cobas KRAS Mutation Test (three laboratories) and the Therascreen KRAS Pyro Kit (one laboratory). More than half of the responding laboratories reported that KRAS mutation testing was carried out on request (e.g. from a pathologist or oncologist); only one laboratory reported routine testing of all CRC samples. In general, there was no clear indication that choice of test method was related to volume of throughput, although both of the laboratories that reported using Sanger sequencing had a low throughput (up to five samples per week). Most respondents reported turnaround times, from receipt of sample to reporting to the clinician, of between 3 and 5 days. The only laboratory to report a turnaround time of < 3 days (24–48 hours) used an in-house pyrosequencing method. Frequency of running the test did not appear to relate to laboratory throughput and only one laboratory reported waiting for a minimum batch size (10 samples) before running the test; this laboratory had a high throughput (> 20 samples per week). The minimum percentage of tumour cells required for testing varied widely across laboratories (< 1% to > 30%), even when the same test method was being used. When reported, the minimum requirement for the cobas KRAS Mutation Test was ≤ 10%. With the exception of those using Sanger sequencing, all laboratories reported a limit of detection for percentage mutation of ≤ 10%. The laboratory that used the Therascreen KRAS Pyro Kit did not provide any data on technical performance. The proportion of samples rejected before analysis was < 2% for all responding laboratories. The rate of failure for analysed samples did not appear to be dependent on test method (3–6% for the cobas KRAS Mutation Test and 0.2–10% for in-house pyrosequencing methods). The majority of responding laboratories reported using microdissection techniques before DNA extraction; however, there was no clear indication that non-use of this technique was associated with a higher rate of sample rejection or test failure. The laboratory that used the Therascreen KRAS Pyro Kit did not provide any data on failure rates. Although most respondents included cost in their reasons for choosing a particular test, it is worth noting that a relatively narrow range of costs was reported across all tests (£100–150), with one laboratory reporting a higher cost (£273) for running a single sample. Prices charged, to both Merck Serono and the NHS, ranged from £99 to £150.
When contacted by NICE in relation to a previous diagnostic assessment of EGFR mutation testing in non-small-cell lung cancer, UK NEQAS stated that ‘Error rates are not always method related and it is not always possible to obtain data from all the labs committing critical genotyping errors. Therefore, any data which could be provided would be skewed with processing and reporting issues rather than being method related.’ Only one KRAS mutation testing method is currently approved by the US FDA; this is the Therascreen KRAS RGQ PCR Kit when used with the QIAamp DSP DNA FFPE Tissue Kit and the QIAGEN Rotor-Gene Q MDx (software version 2.1.0) and KRAS Assay Package. 16 The clinical trial67 used to support FDA approval was not included in this assessment as it did not match our inclusion criteria; it compared treatment with cetuximab and BSC to treatment with BSC alone in patients with mCRC who had previously failed all available chemotherapy. It should be noted that none of the laboratories participating in the UK NEQAS scheme who responded to our survey reported using the Therascreen KRAS RGQ PCR Kit.
Evidence to allow comparison of the accuracy of different KRAS mutation tests was very limited. Only one publication,52 from the CELIM trial, provided sufficient data to allow estimation of the accuracy of a KRAS mutation test (version 1 of the Therascreen KRAS PCR Kit) for predicting response to treatment with cetuximab plus standard chemotherapy. This study reported objective response data and thus did not provide direct information on the value of the KRAS mutation test for predicting resection rate. Because the aim of KRAS mutation testing is to predict likely response to the addition of cetuximab to standard chemotherapy, test positive was defined as a KRAS wild-type tumour. The positive predictive value (70.1%, 95% CI 58.3% to 79.8%), reported in Chapter 3 (see What is the accuracy of KRAS mutation testing for predicting response to treatment with cetuximab plus standard chemotherapy and subsequent resection rates?), indicated that KRAS wild type, as determined using the Therascreen KRAS PCR Kit, may be moderately predictive of tumour response. If the published strong correlation between ORRs and resection rates in patients with isolated liver metastases39 treated with various chemotherapy regimens were assumed to extrapolate to patients with KRAS wild-type tumours treated with standard chemotherapy plus cetuximab, then the expected R0 and R1 resection rates for these patients would be approximately 67%, based on data from the CELIM trial. 52 By contrast, the negative predictive value (40.7%, 95% CI 24.5% to 59.3%) could be interpreted as indicating that the presence of a KRAS mutation, as determined using the Therascreen KRAS PCR Kit, is a relatively poor predictor of non-response. Additional data supplied by the COIN trial investigators54 allowed the calculation of estimates for the accuracy of pyrosequencing and MALDI-TOF (in which both tests were performed on all samples), targeting mutations in codons 12, 13 and 61, for predicting potentially curative resection following treatment with cetuximab plus FOLFOX or XELOX. The positive and negative predictive values derived from these data were 14.9% (95% CI 8.9% to 23.9%) and 83.9% (95% CI 73.8% to 90.5%) respectively; this could be interpreted as indicating that a tumour which is defined as KRAS wild type by this method is a poor predictor of resectability following treatment with cetuximab plus standard chemotherapy, whereas the presence of a KRAS mutation is a good predictor of non-response (tumour remaining unresectable after treatment). The COIN trial reported > 99% concordance on KRAS genotyping between pyrosequencing and MALDI-TOF; it may therefore be assumed that accuracy data from the COIN trial are also representative of the accuracy of both pyrosequencing and MALDI-TOF when used as single tests. It should be noted that any apparent differences in the ability of KRAS mutation tests to predict response to treatment between the CELIM trial and the COIN trial may be caused by other differences between studies (e.g. participant characteristics, in particular the definition of baseline unresectability, and treatment regimens).
Four further studies (six publications27,28,53–56) were included in the review; all were RCTs comparing cetuximab plus standard chemotherapy with standard chemotherapy alone in patients whose tumours were KRAS wild type and all reported data on patients with CRC metastases that were confined to the liver. The standard chemotherapy regimen was different in each of the four trials (FOLFOX4,28,53,56 FOLFIRI,27,53 FOLFIRI or FOLFOX655 and FOLFOX or XELOX54). There was no substantial evidence to indicate a significant difference in treatment effect depending on which of three KRAS mutation tests (LightMix k-ras Gly12, pyrosequencing and MALDI-TOF mass array for mutations in codons 12, 13 and 61, or pyrosequencing for KRAS mutations in codons 12 and 13) was used to identify patients with KRAS wild-type tumours. All three studies that assessed ORR reported a statistically significant higher response rate for participants treated with cetuximab plus standard chemotherapy than for those treated with standard chemotherapy alone; ORs ranged from 3.00 (95% CI 1.49 to 6.03)53 to 4.93 (95% CI 1.42 to 17.06). 28 All four studies reported that the addition of cetuximab to standard chemotherapy was associated with an increase in the rate of R0 resections following treatment. However, it should be noted that the only trial to report a statistically significant treatment effect for R0 resection rate used pyrosequencing to identify KRAS mutations in codons 12 and 13 only. 55 This was also the only trial in which all participants had CRC metastases that were limited to the liver.
Effectiveness data from the CRYSTAL27 and OPUS28 trials were used to inform the technology appraisal underpinning NICE guidance TA1761 on cetuximab for the first-line treatment of mCRC. Data from an interim analysis of the CELIM trial were used as a source of UK data for resection rates following treatment with cetuximab plus standard chemotherapy. 1 Data from the COIN trial54 and the trial by Xu et al. 55 were published subsequently to TA176.
Cost-effectiveness
The review of economic analyses of different methods for KRAS mutation testing to decide between standard chemotherapy and cetuximab in combination with standard chemotherapy in adults with mCRC found four full papers62–65 and one HTA report. 61 Based on all of these publications it can be said that, in general, although KRAS testing is obviously a more cost-effective option than administering cetuximab to all patients, the ICER of KRAS testing and treating only patients with KRAS wild-type tumours with cetuximab compared with treating all patients with standard chemotherapy alone seems rather high.
In the health economic analysis, the cost-effectiveness of different methods of KRAS mutation testing to decide between standard chemotherapy and cetuximab in combination with standard chemotherapy in adults with mCRC was assessed. In light of the limited amount of evidence available, two analyses were performed: ‘linked evidence’ and ‘assumption of equal prognostic value’. All analyses took a ‘no comparator’ approach.
In the linked evidence analysis, the Therascreen KRAS RGQ PCR Kit was compared with pyrosequencing, using the available objective response and resection rate, respectively, to estimate lifetime costs and QALYs. The results of this analysis suggested that the Therascreen KRAS RGQ PCR Kit was more costly and more effective than pyrosequencing, with an ICER of £17,019 per QALY gained. Sensitivity analyses did not show substantial differences compared with the base case. The key driver behind the outcome was the difference in resection rate between treatment with and treatment without cetuximab and the proportions of patients with KRAS wild-type, KRAS mutant and unknown tumours. This was determined by test accuracy and therefore, for the most part, was dependent on ORR (for the Therascreen KRAS RGQ PCR Kit) or resection rate (for pyrosequencing).
It should be noted that this analysis was based on a number of substantial assumptions, which are outlined in Chapter 4 (see Overview of main model assumptions). The following assumptions used were particularly problematic as they are open to doubt and probably have a considerable impact on the model results:
-
The differences between ORRs and resection rates for cetuximab plus chemotherapy compared with chemotherapy alone as reported in the CELIM trial52 combined with the GERCOR trial68 and the COIN trial54 are solely the result of the different tests used (Therascreen KRAS RGQ PCR Kit and pyrosequencing respectively) to distinguish between patients whose tumours are KRAS wild type (and who receive cetuximab) and patients whose tumours are KRAS mutant (and who receive chemotherapy).
-
To calculate the sensitivity and specificity of the tests, required to calculate the proportion of KRAS wild-type and KRAS mutant test results (see Table 8), patients with a tumour tested as KRAS wild type were categorised as FP if no objective response was observed (for the Therascreen KRAS RGQ PCR Kit) or no liver resection was performed (for pyrosequencing) after treatment with cetuximab, whereas patients were categorised as TP if an objective response was observed (Therascreen KRAS RGQ PCR Kit) or a liver resection was performed (pyrosequencing). Similarly, patients with a tumour tested as KRAS mutant were categorised as FN if an objective response was observed (for the Therascreen KRAS RGQ PCR Kit) or a liver resection was performed (for pyrosequencing) after treatment with cetuximab, whereas patients were categorised as TN if no objective response was observed (Therascreen KRAS RGQ PCR Kit) or no liver resection was performed (pyrosequencing).
The results of the linked evidence analysis should therefore be interpreted on the condition that these assumptions hold. Moreover, the uncertainty presented surrounding the results is an underestimation of the true uncertainty as the uncertainty associated with the assumptions was not parameterised in the model and is therefore not reflected in the PSA.
The assumption of equal prognostic value analysis included all tests for which information on technical performance was available from the online survey of NHS laboratories in England and Wales. This includes the tests for which accuracy data based on either response rates or resection rates were not available. Therefore, this analysis assessed whether the tests were likely to be cost-effective given an assumption of equal prognostic value based on the prognostic value of testing with pyrosequencing (as this was the only test for which full data were available on resection rates following treatment with chemotherapy, with and without cetuximab, for patients with initially inoperable liver metastases and both KRAS mutant and KRAS wild-type tumours) and test-specific information on technical failures within the laboratory only, which implies that strategies can differ only with respect to costs. The results of the assumption of equal prognostic value analysis indicated that the strategies were almost equal. The first sensitivity analysis confirmed this. The second sensitivity analysis, in which the proportion of patients with unknown mutation status was taken from the survey instead of the literature, was slightly different in the sense that for this analysis the effectiveness was not assumed equal among all tests and therefore ICERs were available. The results showed that the cobas KRAS Mutation Test was the least expensive and least effective strategy and that Sanger sequencing and HRM analysis were equally the most costly and most effective, with an ICER of £69,815 per QALY gained compared with the cobas KRAS Mutation Test. The other two strategies included in this analysis, that is, the Therascreen KRAS RGQ PCR Kit and pyrosequencing, are ruled out by extended dominance.
Strengths and limitations of the assessment
Clinical effectiveness
Extensive literature searches were conducted in an attempt to maximise retrieval of relevant studies. These included electronic searches of a variety of bibliographic databases as well as screening of clinical trial registers and conference proceedings to identify unpublished studies. Because of the known difficulties in identifying test accuracy studies using study design-related search terms,76 and the potential need to include non-RCTs, search strategies were developed to maximise sensitivity at the expense of specificity. Thus, large numbers of citations were identified and screened, very few of which met the inclusion criteria of the review. The specificity of searches was further reduced as it was not possible to target publications focusing on patients whose metastases were limited to the liver only; these patients were a subgroup in the majority of included studies.
The possibility of publication bias remains a potential problem for all systematic reviews. Considerations may differ for systematic reviews of test accuracy studies. It is relatively simple to define a positive result for studies of treatment, for example a significant difference between the treatment group and the control group that favours treatment. This is not the case for test accuracy studies, which measure agreement between the index test and the reference standard. It would seem likely that studies finding greater agreement (high estimates of sensitivity and specificity) will be published more often. This distinction may be less applicable to studies in this review that provided accuracy data, as these studies aimed to assess the effectiveness of treatment with cetuximab plus standard chemotherapy in different patient groups (CELIM trial52) or compare the effectiveness of cetuximab plus standard chemotherapy and standard chemotherapy alone (COIN trial54); neither study was primarily focused on test performance. Our review included a very small number of clinically heterogeneous studies, both for the accuracy of KRAS mutation testing to predict response to treatment with cetuximab plus standard chemotherapy and for the relative effectiveness of cetuximab plus standard chemotherapy and standard chemotherapy alone in populations with KRAS wild-type tumours, selected using different KRAS mutation test methods. We were therefore unable to undertake any meta-analyses or formal assessment of publication bias. However, our search strategy included a variety of routes to identify unpublished studies and resulted in the inclusion of a number of conference abstracts.
Clear inclusion criteria were specified in the protocol for this review and only one protocol modification occurred during the assessment. The eligibility of studies for inclusion is therefore transparent. In addition, we have provided specific reasons for excluding any studies considered potentially relevant at initial citation screening (see Appendix 5). The review process followed recommended methods to minimise the potential for error and/or bias. 40 Studies were independently screened for inclusion by two reviewers and data extraction and quality assessment were carried out by one reviewer and checked by a second (MW and PW). Any disagreements were resolved by consensus.
Studies included in this review were assessed for risk of bias using published tools appropriate to the study design and/or the type of data extracted. Studies that provided data on the accuracy of KRAS mutation testing for predicting response to treatment with cetuximab plus standard chemotherapy were assessed using a modification of the QUADAS-2 tool. 48 The QUADAS-2 tool is structured into four key domains covering participant selection, index test, reference standard and the flow of patients through the study (including timing of tests). Each domain is rated for risk of bias (low, high or unclear); the participant selection, index test and reference standard domains are also separately rated for concerns regarding the applicability of the study to the review question (low, high or unclear). Studies that provided data on the effectiveness of treatment with cetuximab plus standard chemotherapy compared with standard chemotherapy alone in patients with KRAS wild-type tumours were all RCTs or subgroup analyses from RCTs. These studies were therefore assessed using The Cochrane Collaboration’s tool for assessing risk of bias in randomised trials. 42,47 The results of the risk of bias assessment are reported in full for all included studies in Appendix 3 and in summary form in Chapter 3 (see What is the accuracy of KRAS mutation testing for predicting response to treatment with cetuximab plus standard chemotherapy and subsequent resection rates? and How do outcomes from treatment with cetuximab plus standard chemotherapy vary according to which test is used to select patients for treatment?). The main potential sources of bias identified were exclusion of withdrawals from the analyses (for studies providing data on the accuracy of KRAS mutation tests for predicting response to treatment with cetuximab plus standard chemotherapy) and blinding of participants and personnel in treatment trials. Both of the studies that provided data on the accuracy of KRAS mutation testing for predicting response to treatment had some limitations in their applicability to the target population for this assessment. In the case of the CELIM trial52 data were available to calculate accuracy for the prediction of objective response only rather than for the preferred direct measure resection of liver, and in the case of the COIN trial54 the standard chemotherapy regimen did not fully match that in the inclusion criteria for this assessment. In addition, participants in the CELIM trial52 were described as having technically non-resectable or five or more liver metastases from CRC and it was therefore unclear whether some participants may have had potentially resectable metastases at baseline.
All of the studies included in this review have some limitations with regard to their ability to address the overall aim of comparing the clinical effectiveness of different KRAS mutation tests to determine which patients may benefit from the addition of cetuximab to standard chemotherapy and which should receive standard chemotherapy alone. The COIN trial54 is likely to represent the closest approximation to the ideal study in that, when additional data supplied by the trial investigators are also considered, it provides full information on the comparative treatment effect (cetuximab plus standard chemotherapy vs. standard chemotherapy alone) for patients with both KRAS wild-type tumours and KRAS mutant tumours. In addition, the trial was conducted in the UK and hence provides data that are likely to be directly applicable to practice in the NHS in England and Wales. However, data included in this assessment were derived from subgroup analyses of patients included in the original trial; not all patients included in the original trial had samples available for KRAS mutation testing and, in addition, a much smaller subgroup of patients had metastases that were limited to the liver. Further, the standard chemotherapy regimen used in the COIN trial54 allowed a choice between FOLFOX or XELOX (depending on local hospital practice and patient preference); the use of XELOX as standard chemotherapy does not match the standard chemotherapy described in the inclusion criteria for this assessment, as determined by the recommendations of NICE technology appraisal TA176. 1 The COIN trial used a combination of pyrosequencing and MALDI-TOF, targeting mutations in codons 12, 13 and 61, to determine KRAS mutation status; in common with all other studies included in this assessment, the study was not designed to assess KRAS mutation testing and did not provide any comparative data for other testing methods.
Because methods of testing for KRAS mutation status can differ in terms of both the mutations targeted and the limit of detection (the lowest proportion of tumour cells with a mutation that can be detected), the definitions of KRAS wild type and KRAS mutant vary according to which test is used. All testing methods are essentially reference standard methods for classifying mutation status, as defined by the specific test characteristics. The essential clinical question is to determine which testing method is best at classifying patients such that the maximum treatment effect is achieved both for patients whose tumours are classified as KRAS wild type, who receive cetuximab in addition to standard chemotherapy, and for those whose tumours are classified as KRAS mutant, who receive standard chemotherapy alone. To fully address this question, data of the type supplied by the COIN trial investigators (i.e. treatment effectiveness data for the addition of cetuximab to standard chemotherapy in both patients whose tumours are classified as KRAS wild type and those whose tumours are classified as KRAS mutant) would be required for each proposed KRAS mutation testing method. Ideally, data for all tests would be derived from the same study population, to allow meaningful comparison of the performance of tests for predicting treatment response without confounding by between-study variations in key participant characteristics. Following the recommendations made in NICE technology appraisal TA176,1 obtaining these data may be problematic as it could be argued that a trial in which patients are randomised to receive cetuximab in addition to standard chemotherapy or standard chemotherapy alone, regardless of tumour KRAS mutation status, would be unethical. Although the COIN trial54 was published after TA176,1 more recent UK trials such as New EPOC77 have tended to focus on determining the effectiveness of adding cetuximab to standard chemotherapy in patients with KRAS wild-type tumours. The recently complete, but as yet unpublished, New EPOC trial was a randomised open-label comparison between oxaliplatin/irinotecan plus fluorouracil plus cetuximab and oxaliplatin/irinotecan plus fluorouracil. The trial aimed to assess the effect on PFS of adding cetuximab to standard chemotherapy in patients with KRAS-wild type resectable CRC liver metastases who require chemotherapy. Trials of this type are not primarily concerned with the method used to establish mutation status. An alternative approach to this problem is provided by studies that report sufficient data to calculate the accuracy of different KRAS mutation tests for predicting response to treatment with cetuximab plus standard chemotherapy. These studies can potentially provide information on the extent to which KRAS mutation tests are able to predict resectability of liver metastases following treatment with cetuximab plus standard chemotherapy; outcome data (resection rates or ORRs) are reported for both patients with KRAS wild-type tumours and patients with KRAS mutant tumours. However, we were able to identify only one study of this type, the CELIM trial,52 which used an older version of the Therascreen RGQ PCR Kit. Neither the CELIM trial52 or the COIN trial54 were intended to assess KRAS mutation testing and neither reported comparative data for more than one KRAS mutation test; hence, any apparent differences in test performance observed between the two studies may have arisen as a result of differences in study populations. Of particular note is the way in which unresectable liver metastases were defined in the two studies: participants in the CELIM trial52 were described as having technically non-resectable or five or more liver metastases from CRC and it was therefore unclear whether some participants may have had potentially resectable metastases at baseline, whereas the COIN trial54 explicitly excluded patients receiving combination chemotherapy before resection of operable liver metastases. This difference may partially account for the marked difference in resection rates observable between the two studies for patients with KRAS wild-type tumours who were treated with cetuximab plus standard chemotherapy; (Commercial-in-confidence information has been removed),59 compared with a resection rate of 13/87 (15%) from the CELIM trial. 52 (Commercial-in-confidence information has been removed),59 whereas the COIN trial focused on ‘potentially curative liver resections’,54 and the standard chemotherapy regimens were different in the two trials.
Trials that compared the effectiveness of cetuximab plus standard chemotherapy with that of standard chemotherapy alone in patients with unresectable liver-limited metastases from CRC and whose tumours were KRAS wild type were also included in this review. These trials were included with the aim of providing some indication of how the favourable effects from the addition of cetuximab in these patients may vary according to how patients are selected for treatment (which KRAS mutation test is used). However, it should be noted that differences between these studies, other than the way in which KRAS wild-type mutation status is defined, particularly in relation to the baseline participant characteristics, are likely to contribute to any differences in treatment effects observed. In addition, these trials can provide no information about the relative effectiveness of cetuximab and standard chemotherapy compared with standard chemotherapy alone in patients whose tumours are classified as KRAS mutant.
The effectiveness data available to inform this assessment were very limited. In anticipation of this problem, our assessment included a survey of UK laboratories participating in the NEQAS scheme. This survey aimed to provide additional data on the technical performance of KRAS mutation tests, as seen in routine practice in the UK. We consider that data of this type are potentially more informative than data on the technical performance characteristics of tests obtained under research conditions, using non-clinical samples.
Cost-effectiveness
The review of economic analyses of different methods for KRAS mutation testing included only full economic analyses. Hence, economic studies that had information on test costs may be excluded from the review.
A de novo probabilistic model was developed to assess the cost-effectiveness of different methods for KRAS mutation testing to decide between standard chemotherapy and cetuximab in combination with standard chemotherapy in adults with mCRC in whom metastases are confined to the liver and are initially unresectable. To be consistent with related assessments/appraisals, it was first ensured that the model structure, model assumptions and input parameters in the de novo model were consistent with the manufacturer’s model used in NICE technology appraisal 176. 1,59,69 Model results were also consistent for patients with KRAS wild-type tumours in the sense that the use of cetuximab would still be considered cost-effective according to the de novo model.
In the assessment of the economic value of different tests, a link has to be established between test accuracy, clinical value (e.g. ORR, resection rate) and relative cost-effectiveness. Ideally, the performance of KRAS mutation tests would be assessed against an objective measure of the true presence/absence of a clinically relevant KRAS mutation (the ‘reference standard’), and the comparative effectiveness of treatment (chemotherapy plus cetuximab vs. chemotherapy alone) conditional on the true presence/absence of the KRAS mutation would be determined. However, different testing methods target different ranges of mutations and have different limits of detection (the lowest proportion of mutation detectable in tumour cells) and the optimal combination of mutation location and level for treatment selection remains unclear. For this reason, assessment of test performance based on comparison with a conventional ‘reference standard’ is currently not possible. An alternative way to determine the relative value of diagnostic methods for KRAS mutation testing is to use studies that report on the comparative treatment effect (or a substitute) in patients with both wild-type and mutant KRAS tumours. Thus, ORR or liver resection rate after treatment with cetuximab was assumed to correlate perfectly with the ‘true’ absence/presence of the KRAS mutation. The use of alternative outcome measures to determine test accuracy for the assessment of cost-effectiveness might impact the proportion of KRAS wild types to KRAS mutations and thus might substantially impact the assessment of cost-effectiveness (in either direction) as division of patients over the tumour mutation status categories is a major driver of cost-effectiveness. In the absence of an objective measure of the ‘true’ presence/absence of a clinically significant KRAS mutation, the current cost-effectiveness assessment is, at best, an approximation of the ‘true’ cost-effectiveness of test–treat combinations.
Evidence on test accuracy was available for only two tests (Therascreen KRAS RGQ PCR Kit and pyrosequencing); this was derived from ORR for the Therascreen KRAS RGQ PCR Kit and from resection rate for pyrosequencing. A major assumption underpinning our analyses was that the differences in liver resection rates observed in the two included studies from which these data were derived,52,54 and therefore also differences in the subsequent PFS and OS, can be attributed exclusively to the specific test used. In practice, this assumption would seem unlikely to hold true. These differences could also be caused by, for instance, differences in the characteristics of the respective study populations (i.e. with respect to the type of metastases) or differences in the standard chemotherapy regimen used. In addition, if the assumption of comparability of accuracy rates based on different measures (i.e. ORR and resection rate) holds true,39 this would reduce the likelihood that the main assumption holds.
Uncertainties
Clinical effectiveness
As noted in Strengths and limitations, one important consideration when selecting a KRAS mutation testing method is the variation between tests in the limit of detection (i.e. the minimum percentage of mutation in tumour cells required to produce a positive result). A lower limit of detection can enhance the ability of laboratories to produce results from poor-quality samples. However, it should not be assumed that a lower limit of detection will necessarily result in a more clinically effective test, as it is possible that the addition of cetuximab to standard chemotherapy may still be effective in patients with KRAS mutant tumours in which mutations are present at a very low level (a low proportion of tumour cells harbouring a mutation). None of the studies that met the inclusion criteria for this review reported any data on variation in treatment effect according to the limit of detection used to define a KRAS mutant tumour.
A further area of uncertainty concerns the clinical value of detecting rarer KRAS mutations. The majority of the evidence on the effectiveness of first-line treatment with cetuximab plus standard chemotherapy in patients with liver-limited colorectal metastases whose tumours are KRAS wild type was derived from patients selected using tests that target mutations in codons 12 and 13; only the COIN trial54 used a test method that also targeted mutations in codon 61. Indeed, although no testing method was specified, the ASCO PCO published in 200915 recommended universal KRAS mutation testing in patients with mCRC in whom treatment with EGFR inhibitors is being considered. The recommendation also stated that testing should be carried out in an accredited laboratory and that patients whose tumours have KRAS mutations in codons 12 or 13 should not be treated with EGFR receptor inhibitors. The PCO also highlighted the uncertainty around the clinical relevance of detecting rare mutations in codons 61 and 146. 15 The COIN trial54 reported detection of the following mutations in codon 61, for all samples successfully analysed: Q61H (13/1059, 1.2%), Q61L (5/1289, 0.4%) and Q61R (6/1289, 0.5%); it was not clear whether any of these mutations were detected in patients with liver-limited metastases. The additional clinical value of using tests that target a wider range of mutations remains uncertain, as the low frequency of most KRAS mutations makes it very difficult to adequately assess treatment effects or resistance to EGFR inhibitors in patients with these mutations. A large multicentre observational study conducted in Italy by the KRAS aKtive network78 (a programme promoted by the Italian Association of Medical Oncology and the Italian Society of Surgical Pathology and Cytopathology to support the activity of oncologists and pathologists involved in the management of mCRC patients who require KRAS mutation testing) has collected data on a total of 7432 KRAS mutation analyses. The majority (77%) of testing was conducted using Sanger sequencing, and mutations other than those in codons 12 and 13 represented approximately 5% of the total detected. In addition to the issue of rare mutations, questions have been raised whether all codon 13 mutations predict lack of benefit from treatment with EGFR inhibitors; De Roock et al. 79 suggested that the KRAS G13D mutation may not predict lack of benefit. The COIN trial54 identified 110 participants with this mutation and reported no difference in outcome with the addition of cetuximab to standard chemotherapy; the HR for PFS was 1.11 (95% CI 0.76 to 1.63) in patients with the KRAS G13D mutation and 1.05 (95% CI 0.87 to 1.27) for all other mutations (data for the whole trial population, not the liver-limited metastases subgroup).
As discussed in Strengths and limitations, when assessing the performance of different KRAS mutation tests for the prediction of response to treatment, it is important to have information on the relative effectiveness of different treatment options in patients whose tumours are KRAS mutant as well as in those whose tumours are KRAS wild type. This is because, even when the benefits of adding cetuximab to standard chemotherapy in patients with KRAS wild-type tumours have been established, it is important to determine whether there are any negative effects associated with adding cetuximab to the treatment of patients with KRAS mutant tumours. If there are no negative effects associated with ‘overtreatment’ of patients with KRAS mutant tumours with cetuximab, then a conservative classification of patients with rare or low-level mutations as ‘wild type’ for treatment purposes may be considered clinically appropriate. Similarly, the ability of a test to detect rare mutations and/or a low limit of detection may be considered less important. None of the studies included in this assessment reported any difference in OS between patients with KRAS mutant tumours treated with cetuximab plus standard chemotherapy and those treated with standard chemotherapy alone. The CRYSTAL trial27 also reported no difference in ORRs or PFS, whereas the OPUS trial28 reported a lower ORR (OR 0.46, 95% CI 0.23 to 0.92) and shorter PFS (HR 1.72, 95% CI 1.10 to 2.68) for patients with KRAS mutant tumours who were treated with cetuximab plus standard chemotherapy than for those treated with standard chemotherapy alone. 27 These data were for all patients in the trials with KRAS mutant tumours; for both the CYSTAL and OPUS trials, data on treatment effectiveness in patients with KRAS mutant tumours were not available for the subgroup of patients with liver-limited metastases. Additional data supplied by the COIN trial investigators that were specific to patients with inoperable liver-limited metastases showed no significant difference in PFS or potentially curative resection rates between patients with KRAS mutant tumours who were treated with cetuximab plus standard chemotherapy and patients with KRAS mutant tumours who were treated with standard chemotherapy alone (D Fisher, personal communication).
The timing of KRAS mutation testing can vary, with some clinicians/hospitals undertaking routine testing of all CRC patients at diagnosis, potentially before the disease becomes metastatic, and others waiting until metastases have been detected. It should be noted that only one of the UK laboratories responding to our survey reported routine KRAS testing in all CRC patients. It could be argued that routine testing avoids potential delays in the start of treatment; however, clinical opinion suggested that any such delays would be unlikely to have measurable effects on clinical outcomes. Also, because cetuximab is added to standard chemotherapy in patients with KRAS wild-type tumours, standard chemotherapy can be commenced whilst awaiting the results of KRAS testing so that only the potential additional benefit of cetuximab is subject to delay. A related question is whether a stored biopsy sample from the primary tumour is adequate for KRAS mutation testing once metastases have been detected, or whether potential heterogeneity between tumour sites means that a sample from the metastatic site is preferable. Use of the primary tumour sample is likely to be considered preferable because all patients should have already undergone a biopsy at diagnosis for histological typing; thus, the risks and discomfort of a further invasive procedure (liver biopsy) could potentially be avoided. None of the studies included in this assessment considered the potential impact of sample site on the results of KRAS mutation testing. A systematic review80 identified by our searches, which did not meet the inclusion criteria for this assessment, assessed the concordance of KRAS mutations between primary CRC tissue and mCRC tissue. This review included 19 publications reporting data on a total of 986 paired samples from primary tumours and distant metastases (including, but not limited to, the liver) and reported a pooled concordancy rate of 94.1% (95% CI 88.3% to 95.0%). One of the primary studies included in this review specifically assessed KRAS mutation concordancy between primary colorectal tumours and liver metastases in 305 paired samples;32 KRAS mutation status was determined based on pyrosequencing of codons 12 and 13. This study reported a concordancy rate of 96.4% (95% CI 93.6% to 98.2%), with clinically relevant discordance in six participants (2.0% of the study population); five primary tumours had a KRAS mutation with a wild-type metastasis and one primary tumour was wild type with a KRAS mutation in the metastasis. Although outside the scope of this assessment, these studies could be interpreted as supporting the view that KRAS mutation testing using stored samples from the primary tumour is a valid approach and that testing using liver biopsy samples is unlikely to produce significant clinical benefit.
A variety of KRAS mutation testing methods is currently used by accredited NHS laboratories in England and Wales. None of the methods reported in our survey exactly matched the methods used in any of the studies identified in our systematic review. However, because the COIN trial54 reported > 99% concordance on KRAS genotyping between pyrosequencing and MALDI-TOF, it may be assumed that accuracy data from the COIN trial are also representative of the accuracy of pyrosequencing (used as a single test) for KRAS mutations in codons 12, 13 and 61, the method used by the majority of UK laboratories who responded to our survey. It should be noted that the performance of pyrosequencing methods may vary when different primers are used and that the potential clinical effects of using different KRAS mutation test methods to make decisions on first-line treatment in patients with unresectable liver-limited CRC metastases remain uncertain. The Therascreen KRAS RGQ PCR Kit is the only product currently approved by the FDA;16 however, the clinical study used to support its approval was not conducted in the population specified for this assessment and none of the respondents to our survey of UK laboratories reported using this product. The Therascreen KRAS RGQ PCR Kit, Therascreen KRAS Pyro Kit, cobas KRAS Mutation Test, KRAS LightMix Kit and KRAS StripAssay are all CE marked. No direct data, either from our systematic review or from our survey of UK laboratories, are currently available for the following KRAS mutation testing methods listed in the scope: next-generation sequencing of codons 12, 13 and 61; KRAS StripAssay; MALDI-TOF mass spectrometry of codons 12, 13 and 61 used alone; and HRM analysis of codons 12, 13 and 61 used alone. As was the case for pyrosequencing, concordance between the two KRAS mutation testing methods used in the COIN trial54 means that accuracy data derived from the trial may also be assumed to be representative of the performance of MALDI-TOF when used as a single test for the detection of KRAS mutations in codons 12, 13 and 61.
Cost-effectiveness
Major assumptions were made to be able to model the relative cost-effectiveness of different KRAS mutation tests. It was assumed that the differences in resection rates between the CELIM trial52 and the COIN trial54 and associated subsequent PFS and OS were exclusively attributable to the different mutation tests used (the Therascreen KRAS RGQ PCR Kit and pyrosequencing respectively) to distinguish between patients whose tumours are KRAS wild type and those whose tumours are KRAS mutant. As discussed previously, it is questionable whether this assumption would hold true. Furthermore, to calculate the proportions of patients with KRAS wild-type and KRAS mutant test results, patients with a KRAS wild-type test result were categorised as FP if no objective response was observed on cetuximab (for the Therascreen KRAS RGQ PCR Kit) or when no liver resection was performed (for pyrosequencing) or as TP if a objective response was observed or a resection was performed. Likewise, patients with a KRAS mutant test result were classified as TN when no objective response was observed on cetuximab (for the the Therascreen KRAS RGQ PCR Kit) or no resection was performed (for pyrosequencing) and as FN when an objective response was observed or a liver resection was performed. Ideally, the categorisation of TPs/FP and TNs/FNs should be based on an objective measure of the true presence/absence of a clinically relevant KRAS mutation. However, as previously described, the uncertainty around the exact definition of a clinically relevant mutation is such that, at present, there is no such thing as an objective measure or gold standard.
Moreover, as this model was partially based on the evidence and model structure used in the technology appraisal of cetuximab for the first-line treatment of mCRC,1,59,69 the assumptions underlying that appraisal also apply to this assessment. One example, which applies only to the linked evidence analysis, is the implicit assumption in the manufacturer’s model that, in the absence of a chemotherapy-only arm in the CELIM trial,52 resection rates from the GERCOR trial68 can be applied to patients with KRAS mutant and KRAS unknown status tumours treated with standard chemotherapy, whereas resection rates for patients with KRAS wild-type tumours treated with cetuximab were taken from the CELIM-trial. 52
Finally, it should be emphasised that the uncertainty resulting from the above-mentioned assumptions was not parameterised in the model and is therefore not reflected in the PSA or in the CEACs.
Chapter 6 Conclusions
Implications for service provision
There was no strong evidence that any one method of KRAS mutation testing had greater accuracy than any other for predicting tumour response or potentially curative resection following treatment with cetuximab plus standard chemotherapy in patients with mCRC whose metastases were limited to the liver and were unresectable before chemotherapy. The clinical effectiveness of cetuximab plus standard chemotherapy in patients whose tumours are KRAS wild type did not appear to vary according to which method was used to determine tumour KRAS mutation status. However, it should be noted that the available data were not adequate to fully address either the comparative accuracy of different KRAS mutation testing methods for predicting tumour response or the comparative clinical effectiveness of cetuximab plus standard chemotherapy in patients whose tumours are KRAS wild type, as defined by different testing methods.
The results of the linked evidence analysis indicated that the Therascreen KRAS RGQ PCR Kit was more costly and more effective than pyrosequencing, with an ICER of £17,019 per QALY gained. Sensitivity analyses did not show substantial differences compared with the base case. The key driver behind the outcome was the difference in resection rate between treatment with and treatment without cetuximab and the proportions of patients with KRAS wild-type, KRAS mutant and unknown tumour status, which is determined by test accuracy and therefore, for the most part, is dependent on ORR (for Therascreen KRAS RGQ PCR Kit) or resection rate (for pyrosequencing). It should be noted that some problematic and substantial assumptions were necessary to arrive at the economic results, in particular the assumption that the differences in resection rates observed between the different studies are solely the result of the different tests used. This ignores all other factors that can explain variations in outcomes between the studies. Therefore, these outcomes of the assessment of cost-effectiveness should be interpreted with extreme caution.
The results of the assumption of equal prognostic value analysis (including all tests for which information on technical performance was available from the online survey of NHS laboratories in England and Wales) showed that the cobas KRAS Mutation Test is the least expensive and least effective strategy and that Sanger sequencing and HRM analysis are equally the most costly and most effective, with an ICER of £69,815 per QALY gained compared with the cobas KRAS Mutation Test. The other two strategies included in this analysis, that is, the Therascreen KRAS RGQ PCR Kit and pyrosequencing, are ruled out by extended dominance.
There are no data on the clinical effectiveness or cost-effectiveness of next-generation sequencing of codons 12, 13 and 61; the KRAS StripAssay; MALDI-TOF mass spectrometry of codons 12, 13 and 61 used alone; and HRM analysis of codons 12, 13 and 61 used alone. No published studies were identified for any of these methods and none of these methods are currently in routine clinical use in any of the NHS laboratories in England and Wales that responded to our survey.
Suggested research priorities
The available data have limitations in respect of their ability to address the overall aim of this assessment, that is, to compare the clinical effectiveness of different methods of KRAS mutation testing to determine which patients may benefit from the addition of cetuximab to treatment with standard chemotherapy and which should receive standard chemotherapy alone. Because each different testing method potentially selects a subtly different population, based on the targeting of a different range of mutations and different limits of detection, the most informative studies are those that provide full information on the comparative treatment effect (cetuximab plus standard chemotherapy vs. standard chemotherapy alone) for both patients with KRAS wild-type tumours and patients with KRAS mutant tumours. No published studies of this type were identified. Additional data supplied by the COIN trial investigators meant that these data could be derived for a combination of pyrosequencing and MALDI-TOF mass spectrometry (both methods used for all samples) (D FIsher, personal communication). The very high concordance (> 99%) between the two KRAS mutation testing methods used in the COIN trial means that data from this trial may be assumed to also be representative of the expected values when pyrosequencing or MALDI-TOF are used as single tests to define tumour KRAS mutation status. However, further similar trials are unlikely as randomisation of patients to cetuximab plus standard chemotherapy or standard chemotherapy alone, regardless of tumour KRAS mutation status, would be against current clinical guidance and would be likely to be considered unethical. One possible solution to this problem would be to retest stored samples from previous studies in which patient outcomes are already known, using those KRAS mutation testing methods for which adequate data are currently unavailable. This approach could provide a ‘black box’ answer whereby the relative effectiveness of cetuximab plus standard chemotherapy and standard chemotherapy alone in patients with KRAS wild-type and KRAS mutant tumours could be determined for each testing method. However, it would not provide any information on the underlying reason(s) for any observed differences between tests. As they are likely to represent the most practical approach to obtaining informative data, retrospective comparative accuracy studies, using stored samples for which the patient outcome is already known, should be given priority. The application of this approach to the evaluation of next-generation sequencing might be considered a particular priority, given the likely adoption of next-generation sequencing techniques by laboratories in the near future.
Some methods of KRAS mutation testing, for example the Therascreen KRAS Pyro Kit, can provide quantitative results. Should quantitative testing become part of routine practice, longitudinal follow-up studies relating the level of mutation and/or the presence of rarer mutations to patient outcomes would become possible. Studies of this type could help to assess which features of KRAS mutation tests are likely to be important in determining their clinical effectiveness and should be considered going forward.
Building on information gained from the two study types described above, preliminary research to develop a multifactorial prediction model should be considered. Initially, research of this type is likely to be exploratory in nature; however, models developed could form the basis of tools that will eventually help determine more accurately which patients are most likely to benefit from the addition of cetuximab to treatment with standard chemotherapy.
As the uncertainties associated with clinical effectiveness forced the major assumptions in the economic evaluation, this type of research would also facilitate economic analyses of KRAS mutation testing.
Acknowledgements
The authors acknowledge the clinical advice and expert opinion provided by Dr Mark Harrison, Consultant Oncologist, Mount Vernon Cancer Centre, East and North Hertfordshire NHS Trust; Dr Newton Wong, Consultant Histopathologist, University Hospitals Bristol NHS Foundation Trust; Miss Jennie Bell, Consultant Molecular Geneticist, Birmingham Women’s NHS Foundation Trust; and Dr Phil Chambers, Genomics Facility, Leeds Cancer Research UK Centre. The authors would like to thank the COIN trial investigators, in particular Dr Richard Wilson and Professor Timothy Maughan, for providing additional data. The authors would like to thank the representatives of the NHS laboratories providing KRAS mutation testing who kindly completed our online survey. The authors would also like to thank Merck Serono UK Ltd for providing access to the cost-effectiveness model used in NICE technology appraisal 176. Finally, the authors would like to thank the lay members of the NICE Diagnostics Advisory Committee and Assessment Subgroup for providing input on the patients’ perspective at key stages of the assessment process.
Contributions of authors
Marie Westwood and Penny Whiting planned and performed the systematic review and interpretation of the evidence.
Manuela Joore, Thea van Asselt and Bram Ramaekers planned and performed the cost-effectiveness analyses and interpreted the results.
Nigel Armstrong contributed to the planning and interpretation of the cost-effectiveness analyses and the acquisition of input data for modelling.
Caro Noake and Janine Ross devised and performed the literature searches and provided information support to the project.
Jos Kleijnen and Johan Severens provided senior advice and support to the systematic review and cost-effectiveness analyses respectively.
All parties were involved in drafting and/or commenting on the report.
Disclaimers
This report presents independent research funded by the National Institute for Health Research (NIHR). The views and opinions expressed by authors in this publication are those of the authors and do not necessarily reflect those of the NHS, the NIHR, NETSCC, the HTA programme or the Department of Health. If there are verbatim quotations included in this publication the views and opinions expressed by the interviewees are those of the interviewees and do not necessarily reflect those of the authors, those of the NHS, the NIHR, NETSCC, the HTA programme or the Department of Health.
References
- Cetuximab for the First-Line Treatment of Metastatic Colorectal Cancer. London: NICE; 2009.
- Cancer Registrations in England, 2010: Number of New Cases Diagnosed By Site. London: Office for National Statistics; 2012.
- Bowel Cancer Incidence Statistics. London: Cancer Research UK; 2012.
- Bowel Cancer Mortality Statistics. London: Cancer Research UK; 2012.
- Bowel Cancer Survival Statistics. London: Cancer Research UK; 2012.
- Cancer Incidence by Deprivation: England, 1995–2004. London: National Cancer Intelligence Network Coordinating Centre; 2009.
- Finan P, Smith J, Greenaway K, Yelland A, Scott N, Henderson J, et al. National Bowel Cancer Audit Report 2011. London: NHS Information Centre for Health and Social Care; 2011.
- Neumann J, Zeindl-Eberhart E, Kirchner T, Jung A. Frequency and type of KRAS mutations in routine diagnostic analysis of metastatic colorectal cancer. Pathol Res Pract 2009;205:858-62. http://dx.doi.org/10.1016/j.prp.2009.07.010.
- Cejas P, Lopez-Gomez M, Aguayo C, Madero R, de Castro Carpeno J, Belda-Iniesta C, et al. KRAS mutations in primary colorectal cancer tumors and related metastases: a potential role in prediction of lung metastasis. PLoS One 2009;4. http://dx.doi.org/10.1371/journal.pone.0008199.
- Baldus SE, Schaefer KL, Engers R, Hartleb D, Stoecklein NH, Gabbert HE. Prevalence and heterogeneity of KRAS, BRAF, and PIK3CA mutations in primary colorectal adenocarcinomas and their corresponding metastases. Clin Cancer Res 2010;16:790-9. http://dx.doi.org/10.1158/1078-0432.CCR-09-2446.
- Vaughn CP, Zobell SD, Furtado LV, Baker CL, Samowitz WS. Frequency of KRAS, BRAF, and NRAS mutations in colorectal cancer. Genes Chromosomes Cancer 2011;50:307-12. http://dx.doi.org/10.1002/gcc.20854.
- KRAS Pilot EQA 2012–13. Newcastle upon Tyne: UK NEQAS for Molecular Genetics; 2012.
- Liu P, Liang H, Xue L, Yang C, Liu Y, Zhou K, et al. Potential clinical significance of plasma-based KRAS mutation analysis using the COLD-PCR/TaqMan((R)) – MGB probe genotyping method. Exp Ther Med 2012;4:109-12.
- Morgan SR, Whiteley J, Donald E, Smith J, Eisenberg MT, Kallam E, et al. Comparison of KRAS mutation assessment in tumor DNA and circulating free DNA in plasma and serum samples. Clin Med Insights Pathol 2012;5:15-22. http://dx.doi.org/10.4137/CPath.S8798.
- Allegra CJ, Jessup JM, Somerfield MR, Hamilton SR, Hammond EH, Hayes DF, et al. American Society of Clinical Oncology provisional clinical opinion: testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol 2009;27:2091-6. http://dx.doi.org/10.1200/JCO.2009.21.9170.
- US Food and Drug Administration . Summary of Safety and Effectiveness Data: Therascreen® KRAS RGQ PCR Kit 2012. www.accessdata.fda.gov/cdrh_docs/pdf11/P110030b.pdf (accessed 21 May 2013).
- QIAGEN . Therascreen KRAS RGQ PCR Kit Handbook (Version 1) 2011. www.nice.org.uk/nicemedia/live/13937/62336/62336.pdf (accessed 5 June 2014).
- QIAGEN . Therascreen KRAS Pyro Kit Handbook (Version 1) 2011. www.nice.org.uk/nicemedia/live/13937/62336/62336.pdf (accessed 5 June 2014).
- TIB MOLBIOL . LightMix Kit K-RAS Mutation Codons 12 13 (Version 110621) 2011. www.nice.org.uk/nicemedia/live/13937/62336/62336.pdf (accessed 5 June 2014).
- ViennaLab Diagnostics . KRAS StripAssay Instructions for Use 2010. www.nice.org.uk/nicemedia/live/13937/62336/62336.pdf (accessed 5 June 2014).
- QIAGEN . Principle of Pyrosequencing Technology 2012. www.pyrosequencing.com/DynPage.aspx?id=7454&mn1=1366&mn2=1367.
- Applied Biosystems . Overview of DNA Sequencing 2011. www.appliedbiosystems.com/absite/us/en/home/applications-technologies/dna-sequencing-fragment-analysis/overview-of-dna-sequencing.html.
- Illumina . An Introduction to Next Generation Sequencing Technology 2012. www.nice.org.uk/nicemedia/live/13937/62336/62336.pdf (accessed 5 June 2014).
- PrimerDesign . Beginners Guide to High Resolution Melt Analysis 2009. www.nice.org.uk/nicemedia/live/13937/62336/62336.pdf (accessed 5 June 2014).
- Elso C, Toohey B, Reid GE, Poetter K, Simpson RJ, Foote SJ. Mutation detection using mass spectrometric separation of tiny oligonucleotide fragments. Genome Res 2002;12:1428-33. http://dx.doi.org/10.1101/gr.GR-1578R.
- Molinari F, Felicioni L, Buscarino M, De Dosso S, Buttitta F, Malatesta S, et al. Increased detection sensitivity for KRAS mutations enhances the prediction of anti-EGFR monoclonal antibody resistance in metastatic colorectal cancer. Clin Cancer Res 2011;17:4901-14. http://dx.doi.org/10.1158/1078-0432.CCR-10-3137.
- Van Cutsem E, Kohne CH, Hitre E, Zaluski J, Chang Chien CR, Makhson A, et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med 2009;360:1408-17. http://dx.doi.org/10.1056/NEJMoa0805019.
- Bokemeyer C, Bondarenko I, Hartmann JT, De Braud F, Schuch G, Zubel A, et al. Efficacy according to biomarker status of cetuximab plus FOLFOX-4 as first-line treatment for metastatic colorectal cancer: the OPUS study. Ann Oncol 2011;22:1535-46. http://dx.doi.org/10.1093/annonc/mdq632.
- Colorectal Cancer: the Diagnosis and Management of Colorectal Cancer. London: NICE; 2011.
- Labianca R, Nordlinger B, Beretta GD, Brouquet A, Cervantes A. Primary colon cancer: ESMO clinical practice guidelines for diagnosis, adjuvant treatment and follow-up. Ann Oncol 2010;21:70-7. http://dx.doi.org/10.1093/annonc/mdq168.
- Royal College of Pathologists . The Future Provision of Molecular Diagnostic Services for Aquired Diseases in the UK 2010. www.nice.org.uk/nicemedia/live/13937/62336/62336.pdf (accessed 5 June 2014).
- Knijn N, Mekenkamp LJ, Klomp M, Vink-Borger ME, Tol J, Teerenstra S, et al. KRAS mutation analysis: a comparison between primary tumours and matched liver metastases in 305 colorectal cancer patients. Br J Cancer 2011;104:1020-6. http://dx.doi.org/10.1038/bjc.2011.26.
- Richman SD, Chambers P, Seymour MT, Daly C, Grant S, Hemmings G, et al. Intra-tumoral heterogeneity of KRAS and BRAF mutation status in patients with advanced colorectal cancer (aCRC) and cost-effectiveness of multiple sample testing. Anal Cell Pathol (Amst) 2011;34:61-6.
- Guidance on the Use of Capecitabine and Tegafur with Uracil for Metastatic Colorectal Cancer. London: NICE; 2003.
- Summary of Product Characteristics: Erbitux 5 mg/ml Solution for Infusion. London: EMEA; 2009.
- Cetuximab, Bevacizumab and Panitumumab for the Treatment of Metastatic Colorectal Cancer after First-Line Chemotherapy. London: NICE; 2012.
- World Health Organization . WHO Handbook for Reporting Results of Cancer Treatment 1979. http://whqlibdoc.who.int/offset/WHO_OFFSET_48.pdf (accessed 4 December 2012).
- Therasse P, Arbuck S, Eisenhauer E, Wanders J, Kaplan R, Rubinstein L, et al. New guidelines to evaluate the response to treatment in solid tumors. J Natl Cancer Inst 2000;92:205-16. http://dx.doi.org/10.1093/jnci/92.3.205.
- Folprecht G, Grothey A, Alberts S, Raab HR, Kohne CH. Neoadjuvant treatment of unresectable colorectal liver metastases: correlation between tumour response and resection rates. Ann Oncol 2005;16:1311-19. http://dx.doi.org/10.1093/annonc/mdi246.
- Systematic Reviews: CRD’s Guidance for Undertaking Reviews in Health Care. York: University of York; 2009.
- Diagnostics Assessment Programme Manual. Machester: NICE; 2011.
- Cochrane Diagnostic Test Accuracy Working Group . Handbook for DTA Reviews 2009. http://srdta.cochrane.org/handbook-dta-reviews (accessed 29 August 2013).
- McGowan J, Sampson M, Lefebvre C. An evidence based checklist for the peer review of electronic search strategies (PRESS EBC). Evidence Based Libr Inform Prac 2010;5:1-6.
- Royle P, Waugh N. Should systematic reviews include searches for published errata?. Health Info Libr J 2004;21:14-20. http://dx.doi.org/10.1111/j.1471-1842.2004.00459.x.
- Wright K, McDaid C. Is the Retraction of Journal Articles in Electronic Journals and Databases Consistent and Timely? A Case Study n.d.
- Wright K, McDaid C. Reporting of article retractions in bibliographic databases and online journals. J Med Libr Assoc 2011;99:164-7. http://dx.doi.org/10.3163/1536-5050.99.2.010.
- Higgins JP, Altman DG, Gotzsche PC, Juni P, Moher D, Oxman AD, et al. The Cochrane Collaboration’s tool for assessing risk of bias in randomised trials. BMJ 2011;343. http://dx.doi.org/10.1136/bmj.d5928.
- Whiting PF, Rutjes AWS, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med 2011;155:529-36. http://dx.doi.org/10.7326/0003-4819-155-8-201110180-00009.
- Reitsma JB, Glas AS, Rutjes AWS, Scholten RJPM, Bossuyt PMM, Zwinderman AH. Bivariate analysis of sensitivity and specificity produces informative summary measures in diagnostic reviews. J Clin Epidemiol 2005;58:982-90. http://dx.doi.org/10.1016/j.jclinepi.2005.02.022.
- Harbord RM, Deeks JJ, Egger M, Whiting P, Sterne JA. A unification of models for meta-analysis of diagnostic accuracy studies. Biostatistics 2007;8:239-51. http://dx.doi.org/10.1093/biostatistics/kxl004.
- Harbord RM, Whiting P, Sterne JA, Egger M, Deeks JJ, Shang A, et al. An empirical comparison of methods for meta-analysis of diagnostic accuracy showed hierarchical models are necessary. J Clin Epidemiol 2008;61:1095-103. http://dx.doi.org/10.1016/j.jclinepi.2007.09.013.
- Folprecht G, Gruenberger T, Bechstein WO, Raab HR, Lordick F, Hartmann JT, et al. Tumour response and secondary resectability of colorectal liver metastases following neoadjuvant chemotherapy with cetuximab: the CELIM randomised phase 2 trial. Lancet Oncol 2010;11:38-47. http://dx.doi.org/10.1016/S1470-2045(09)70330-4.
- Kohne C, Bokemeyer C, Heeger S, Sartorius U, Rougier P, Van Cutsem E. Efficacy of chemotherapy plus cetuximab according to metastatic site in KRAS wild-type metastatic colorectal cancer (mCRC): analysis of CRYSTAL and OPUS studies. J Clin Oncol 2011;29.
- Maughan TS, Adams RA, Smith CG, Meade AM, Seymour MT, Wilson RH, et al. Addition of cetuximab to oxaliplatin-based first-line combination chemotherapy for treatment of advanced colorectal cancer: results of the randomised phase 3 MRC COIN trial. Lancet 2011;377:2103-14. http://dx.doi.org/10.1016/S0140-6736(11)60613-2.
- Xu J, Ye L, Ren L, Wei Y. A randomized, controlled trial of cetuximab plus chemotherapy for patients with kras wild-type unresectable colorectal liver-limited metastases. Ann Oncol 2012;23.
- Bokemeyer C, Bondarenko I, Makhson A, Hartmann JT, Aparicio J, De Braud F, et al. Fluorouracil, leucovorin, and oxaliplatin with and without cetuximab in the first-line treatment of metastatic colorectal cancer. J Clin Oncol 2009;27:663-71. http://dx.doi.org/10.1200/JCO.2008.20.8397.
- Ocvirk J, Brodowicz T, Wrba F, Ciuleanu TE, Kurteva G, Beslija S, et al. Cetuximab plus FOLFOX6 or FOLFIRI in metastatic colorectal cancer: CECOG trial. World J Gastroenterol 2010;16:3133-43. http://dx.doi.org/10.3748/wjg.v16.i25.3133.
- Tveit KM, Guren T, Glimelius B, Pfeiffer P, Sorbye H, Pyrhonen S, et al. Phase III trial of cetuximab with continuous or intermittent fluorouracil, leucovorin, and oxaliplatin (Nordic FLOX) versus FLOX alone in first-line treatment of metastatic colorectal cancer: the NORDIC-VII study. J Clin Oncol 2012;30:1755-62. http://dx.doi.org/10.1200/JCO.2011.38.0915.
- Erbitux (Cetuximab) for the First-Line Treatment of Metastatic Colorectal Cancer: Submission to National Institute for Health and Clinical Excellence. Merck Serono Ltd; 2008.
- Xu J. Cetuximab in Combination with Chemotherapy for the Treatment of Metastatic Colorectal Cancer. Bethesda, MD: National Library of Medicine; 2012.
- Health Quality Ontario . KRAS testing for anti-EGFR therapy in advanced colorectal cancer: an evidence-based and economic analysis. Ont Health Technol Assess Ser 2010;10:1-49.
- Vijayaraghavan A, Efrusy MB, Goke B, Kirchner T, Santas CC, Goldberg RM. Cost-effectiveness of KRAS testing in metastatic colorectal cancer patients in the United States and Germany. Int J Cancer 2012;131:438-45. http://dx.doi.org/10.1002/ijc.26400.
- Behl AS, Goddard KAB, Flottemesch TJ, Veenstra D, Meenan RT, Lin JS, et al. Cost-effectiveness analysis of screening for KRAS and BRAF mutations in metastatic colorectal cancer. J Natl Cancer Inst 2012;104:1785-95. http://dx.doi.org/10.1093/jnci/djs433.
- Shiroiwa T, Motoo Y, Tsutani K. Cost-effectiveness analysis of KRAS testing and cetuximab as last-line therapy for colorectal cancer. Mol Diagn Ther 2010;14:375-84. http://dx.doi.org/10.1007/BF03256395.
- Blank PR, Moch H, Szucs TD, Schwenkglenks M. KRAS and BRAF mutation analysis in metastatic colorectal cancer: a cost-effectiveness analysis from a Swiss perspective. Clin Cancer Res 2011;17:6338-46. http://dx.doi.org/10.1158/1078-0432.CCR-10-2267.
- Drummond MF, Jefferson TO. Guidelines for authors and peer reviewers of economic submissions to the BMJ. The BMJ Economic Evaluation Working Party. BMJ 1996;313:275-83. http://dx.doi.org/10.1136/bmj.313.7052.275.
- Karapetis CS, Khambata-Ford S, Jonker DJ, O’Callaghan CJ, Tu D, Tebbutt NC, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med 2008;359:1757-65. http://dx.doi.org/10.1056/NEJMoa0804385.
- Tournigand C, André T, Achille E, Lledo G, Flesh M, Mery-Mignard D, et al. FOLFORI followed by FOLFOX6 or the reverse sequence in advanced colorectal cancer: a randomized GERCOR study. J Clin Oncol 2004;22:229-37. http://dx.doi.org/10.1200/JCO.2004.05.113.
- Erbitux (Cetuximab) for the First-Line Treatment of Metastatic Colorectal Cancer: Submission to National Institute for Health and Clinical Excellence. Merck Serono Ltd; 2008.
- Andreyev HJN, Norman AR, Cunningham D, Oates J, Dix BR, Iacopetta BJ, et al. Kirsten ras mutations in patients with colorectal cancer: the ‘RASCAL II’ study. Br J Cancer 2001;85:692-6. http://dx.doi.org/10.1054/bjoc.2001.1964.
- Nordlinger B, Sorbye H, Glimelius B, Poston GJ, Schlag PM, Rougier P, et al. Perioperative chemotherapy with FOLFOX4 and surgery versus surgery alone for resectable liver metastases from colorectal cancer (EORTC Intergroup trial 40983): a randomised controlled trial. Lancet 2008;371:1007-16. http://dx.doi.org/10.1016/S0140-6736(08)60455-9.
- Simmonds PC, Primrose JN, Colquitt JL, Garden OJ, Poston GJ, Rees M. Surgical resection of hepatic metastases from colorectal cancer: a systematic review of published studies. Br J Cancer 2006;94:982-99. http://dx.doi.org/10.1038/sj.bjc.6603033.
- Jonker DJ, O’Callaghan CJ, Karapetis CS, Zalcberg JR, Tu D, Au HJ, et al. Cetuximab for the treatment of colorectal cancer. N Engl J Med 2007;357:2040-8. http://dx.doi.org/10.1056/NEJMoa071834.
- Mittmann N, Au HJ, Tu D, O’Callaghan CJ, Karapetis CS, Moore MJ, et al. A prospective economic analysis of cost-effectiveness of cetuximab for metastatic colorectal cancer patients from the NCIC CTG and AGITG CO.17 trial. J Clin Oncol 2008;26.
- Adam R, Delvart V, Pascal G, Valeanu A, Castaing D, Azoulay D, et al. Rescue surgery for unresectable colorectal liver metastases downstaged by chemotherapy: a model to predict long-term survival. Ann Surg 2004;240:644-57.
- Whiting P, Westwood M, Beynon R, Burke M, Sterne JA, Glanville J. Inclusion of methodological filters in searches for diagnostic test accuracy studies misses relevant studies. J Clin Epidemiol 2011;64:602-7. http://dx.doi.org/10.1016/j.jclinepi.2010.07.006.
- Combination Chemotherapy with or without Cetuximab before and after Surgery in Treating Patients with Resectable Liver Metastases Caused by Colorectal Cancer. Bethesda, MD: National Library of Medicine; 2013.
- Marchetti A, Pinto C, Taddei G, Clemente C, Troncone G, Russo A, et al. KRAS aKtive, an Italian network for assessment of KRAS mutations in colorectal cancer patients: results on 7,432 cases. ASCO Meeting Abstr 2012;30.
- De Roock W, Jonker DJ, Di Nicolantonio F, Sartore-Bianchi A, Tu D, Siena S, et al. Association of KRAS p.G13D mutation with outcome in patients with chemotherapy-refractory metastatic colorectal cancer treated with cetuximab. JAMA 2010;304:1812-20. http://dx.doi.org/10.1001/jama.2010.1535.
- Han CB, Li F, Ma JT, Zou HW. Concordant KRAS mutations in primary and metastatic colorectal cancer tissue specimens: a meta-analysis and systematic review. Cancer Invest 2012;30:741-7. http://dx.doi.org/10.3109/07357907.2012.732159.
- Adams R, Maughan T, Smith C, Seymour M, Wilson R, Meade A, et al. Addition of cetuximab to oxalplatin-based combination chemotherapy (CT) in patients with K-ras wild-type advanced colorectal cancer (ACRC): effects on overall survival. MRC COIN (CR10) trial results. Ann Oncol 2009;20.
- Adams R, Wilson RH, Seymour MT, Meade AM, Madi A, Cassidy J, et al. Intermittent versus continuous oxaliplatin-fluoropyrimidine (Ox-Fp) chemotherapy (CT) in first-line treatment of patients (pts) with advanced colorectal cancer (aCRC): predictive factors (PF), quality of life (QL), and final efficacy results from the MRC COIN trial. J Clin Oncol 2010;28.
- Adams R, Wilson R, Seymour M, Meade A, Madi A, Cassidy J, et al. Intermittent vs. continuous oxaliplatinfluoropyrimidine chemotherapy in the MRC COIN trial in advanced colorectal cancer: updated efficacy results, quality of life and potential predictive factors. 12th World Congress on Gastrointestinal Cancer, ESMO Conference 2010; 30 June–3 July 2010; Barcelona, Spain. Ann Oncol 2010;21.
- Alberts SR, Sargent DJ, Smyrk TC, Shields AF, Chan E, Goldberg RM, et al. Adjuvant mFOLFOX6 with or without cetuxiumab (Cmab) in KRAS wild-type (WT) patients (pts) with resected stage III colon cancer (CC): results from NCCTG Intergroup phase III trial N0147. J Clin Oncol 2010;28.
- Assenat E, Desseigne F, Thezenas S, Viret F, Mineur L, Kramar A, et al. Cetuximab plus FOLFIRINOX (ERBIRINOX) as first-line treatment for unresectable metastatic colorectal cancer: a phase II trial. Oncologist 2011;16:1557-64. http://dx.doi.org/10.1634/theoncologist.2011-0141.
- Baker JB, Dutta D, Watson D, Maddala T, Shak S, Rowinsky EK, et al. Evaluation of tumor gene expression and K-RAS mutations in FFPE tumor tissue as predictors of response to cetuximab in metastatic colorectal cancer. J Clin Oncol 2008;26.
- Baloglu H, Yilmaz I, Kucukodaci Z. A KRAS mutation profile in colorectal carcinomas: mutation detection technique may affect patient selection for anti-EGFR therapy. Lab Invest 2012;92.
- Bokemeyer C, Bondarenko I, Hartmann JT, De Braud FG, Volovat C, Nippgen J, et al. KRAS status and efficacy of first-line treatment of patients with metastatic colorectal cancer (mCRC) with FOLFOX with or without cetuximab: the OPUS experience. J Clin Oncol 2008;26.
- Bokemeyer C, Bondarenko I, Hartmann JT, De Braud F, Schuch G, Zubel A, et al. Overall survival of patients with KRAS wild-type tumours treated with FOLFOX4 ± cetuximab as 1st-line treatment for metastatic colorectal cancer: the OPUS study. Ejc Suppl 2009;7. http://dx.doi.org/10.1016/S1359-6349(09)71174-7.
- Bokemeyer C, Kohne C, Rougier P, Stroh C, Schlichting M, Van Cutsem E. Cetuximab with chemotherapy (CT) as first-line treatment for metastatic colorectal cancer (mCRC): analysis of the CRYSTAL and OPUS studies according to KRAS and BRAF mutation status. J Clin Oncol 2010;28.
- Bokemeyer C, Van Cutsem E, Rougier P, Ciardiello F, Heeger S, Schlichting M, et al. Addition of cetuximab to chemotherapy as first-line treatment for KRAS wild-type metastatic colorectal cancer: pooled analysis of the CRYSTAL and OPUS randomised clinical trials. Eur J Cancer 2012;48:1466-75. http://dx.doi.org/10.1016/j.ejca.2012.02.057.
- Chuko J, Yeh MK, Chen BJ, Hu KY. Efficacy of cetuximab on wild-type and mutant KRAS in colorectal cancer: systematic review and meta-analysis. J Med Sci 2010;30:189-98.
- Cohen D, Ma H, Yuan Y, Hochman T, Hochster HS. Biweekly cetuximab (C), oxaliplatin (Ox), and capecitabine (Cape) in first line therapy of metastatic colorectal cancer (mCRC): preliminary results on toxicity and K-RAS status. J Clin Oncol 2008;26.
- Colucci G, Giuliani F, Garufi C, Mattioli R, Manzione L, Russo A, et al. Cetuximab plus FOLFOX-4 in untreated patients with advanced colorectal cancer: a Gruppo Oncologico dell’Italia Meridionale multicenter phase II study. Oncology 2010;79:415-22. http://dx.doi.org/10.1159/000323279.
- Di Salvatore M, Inno A, Orlandi A, Martini M, Nazzicone G, Ferraro D, et al. KRAS and BRAF mutational status and PTEN, cMET, and IGF1R expression as predictive markers of response to cetuximab plus chemotherapy in metastatic colorectal cancer (mCRC). J Clin Oncol 2010;28.
- Dubus P, Berhouet S, Carrere N, Merlio JP. Predicting response to anti-EGFR therapy for colorectal carcinoma by KRAS mutation testing. Cytometry Part B 2009;76B:420-1.
- Folprecht G, Kohne CH. Cetuximab in combination with 5-fluorouracil, leucovorin and irinotecan as a neoadjuvant chemotherapy in patients with initially unresectable colorectal liver metastases. Adv Gastrointest Cancers 2008;6:13-4.
- Folprecht G, Nowacki M, Lang I, Cascinu S, Shchepotin I, Maurel J, et al. Cetuximab plus FOLFIRI first line in patients (pts) with metastatic colorectal cancer (mCRC): a quality-of-life (QoL) analysis of the CRYSTAL trial. J Clin Oncol 2009;27.
- Folprecht G, Kohne C, Bokemeyer C, Rougier P, Schlichting M, Heeger S, et al. Cetuximab and 1st-line chemotherapy in elderly and younger patients with metastatic colorectal cancer (MCRC): a pooled analysis of the crystal and opus studies. Ann Oncol 2010;21.
- Gajate P, Sastre J, Bando I, Alonso T, Cillero L, Sanz J, et al. Influence of KRAS p.G13D mutation in patients with metastatic colorectal cancer treated with cetuximab. Clin Colorectal Cancer 2012;11:291-6. http://dx.doi.org/10.1016/j.clcc.2012.02.003.
- Gao J, Wang TT, Yu JW, Li YY, Shen L. KRAS and BRAF could predict response to cetuximab in Chinese colorectal cancer patients. Chinese J Cancer Res 2011;23:271-5. http://dx.doi.org/10.1007/s11670-011-0271-4.
- Garufi C, Torsello A, Tumolo S, Mottolese M, Melucci E, Campanella C, et al. Induction of resectability of colorectal liver metastases (CLM) with cetuximab (Cmab) plus CPT-11/5-fluorouracil (5-FU)/leucovorin (FA)/oxaliplatin (L-OHP) (CPT-11-FFL) (POCHER trial). Eur J Cancer 2009;7. http://dx.doi.org/10.1016/S1359-6349(09)71181-4.
- Goldberg RM, Sargent DJ, Thibodeau SN, Mahoney MR, Shields AF, Chan E, et al. Adjuvant mFOLFOX6 plus or minus cetuximab (Cmab) in patients (pts) with KRAS mutant (m) resected stage III colon cancer (CC): NCCTG Intergroup Phase III Trial N0147. J Clin Oncol 2010;28.
- Griebsch I, Lang I, Sartorius U, Van Cutsem E. The effect of tumor response on quality of life (QoL) in patients with KRAS wild-type metastatic colorectal cancer (mCRC): analysis from the CRYSTAL study. J Clin Oncol 2011;29.
- Harbison CT, Horak CE, Ledeine JM, Mukhopadhyay P, Malone DP, O’Callaghan C, et al. Validation of companion diagnostic for detection of mutations in codons 12 and 13 of the KRAS gene in patients with metastatic colorectal cancer: analysis of the NCIC CTG CO.17 trial. Arch Pathol Lab Med 2012. doi: 10.5858/arpa.2012-0367-OA.
- Huang J, Sargent DJ, Mahoney MR, Thibodeau SN, Smyrk TC, Sinicrope F, et al. Adjuvant FOLFIRI with or without cetuximab in patients with resected stage III colon cancer: NCCTG Intergroup phase III trial N0147. J Clin Oncol 2011;29.
- Ibrahim EM, Zekri JM, Bin Sadiq BM. Cetuximab-based therapy for metastatic colorectal cancer: a meta-analysis of the effect of K-RAS mutations. Int J Colorectal Dis 2010;25:713-21. http://dx.doi.org/10.1007/s00384-010-0927-4.
- Jones C, Taylor MA, McWilliams B. The role of cetuximab as first-line treatment of colorectal liver metastases. HPB (Oxford) 2013;15:11-7. http://dx.doi.org/10.1111/j.1477-2574.2012.00591.x.
- Jonker DJ, Karapetis C, Harbison C, O’Callaghan CJ, Tu D, Simes RJ, et al. High epiregulin (EREG) gene expression plus K-RAS wild-type (WT) status as predictors of cetuximab benefit in the treatment of advanced colorectal cancer (ACRC): results from NCIC CTG CO.17-A phase III trial of cetuximab versus best supportive care (BSC). J Clin Oncol 2009;27.
- Kimura T, Okamoto K, Miyamoto H, Kimura M, Kitamura S, Takenaka H, et al. Clinical benefit of high-sensitivity KRAS mutation testing in metastatic colorectal cancer treated with anti-EGFR antibody therapy. Oncology (Switzerland) 2012;82:298-304.
- Kohne C, Stroiakovski D, Chang-chien C, Lim R, Pinter T, Bodoky G, et al. Predictive biomarkers to improve treatment of metastatic colorectal cancer (mCRC): outcomes with cetuximab plus FOLFIRI in the CRYSTAL trial. J Clin Oncol 2009;27.
- Ku GY, Haaland BA, de Lima Lopes G. Cetuximab in the first-line treatment of K-RAS wild-type metastatic colorectal cancer: the choice and schedule of fluoropyrimidine matters. Cancer Chemother Pharmacol 2012;70:231-8. http://dx.doi.org/10.1007/s00280-012-1898-7.
- Lang I, Kohne CH, Folprecht G, Nowacki MP, Cascinu S, Shchepotin I, et al. Cetuximab plus FOLFIRI in 1st-line treatment of metastatic colorectal cancer: quality of life (QoL) analysis of patients (pts) with KRAS wild-type (wt) tumours in the CRYSTAL trial. Eur J Cancer 2009;7. http://dx.doi.org/10.1016/S1359-6349(09)71173-5.
- Lievre A, Bachet JB, Le Corre D, Boige V, Landi B, Emile JF, et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res 2006;66:3992-5. http://dx.doi.org/10.1158/0008-5472.CAN-06-0191.
- Lin OSR. Activating KRAS mutations as an independent predictor in metastatic colorectal cancer patients treated with cetuximab. Ann Oncol 2010;21.
- Lin AY, Buckley NS, Lu A-TT, Kouzminova NB, Salpeter SR. Effect of KRAS mutational status in advanced colorectal cancer on the outcomes of anti-epidermal growth factor receptor monoclonal antibody therapy: a systematic review and meta-analysis. Clin Colorectal Cancer 2011;10:63-9. http://dx.doi.org/10.3816/CCC.2011.n.009.
- Linardou H, Dahabreh IJ, Kanaloupiti D, Siannis F, Bafaloukos D, Kosmidis P, et al. Assessment of somatic K-RAS mutations as a mechanism associated with resistance to EGFR-targeted agents: a systematic review and meta-analysis of studies in advanced non-small-cell lung cancer and metastatic colorectal cancer. Lancet Oncol 2008;9:962-2. http://dx.doi.org/10.1016/S1470-2045(08)70206-7.
- Loupakis F, Cremolini C, Salvatore L, Schirripa M, Lonardi S, Vaccaro V, et al. Clinical impact of anti-epidermal growth factor receptor monoclonal antibodies in first-line treatment of metastatic colorectal cancer: meta-analytical estimation and implications for therapeutic strategies. Cancer 2012;118:1523-32. http://dx.doi.org/10.1002/cncr.26460.
- Malapelle U, Bellevicine C, Salatiello M, De Luca C, Rispo E, Riccio P, et al. Sanger sequencing in routine KRAS testing: a review of 1720 cases from a pathologist’s perspective. J Clin Pathol 2012;65:940-4. http://dx.doi.org/10.1136/jclinpath-2012-200773.
- Malapelle U, Carlomagno C, Salatiello M, De Stefano A, De Luca C, Bianco R, et al. KRAS mutation detection by high-resolution melting analysis significantly predicts clinical benefit of cetuximab in metastatic colorectal cancer. Br J Cancer 2012;107:626-31. http://dx.doi.org/10.1038/bjc.2012.275.
- Mancuso A, Leone A, Vigna L, Calabro F, Giuliani R, De Marco S, et al. EGFR, DCC, and K-RAS mutations as predictive factors for cetuximab sensitivity in metastatic colorectal cancer (mCRC). J Clin Oncol 2008;26.
- Maughan T, Adams RA, Smith CG, Seymour MT, Wilson R, Meade AM, et al. Addition of cetuximab to oxaliplatin-based combination chemotherapy (CT) in patients with KRAS wild-type advanced colorectal cancer (ACRC): a randomised superiority trial (MRC COIN). Eur J Cancer 2009;7:4-5. http://dx.doi.org/10.1016/S1359-6349(09)72034-8.
- Maughan TS, Adams R, Smith CG, Seymour MT, Wilson RH, Meade AM, et al. Identification of potentially responsive subsets when cetuximab is added to oxaliplatin-fluoropyrimidine chemotherapy (CT) in first-line advanced colorectal cancer (aCRC): mature results of the MRC COIN trial. J Clin Oncol 2010;28.
- Maughan T, Adams R, Smith C, Seymour M, Wilson R, Meade A, et al. The addition of cetuximab to oxaliplatinfluoropyrimidine chemotherapy in first-line advanced colorectal cancer in the MRC COIN trial: identification of potentially responsive subsets of patients. Ann Oncol 2010;21:vi17-18.
- Mayer B, Bogner A, Jauch KW, Singer T. KRAS/BRAF mutation status as a predictive biomarker for anti-EGFR antibody therapy in primary and metastatic colorectal carcinoma. Onkologie 2010;33.
- Cetuximab Combined with Irinotecan in First-Line Therapy for Metastatic Colorectal Cancer (CRYSTAL). Bethesda, MD: National Library of Medicine; 2011.
- Modest DP, Brodowicz T, Stintzing S, Jung A, Neumann J, Laubender RP, et al. Impact of the specific mutation in KRAS codon 12 mutated tumors on treatment efficacy in patients with metastatic colorectal cancer receiving cetuximab-based first-line therapy: a pooled analysis of three trials. Oncology (Switzerland) 2012;83:241-7.
- Molinari FM, Felicioni LF, Buscarino MB, De Dosso SD, Buttitta FB, Malatesta SM, et al. High sensitive methods to identify K-RAS mutations increase the detection of non-responder metastatic colorectal cancer patients treated with anti-EGFR monoclonal antibodies. Ann Oncol 2010;21:I33-4.
- Moosmann N, von Weikersthal LF, Vehling-Kaiser U, Stauch M, Hass HG, Dietzfelbinger H, et al. Cetuximab plus capecitabine and irinotecan compared with cetuximab plus capecitabine and oxaliplatin as first-line treatment for patients with metastatic colorectal cancer: AIO KRK-0104 – a randomized trial of the German AIO CRC study group. J Clin Oncol 2011;29:1050-8. http://dx.doi.org/10.1200/JCO.2010.31.1936.
- Ocvirk J, Vrbanec D, Beslija S, Koza I, Ciuleanu T, Papamichael D, et al. Relationship between KRAS status and efficacy of FOLFOX6 + cetuximab or FOLFIRI + cetuximab in first-line treatment of patients with metastatic colorectal cancer (MCRC): the CECOG/CORE1.2.001 trial. Ann Oncol 2009;20.
- Passardi A, Ulivi P, Valgiusti M, Scarpi E, Moscati R, Chiadini E, et al. The role of KRAS, BRAF, and PI3K mutations as markers of resistance to cetuximab in metastatic colorectal cancer. J Clin Oncol 2011;29.
- Petrelli F, Borgonovo K, Cabiddu M, Ghilardi M, Barni S. Cetuximab and panitumumab in KRAS wild-type colorectal cancer: a meta-analysis. Int J Colorectal Dis 2011;26:823-33. http://dx.doi.org/10.1007/s00384-011-1149-0.
- Petrelli F, Barni S. Resectability and outcome with anti-EGFR agents in patients with KRAS wild-type colorectal liver-limited metastases: a meta-analysis. Int J Colorectal Dis 2012;27:997-1004. http://dx.doi.org/10.1007/s00384-012-1438-2.
- Piessevaux H, Schlichting M, Heeger S, Van Cutsem E, Tejpar S. Early tumor shrinkage for the prediction of efficacy of cetuximab in metastatic colorectal cancer (MCRC): analysis from the CRYSTAL study. Ann Oncol 2010;21:viii193-4.
- Piessevaux H, Van Cutsem E, Bokemeyer C, Schlichting M, Heeger S, Tejpar S. Early tumor shrinkage and long-term outcome in metastatic colorectal cancer (mCRC): assessment of predictive utility across treatment arms in the CRYSTAL and OPUS studies. J Clin Oncol 2011;29.
- Piessevaux H, Bokemeyer C, Schlichting M, Heeger S, Tejpar S. Impact of early tumor shrinkage on long-term outcome in metastatic colorectal cancer (mCRC) treated with FOLFOX4 with or without cetuximab: lessons from the OPUS trial. J Clin Oncol 2011;29.
- Qiu LX, Mao C, Zhang J, Zhu XD, Liao RY, Xue K, et al. Predictive and prognostic value of KRAS mutations in metastatic colorectal cancer patients treated with cetuximab: a meta-analysis of 22 studies. Eur J Cancer 2010;46:2781-7. http://dx.doi.org/10.1016/j.ejca.2010.05.022.
- Raoul JL, Van Laethem JL, Peeters M, Brezault C, Husseini F, Cals L, et al. Cetuximab in combination with irinotecan/5-fluorouracil/folinic acid (FOLFIRI) in the initial treatment of metastatic colorectal cancer: a multicentre two-part phase I/II study. BMC Cancer 2009;9. http://dx.doi.org/10.1186/1471-2407-9-112.
- Rivera F, Gravalos C, Massuti B, Puente J, Marcuello E, Valladares M, et al. Cetuximab plus capecitabine as first-line treatment for elderly patients (pts) with advanced colorectal cancer (mCRC). Final analysis of activity and survival according to KRAS status – the TTD-06–01 Spanish Cooperative Group trial. Eur J Cancer 2009;7. http://dx.doi.org/10.1016/S1359-6349(09)70738-4.
- Rose JS, Serna DS, Martin LK, Li X, Weatherby LM, Abdel-Misih S, et al. Influence of KRAS mutation status in metachronous and synchronous metastatic colorectal adenocarcinoma. Cancer 2012;118:6243-52. http://dx.doi.org/10.1002/cncr.27666.
- Salazar R, Mini E, Folprecht G, Subtil F, van Laethem J, Thaler J, et al. Adjuvant FOLFOX4 plus or minus cetuximab (CMAB) in patients with KRAS mutant (MKRAS) resected stage III colon cancer (CC). Results from the PETACC8 Intergroup trial. Ann Oncol 2012;23:178-9.
- Schuch G, Bondarenko I, Hartmann J, De Braud F, Volovat C, Nippgen J, et al. The OPUS study: predictive value of KRAS status on clinical outcome in patients with metastatic colorectal cancer treated with either FOLFOX4 or FOLFOX4 plus cetuximab. Ann Oncol 2008;19.
- Serna DS, Martin LK, Li X, Weatherby LM, Rose JS, Bekaii-Saab T. Influence of KRAS mutation status in metachronous and synchronous metastatic colorectal adenocarcinomas. J Clin Oncol 2011;29.
- Shinozaki E, Bando H, Nishina T, Yamazaki K, Kadowaki S, Yuki S, et al. Clinical outcome of cetuximab for metastatic colorectal cancer patients harboring KRAS codon61, KRAS codon146, BRAF, NRAS OR PIK3CA mutations. Ann Oncol 2012;23:196-7.
- Simon I, Tian S, Moreno V, Roepman P, Tabernero J, Snel M, et al. The role of activating mutations of KRAS, BRAF, and PIK3CA pathway convergence at the transcriptional level and prediction of treatment response to cetuximab in colorectal cancer. J Clin Oncol 2011;29.
- Stenger M. Cetuximab plus FOLFIRI in first-line treatment of KRAS mutation-negative, EGFR-positive metastatic colorectal cancer. Community Oncol 2012;9:304-6. http://dx.doi.org/10.1016/j.cmonc.2012.09.006.
- Stintzing S, Jung A, Vehling-Kaiser U, Stauch M, Hass H, Dietzfelbinger H, et al. Cetuximab plus XELIRI versus cetuximab plus XELOX as first-line treatment for patients with metastatic colorectal cancer (MCRC): analysis of the randomized trial of the German AIO CRC study group: KRK-0204. Ann Oncol 2010;21:viii189-90.
- Taieb J, Tabernero J, Mini E, Subtil F, Folprecht G, Van Laethem JL, et al. Adjuvant FOLFOX4 ± cetuximab in KRAS wild-type patients with resected stage III colon cancer results from the PETACC8 Intergroup Trial. Ann Oncol 2012;23.
- Tejpar S, Bokemeyer C, Celik I, Schlichting M, Sartorius U, Van Cutsem E. Influence of KRAS G13D mutations on outcome in patients with metastatic colorectal cancer (mCRC) treated with first-line chemotherapy with or without cetuximab. J Clin Oncol 2011;29.
- Tejpar S, Bokemeyer C, Celik I, Schlichting M, Van Cutsem E. The role of the KRAS G13D mutation in patients with metastatic colorectal cancer (mCRC) treated with first-line chemotherapy plus cetuximab. J Clin Oncol 2011;29.
- Tejpar S, Bokemeyer C, Celik I, Schlichting M, Heeger S, Van Cutsem E. KRAS mutations and outcome in patients with metastatic colorectal cancer treated with first-line chemotherapy with or without cetuximab. Ann Oncol 2011;22:v16-17.
- Tejpar S, Celik I, Schlichting M, Sartorius U, Bokemeyer C, Van Cutsem E. Association of KRAS G13D tumor mutations with outcome in patients with metastatic colorectal cancer treated with first-line chemotherapy with or without cetuximab. J Clin Oncol 2012;30:3570-7. http://dx.doi.org/10.1200/JCO.2012.42.2592.
- Tsoukalas N, Bagos P, Hamodrakas S. Multivariant metal-analysis of KRAS mutations’s predictive value for response to cetuximab in colorectal cancer. Ann Oncol 2010;21.
- Tsoukalas N, Tzovaras A, Tolia M, Papakostidi A, Kostakis I, Ardavanis A, et al. The efficacy of cetuximab and mutations of the KRAS gene in colorectal cancer. Arch Hellenic Med 2011;28:674-9.
- Tsoukalas N, Tzovaras AA, Tolia M, Kostakis ID, Papakostidi A, Pistamaltzian N, et al. Meta-analysis of the predictive value of KRAS mutations in treatment response using cetuximab in colorectal cancer. J BUON 2012;17:73-8.
- Tveit K, Guren T, Glimelius B, Pfeiffer P, Sorbye H, Pyrhonen S, et al. Randomized phase III study of 5-fluorouracil/folinate/oxaliplatin given continuously or intermittently with or without cetuximab, as first-line treatment of metastatic colorectal cancer: the Nordic VII study (NCT00145314), by the Nordic Colorectal Cancer Biomodulation Group. Ann Oncol 2010;21.
- Tveit K, Guren T, Glimelius B, Pfeiffer P, Sorbye H, Pyrhonen S, et al. Randomized phase III study of 5-fluorouracil/folinate/oxaliplatin given continuously or intermittently with or without cetuximab, as first-line treatment of metastatic colorectal cancer: the NORDIC VII study (NCT00145314), by the Nordic Colorectal Cancer Biomodulation Group. J Clin Oncol 2011;29.
- Ubago R, Castillo MA, Flores S, Rodriguez R, Beltran C. Cetuximab for the first-line treatment of metastatic colorectal cancer. Value Health 2011;14:A433-4. http://dx.doi.org/10.1016/j.jval.2011.08.1104.
- Vale C, Tierney JF, Meade A, Fisher D, Kaplan R, Adams RA, et al. Impact of K-RAS status on the effects of EGFR-targeted monoclonal antibody (MAb) therapy in advanced colorectal cancer (ACRC): a systematic review and meta-analysis of randomized controlled trials (RCTs). J Clin Oncol 2009;27.
- Van Cutsem E, Lang I, D’Haens G, Moiseyenko V, Zaluski J, Folprecht G, et al. KRAS status and efficacy in the crystal study: 1st-line treatment of patients with metastatic colorectal cancer (MCRC) receiving FOLFIRI with or without cetuximab. Ann Oncol 2008;19.
- Van Cutsem E, Rougier P, Kohne C, Stroh C, Schlichting M, Bokemeyer C. A meta-analysis of the CRYSTAL and OPUS studies combining cetuximab with chemotherapy (CT) as 1st-line treatment for patients (pts) with metastatic colorectal cancer (mCRC): results according to KRAS and BRAF mutation status. Eur J Cancer Suppl 2009;7. http://dx.doi.org/10.1016/S1359-6349(09)71172-3.
- Van Cutsem E, Lang I, Folprecht G, Nowacki M, Barone C, Shchepotin I, et al. Cetuximab plus FOLFIRI: final data from the CRYSTAL study on the association of KRAS and BRAF biomarker status with treatment outcome. J Clin Oncol 2010;28.
- Van Cutsem E, Rougier P, Lang I, Folprecht G, Nowacki M, Barone C, et al. The influence of KRAS and BRAF tumor mutation status on treatment outcome with cetuximab plus FOLFIRI: final data from the CRYSTAL study. Ann Oncol 2010;21.
- Van Cutsem E, Kohne C-H, Lang I, Folprecht G, Nowacki MP, Cascinu S, et al. Cetuximab plus irinotecan, fluorouracil, and leucovorin as first-line treatment for metastatic colorectal cancer: updated analysis of overall survival according to tumor KRAS and BRAF mutation status. J Clin Oncol 2011;29:2011-19. http://dx.doi.org/10.1200/JCO.2010.33.5091.
- Wasan H, Adams RA, Wilson RH, Pugh CA, Fisher D, Madi A, et al. Intermittent chemotherapy (CT) plus continuous or intermittent cetuximab (C) in the first-line treatment of advanced colorectal cancer (aCRC): results of the two-arm phase II randomised MRC COIN-B trial. Eur J Cancer 2011;47. http://dx.doi.org/10.1016/S0959-8049(11)71651-3.
- Whitehall V, Tran K, Umapathy A, Grieu F, Hewitt C, Evans TJ, et al. A multicenter blinded study to evaluate KRAS mutation testing methodologies in the clinical setting. J Mol Diagn 2009;11:543-52. http://dx.doi.org/10.2353/jmoldx.2009.090057.
- Yen L-C, Uen Y-H, Wu D-C, Lu C-Y, Yu F-J, Wu IC, et al. Activating KRAS mutations and overexpression of epidermal growth factor receptor as independent predictors in metastatic colorectal cancer patients treated with cetuximab. Ann Surg 2010;251:254-60. http://dx.doi.org/10.1097/SLA.0b013e3181bc9d96.
- Zhang L, Ma L, Zhou Q. Overall and KRAS-specific results of combined cetuximab treatment and chemotherapy for metastatic colorectal cancer: a meta-analysis. Int J Colorectal Dis 2011;26:1025-33. http://dx.doi.org/10.1007/s00384-011-1197-5.
Appendix 1 Literature search strategies
Clinical effectiveness search strategies
CRC + KRAS (limit: 2000-C)
EMBASE (OvidSP), 2000 to Week 3 2013
Searched 22 January 2013
-
exp colon cancer/ or exp rectum cancer/ or colorectal tumor/ (169,199)
-
((colorect$ or rectal$ or rectum$ or colon$ or sigma$ or sigmo$ or rectosigm$ or bowel$ or anal or anus) adj3 (cancer$ or neoplas$ or oncolog$ or malignan$ or tumo?r$ or carcinoma$ or adenocarcinoma$ or metasta$ or meta-sta$ or sarcoma$ or adenom$ or lesion$)).ti,ab,ot,hw. (245,923)
-
(m-CRC or CRC).ti,ab,ot. (14,043)
-
((cecum or cecal or caecum or caecal or il?eoc?ecal or il?eoc?ecum) adj3 (cancer$ or neoplas$ or oncolog$ or malignan$ or tumo?r$ or carcinoma$ or adenocarcinoma$ or metasta$ or meta-sta$ or sarcoma$ or adenom$ or lesion$)).ti,ab,ot. (2124)
-
(large intestin$ adj3 (cancer$ or neoplas$ or oncolog$ or malignan$ or tumo?r$ or carcinoma$ or adenocarcinoma$ or metasta$ or meta-sta$ or sarcoma$ or adenom$ or lesion$)).ti,ab,ot. (1871)
-
(lower intestin$ adj3 (cancer$ or neoplas$ or oncolog$ or malignan$ or tumo?r$ or carcinoma$ or adenocarcinoma$ or metasta$ or meta-sta$ or sarcoma$ or adenom$ or lesion$)).ti,ab,ot. (26)
-
or/1-6 (249,697)
-
k ras oncogene/ (4953)
-
(k ras or kras or k-ras or V-Ki-ras$ or V-K-ras or V-Ki-ras or v ki ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS).af. (17,025)
-
(Kirsten adj3 (murine or rat) adj3 sarcoma$).ti,ab,ot. (396)
-
(thera?screen$ or therascreen$).af. (67)
-
(Cobas adj3 (k ras or kras or k-ras or V-Ki-ras$ or V-K-ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS)).af. (8)
-
(sanger sequencing adj3 (k ras or kras or k-ras or V-Ki-ras$ or V-K-ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS)).af. (15)
-
(pyrosequencing adj3 (k ras or kras or k-ras or V-Ki-ras$ or V-K-ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS)).af. (25)
-
((HRM or HRMA or dHPLC) adj3 (k ras or kras or k-ras or V-Ki-ras$ or V-K-ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS)).af. (13)
-
(high resolution adj3 melt$ adj3 (k ras or kras or k-ras or V-Ki-ras$ or V-K-ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS)).af. (8)
-
(SNapShot adj3 (k ras or kras or k-ras or V-Ki-ras$ or V-K-ras or V-Ki-ras or v ki ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS)).af. (5)
-
(Next generation sequencing adj3 (k ras or kras or k-ras or V-Ki-ras$ or V-K-ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS)).af. (1)
-
high resolution melting analysis/ (691)
-
19 and (8 or 9 or 10) (62)
-
or/8-18,20 (17,279)
-
7 and 21 (5716)
-
limit 22 to yr=“2000 -Current” (5036)
-
limit 23 to embase (4540)
MEDLINE (OvidSP), 2000 to January Week 2 2013
Searched 22 January 2013
-
exp Colorectal Neoplasms/ (134,723)
-
((colorect$ or rectal$ or rectum$ or colon$ or sigma$ or sigmo$ or rectosigm$ or bowel$ or anal or anus) adj3 (cancer$ or neoplas$ or oncolog$ or malignan$ or tumo?r$ or carcinoma$ or adenocarcinoma$ or metasta$ or meta-sta$ or sarcoma$ or adenom$ or lesion$)).ti,ab,ot,hw. (165,769)
-
(m-CRC or CRC).ti,ab,ot. (8215)
-
((cecum or cecal or caecum or caecal or il?eoc?ecal or il?eoc?ecum) adj3 (cancer$ or neoplas$ or oncolog$ or malignan$ or tumo?r$ or carcinoma$ or adenocarcinoma$ or metasta$ or meta-sta$ or sarcoma$ or adenom$ or lesion$)).ti,ab,ot. (1570)
-
(large intestin$ adj3 (cancer$ or neoplas$ or oncolog$ or malignan$ or tumo?r$ or carcinoma$ or adenocarcinoma$ or metasta$ or meta-sta$ or sarcoma$ or adenom$ or lesion$)).ti,ab,ot. (1541)
-
(lower intestin$ adj3 (cancer$ or neoplas$ or oncolog$ or malignan$ or tumo?r$ or carcinoma$ or adenocarcinoma$ or metasta$ or meta-sta$ or sarcoma$ or adenom$ or lesion$)).ti,ab,ot. (23)
-
or/1-6 (170,682)
-
Genes, ras/ (11,077)
-
(k ras or kras or k-ras or V-Ki-ras$ or V-K-ras or V-Ki-ras or v ki ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS).af. (9538)
-
(Kirsten adj3 (murine or rat) adj3 sarcoma$).ti,ab,ot. (346)
-
(thera?screen$ or therascreen$).af. (16)
-
(Cobas adj3 (k ras or kras or k-ras or V-Ki-ras$ or V-K-ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS)).af. (2)
-
(sanger sequencing adj3 (k ras or kras or k-ras or V-Ki-ras$ or V-K-ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS)).af. (4)
-
(pyrosequencing adj3 (k ras or kras or k-ras or V-Ki-ras$ or V-K-ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS)).af. (12)
-
((HRM or HRMA) adj3 (k ras or kras or k-ras or V-Ki-ras$ or V-K-ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS)).af. (5)
-
(high resolution adj3 melt$ adj3 (k ras or kras or k-ras or V-Ki-ras$ or V-K-ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS)).af. (4)
-
(SNapShot adj3 (k ras or kras or k-ras or V-Ki-ras$ or V-K-ras or V-Ki-ras or v ki ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS)).af. (3)
-
(Next generation sequencing adj3 (k ras or kras or k-ras or V-Ki-ras$ or V-K-ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS)).af. (0)
-
or/8-18 (16,696)
-
7 and 19 (3083)
-
limit 20 to yr=“2000 -Current” (2293)
-
remove duplicates from 21 (2278)
MEDLINE In-Process & Other Non-Indexed Citations (OvidSP) and Daily Update (OvidSP), up to 21 January 2013
Searched 22 January 2013
-
exp Colorectal Neoplasms/ (194)
-
((colorect$ or rectal$ or rectum$ or colon$ or sigma$ or sigmo$ or rectosigm$ or bowel$ or anal or anus) adj3 (cancer$ or neoplas$ or oncolog$ or malignan$ or tumo?r$ or carcinoma$ or adenocarcinoma$ or metasta$ or meta-sta$ or sarcoma$ or adenom$ or lesion$)).ti,ab,ot,hw. (7747)
-
(m-CRC or CRC).ti,ab,ot. (1006)
-
((cecum or cecal or caecum or caecal or il?eoc?ecal or il?eoc?ecum) adj3 (cancer$ or neoplas$ or oncolog$ or malignan$ or tumo?r$ or carcinoma$ or adenocarcinoma$ or metasta$ or meta-sta$ or sarcoma$ or adenom$ or lesion$)).ti,ab,ot. (108)
-
(large intestin$ adj3 (cancer$ or neoplas$ or oncolog$ or malignan$ or tumo?r$ or carcinoma$ or adenocarcinoma$ or metasta$ or meta-sta$ or sarcoma$ or adenom$ or lesion$)).ti,ab,ot. (28)
-
(lower intestin$ adj3 (cancer$ or neoplas$ or oncolog$ or malignan$ or tumo?r$ or carcinoma$ or adenocarcinoma$ or metasta$ or meta-sta$ or sarcoma$ or adenom$ or lesion$)).ti,ab,ot. (0)
-
or/1-6 (7930)
-
Genes, ras/ (8)
-
(k ras or kras or k-ras or V-Ki-ras$ or V-K-ras or V-Ki-ras or v ki ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS).af. (659)
-
(Kirsten adj3 (murine or rat) adj3 sarcoma$).ti,ab,ot. (17)
-
(thera?screen$ or therascreen$).af. (5)
-
(Cobas adj3 (k ras or kras or k-ras or V-Ki-ras$ or V-K-ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS)).af. (0)
-
(sanger sequencing adj3 (k ras or kras or k-ras or V-Ki-ras$ or V-K-ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS)).af. (2)
-
(pyrosequencing adj3 (k ras or kras or k-ras or V-Ki-ras$ or V-K-ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS)).af. (0)
-
((HRM or HRMA) adj3 (k ras or kras or k-ras or V-Ki-ras$ or V-K-ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS)).af. (1)
-
(high resolution adj3 melt$ adj3 (k ras or kras or k-ras or V-Ki-ras$ or V-K-ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS)).af. (0)
-
(SNapShot adj3 (k ras or kras or k-ras or V-Ki-ras$ or V-K-ras or V-Ki-ras or v ki ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS)).af. (1)
-
(Next generation sequencing adj3 (k ras or kras or k-ras or V-Ki-ras$ or V-K-ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS)).af. (0)
-
or/8-18 (667)
-
7 and 19 (269)
Cochrane Database of Systematic Reviews (CDSR) (Wiley), 2000 to Issue 12, 2012; Cochrane Central Register of Controlled Trials (CENTRAL) (Wiley), 2000 to Issue 12, 2012; Database of Abstracts of Reviews of Effects (DARE) (Wiley), 2000 to Issue 4, 2012; and Health Technology Assessment (HTA) database (Wiley), 2000 to Issue 4, 2012
Searched 22 January 2013
#1 MeSH descriptor: [Colorectal Neoplasms] explode all trees (4380)
#2 ((colorect* or rectal* or rectum* or colon* or sigma* or sigmo* or rectosigm* or bowel* or anal or anus) near/3 (cancer* or neoplas* or oncolog* or malignan* or tumour* or tumor* or carcinoma* or adenocarcinoma* or metasta* or meta-sta* or sarcoma* or adenom* or lesion*)) (7773)
#3 (m-CRC or CRC) (715)
#4 ((cecum or cecal or caecum or caecal or ileocecal or ileocaecal or ileocaecum or ileocecum) near/3 (cancer* or neoplas* or oncolog* or malignan* or tumour* or tumor* or carcinoma* or adenocarcinoma* or metasta* or meta-sta* or sarcoma* or adenom* or lesion*)) (24)
#5 (large intestin* near/3 (cancer* or neoplas* or oncolog* or malignan* or tumour* or tumor*or carcinoma* or adenocarcinoma* or metasta* or meta-sta* or sarcoma* or adenom* or lesion*)) (86)
#6 (lower intestin* near/3 (cancer* or neoplas* or oncolog* or malignan* or tumour* or tumor* or carcinoma* or adenocarcinoma* or metasta* or meta-sta* or sarcoma* or adenom* or lesion*)) (114)
#7 #1 or #2 or #3 or #4 or #5 or #6 (8053)
#8 MeSH descriptor: [Genes, ras] this term only (46)
#9 (k ras or kras or K-ras or V-Ki-ras* or V-K-ras or V-Ki-ras or v ki ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS) (386)
#10 (Kirsten near/3 (murine or rat) near/3 sarcoma*) (7)
#11 (thera screen* or thera-screen* or therascreen*) (13)
#12 (Cobas) (115)
#13 (sanger sequencing) (7)
#14 (pyrosequencing) (18)
#15 (HRM or HRMA) (11)
#16 (high resolution near/3 melt*) (1)
#17 (SNapShot) (50)
#18 (“Next generation sequencing”) (2)
#19 #8 or #9 or #10 or #11 or #12 or #13 or #14 or #15 or #16 or #17 or #18 (605)
#20 #7 and #19 from 2000 (98)
The CDSR search retrieved nine references, the CENTRAL search retrieved 65 references, the DARE search retrieved 11 references and the HTA database search retrieved nine references.
National Institute for Health Research (NIHR) Health Technology Assessment (HTA) programme (Internet), up to 25 January 2013
Searched 25 January 2013
Browsed by relevant terms; retrieved two references.
Science Citation Index (SCI-EXPANDED) (Web of Knowledge) and Conference Proceedings Citation Index (CPCI-S) (Web of Knowledge), 2000 to 22 January 2013
Searched 23 January 2013
Databases=SCI-EXPANDED, CPCI-S Timespan=2000-01-01 - 2013-01-23
#18 3597 #17 AND #6
#17 10,477 #16 OR #15 OR #14 OR #13 OR #12 OR #11 OR #10 OR #9 OR #8 OR #7
#16 26 TS=((Next SAME generation SAME sequencing) SAME (k ras or kras or V-Ki-ras* or V-K-ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS))
#15 23 TS=(SNapShot SAME (k ras or kras or V-Ki-ras* or V-K-ras or V-Ki-ras or v ki ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS))
#14 75 TS=((high SAME resolution SAME melt*) SAME (k ras or kras or V-Ki-ras* or V-K-ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS))
#13 59 TS=((HRM or HRMA or dHPLC) SAME (k ras or kras or V-Ki-ras* or V-K-ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS))
#12 94 TS=(pyrosequencing SAME (k ras or kras or V-Ki-ras* or V-K-ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS))
#11 49 TS=((sanger SAME sequencing) SAME (k ras or kras or V-Ki-ras* or V-K-ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS))
#10 2 TS=(Cobas SAME (k ras or kras or V-Ki-ras* or V-K-ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS))
#9 17 TS=(thera$screen* or therascreen*)
#8 96 TS=(Kirsten NEAR/3 (murine or rat) NEAR/3 sarcoma*)
#7 10,467 TS=(k ras or kras or V-Ki-ras* or V-K-ras or V-Ki-ras or v ki ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS)
#6 134,422 #1 or #2 or #3 or #4 or #5
#5 1328 TS=(lower SAME intestin* NEAR/3 (cancer* or neoplas* or oncolog* or malignan* or tumo$r* or carcinoma* or adenocarcinoma* or metasta* or meta-sta* or sarcoma* or adenom* or lesion*))
#4 1113 TS=(large SAME intestin* NEAR/3 (cancer* or neoplas* or oncolog* or malignan* or tumo$r* or carcinoma* or adenocarcinoma* or metasta* or meta-sta* or sarcoma* or adenom* or lesion*))
#3 484 TS=((cecum or cecal or caecum or caecal or il$eoc$ecal or il$eoc$ecum) NEAR/3 (cancer* or neoplas* or oncolog* or malignan* or tumo$r* or carcinoma* or adenocarcinoma* or metasta* or meta-sta* or sarcoma* or adenom* or lesion*))
#2 9622 TS=(m-CRC or CRC)
#1 130,942 TS=((colorect* or rectal* or rectum* or colon* or sigma* or sigmo* or rectosigm* or bowel* or anal or anus) NEAR/3 (cancer* or neoplas* or oncolog* or malignan* or tumo?r* or carcinoma* or adenocarcinoma* or metasta* or meta-sta* or sarcoma* or adenom* or lesion*))
BIOSIS Previews (Web of Knowledge), 2000 to 22 January 2013
Searched 23 January 2013
Databases=BIOSIS Previews Timespan=2000-2013
#18 2641 #17 AND #6
#17 8621 #16 OR #15 OR #14 OR #13 OR #12 OR #11 OR #10 OR #9 OR #8 OR #7
#16 39 TS=((Next SAME generation SAME sequencing) SAME (k-ras or k ras or kras or V-Ki-ras* or V-K-ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS))
#15 36 TS=(SNapShot SAME (k-ras or k ras or kras or V-Ki-ras* or V-K-ras or V-Ki-ras or v ki ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS))
#14 73 TS=((high SAME resolution SAME melt*) SAME (k-ras or k ras or kras or V-Ki-ras* or V-K-ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS))
#13 55 TS=((HRM or HRMA or dHPLC) SAME (k-ras or k ras or kras or V-Ki-ras* or V-K-ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS))
#12 133 TS=(pyrosequencing SAME (k-ras or k ras or kras or V-Ki-ras* or V-K-ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS))
#11 95 TS=((sanger SAME sequencing) SAME (k-ras or k ras or kras or V-Ki-ras* or V-K-ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS))
#10 4 TS=(Cobas SAME (k-ras or k ras or kras or V-Ki-ras* or V-K-ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS))
#9 14 TS=(thera$screen* or therascreen*)
#8 153 TS=(Kirsten NEAR/3 (murine or rat) NEAR/3 sarcoma*)
#7 8611 TS=(k-ras or k ras or kras or V-Ki-ras* or V-K-ras or V-Ki-ras or v ki ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS)
#6 97,980 #1 or #2 or #3 or #4 or #5
#5 1020 TS=(lower SAME intestin* NEAR/3 (cancer* or neoplas* or oncolog* or malignan* or tumo$r* or carcinoma* or adenocarcinoma* or metasta* or meta-sta* or sarcoma* or adenom* or lesion*))
#4 805 TS=(large SAME intestin* NEAR/3 (cancer* or neoplas* or oncolog* or malignan* or tumo$r* or carcinoma* or adenocarcinoma* or metasta* or meta-sta* or sarcoma* or adenom* or lesion*))
#3 424 TS=((cecum or cecal or caecum or caecal or il$eoc$ecal or il$eoc$ecum) NEAR/3 (cancer* or neoplas* or oncolog* or malignan* or tumo$r* or carcinoma* or adenocarcinoma* or metasta* or meta-sta* or sarcoma* or adenom* or lesion*))
#2 7235 TS=(m-CRC or CRC)
#1 95,876 TS=((colorect* or rectal* or rectum* or colon* or sigma* or sigmo* or rectosigm* or bowel* or anal or anus) NEAR/3 (cancer* or neoplas* or oncolog* or malignan* or tumo?r* or carcinoma* or adenocarcinoma* or metasta* or meta-sta* or sarcoma* or adenom* or lesion*))
Latin American and Caribbean Health Sciences (LILACS) (Internet), up to 24 January 2013
Searched 23 January 2013
| Terms | Records |
|---|---|
| (k-ras or “k ras” OR kras OR v-ki-ras$ OR v-k-ras OR v-ki-ras OR “v ki ras” OR c-ki-ras OR c-k-ras OR ki-ras OR “ki ras” OR kras1 OR kras2 OR kras1p OR rask OR rask1 OR rask2 OR “kirsten ras” OR therascreen$ OR thera-screen$ OR cobas OR hrm OR dhplc OR snapshot OR (high AND resolution AND melt) OR prosequencing OR (sanger AND sequencing)) | 213 |
| ((MH:C04.588.274.476.411.307 or MH:C06.301.371.411.307 or MH:C06.405.249.411.307 or MH:C06.405.469.158.356 or MH:C06.405.469.491.307 or MH:C06.405.469.860.180 or MH:C04.588.274.476.411.184 or colorectal neoplasms$ or “neoplasias colorrectales” or “neoplasias colorrectais” or “colorectal cancer” or CRC or m$crc) AND (k-ras or “k ras” or kras or V-Ki-ras$ or V-K-ras or V-Ki-ras or “v ki ras” or c-ki-ras or c-k-ras or ki-ras or “ki ras” or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or “Kirsten RAS” or therascreen$ or thera-screen$ or cobas or HRM or dHPLC or snapshot or (high and resolution and melt) or prosequencing or (sanger and sequencing))) | 123 |
| Total | 336 |
International Prospective Register of Systematic Reviews (PROSPERO) (Internet), up to 25 January 2013
Searched 25 January 2013
Searched for terms in ‘All Fields’
| Terms | Records |
|---|---|
| KRAS or K-RAS | 2 |
| Colorectal Cancer | 2/14 (same records as above) |
| Cobas | 0 |
| Therascreen | 0 |
| Thera-screen | 0 |
| Sequencing | 0/3 |
| Pyrosequencing | 0 |
| HRM or HRMA or dHPLC | 0 |
| High resolution | 0 |
| kirsten | 0 |
| Oncogene | 0 |
| RASK | 0 |
| Snapshot | 0 |
| Colon Cancer | 1/3 (included in KRAS result) |
| Total | 2 |
Clinicaltrials.gov (Internet), 2000 to 23 January 2013
http://clinicaltrials.gov/ct2/search/advanced
Searched 23 January 2013
Advanced search option – search terms box
Limited to results received from 1 January 2000 to 23 January 2013
| Search terms | Condition | Records |
|---|---|---|
| (k ras OR kras OR K-ras OR V-Ki-ras* OR V-K-ras OR V-Ki-ras OR v ki ras OR c-ki-ras OR c-k-ras OR ki-ras OR ki ras OR Kras1 OR Kras2 OR KRAS1P OR RASK OR RASK1 OR RASK2 OR Kirsten RAS) | (colorect* OR rectal* OR rectum* OR colon* OR sigma* OR sigmo* OR rectosigm* OR bowel* OR anal OR anus OR CRC OR m-CRC OR cecum OR cecal OR caecum OR caecal OR ileocecal OR ileocaecal OR ileocaecum OR ileocecum OR large intestin* OR lower intestin*) | 165 |
| (Kirsten murine sarcoma* OR Kirsten rat sarcoma*) | (colorect* OR rectal* OR rectum* OR colon* OR sigma* OR sigmo* OR rectosigm* OR bowel* OR anal OR anus OR CRC OR m-CRC OR cecum OR cecal OR caecum OR caecal OR ileocecal OR ileocaecal OR ileocaecum OR ileocecum OR large intestin* OR lower intestin*) | 13 |
| thera screen OR thera-screen OR therascreen | 0 | |
| Total | 178 |
World Health Organization (WHO) International Clinical Trials Registry Platform (ICTRP) (Internet), 2000 to 25 January 2013
Searched 25 January 2013
Advanced search option
| Title | Condition | Intervention | Records |
|---|---|---|---|
| (KRAS or K-RAS or K ras) | (colon cancer or CRC or colorectal cancer or rectal cancer or rectum cancer) | 67 | |
| (colon cancer or CRC or colorectal cancer or rectal cancer or rectum cancer) | (KRAS or K-RAS or Kras) | 1 | |
| (Kirsten murine sarcoma* OR Kirsten rat sarcoma*) | (colon cancer or CRC or colorectal cancer or rectal cancer or rectum cancer) | 3 | |
| (colon cancer or CRC or colorectal cancer or rectal cancer or rectum cancer) | (Kirsten murine sarcoma* OR Kirsten rat sarcoma*) | Unable to run this line because of error with results screen | |
| thera screen OR thera-screen OR therascreen | 0 | ||
| thera screen OR thera-screen OR therascreen | 0 | ||
| Total | 71 |
Current Controlled Trials (metaRegister of Controlled Trials) (Internet), up to 29 January 2013
Searched 29 January 2013
| Search terms | Results |
|---|---|
| (Kirsten murine sarcoma* OR Kirsten rat sarcoma*) | 7 |
| (KRAS or K-RAS or K ras) | 146 |
| (thera screen OR thera-screen OR therascreen) | 0 |
| Total | 153 |
Conference searches
European Society for Medical Oncology conference proceedings (Internet), 2007–13
Searched 5 February 2013
2012 37th ESMO Congress, Vienna – http://annonc.oxfordjournals.org/content/23/suppl_9.
2011 ECCO 16 and 36th ESMO Multidisciplinary Congress, Brussels – www.ejcancer.info/issues.
2010 35th ESMO Congress, Milan – http://annonc.oxfordjournals.org/content/21/suppl_8.
2009 ECCO 15 and 34th ESMO Multidisciplinary Congress – www.ejcancer.info.
2008 33rd ESMO Congress, Stockholm – http://annonc.oxfordjournals.org/content/vol19/suppl_8/.
2007 ESMO Conference, Lugano – http://annonc.oxfordjournals.org/content/18/suppl_9.toc.
| Intervention | 2007 | 2008 | 2009a | 2010 | 2011a | 2012 |
|---|---|---|---|---|---|---|
| KRAS | 3/4 | 8 | 10 | 15 | 99 | 22 |
| K-RAS | 3/4 | 7 | 22 | 7 | 30 | 11 |
| K RAS | 22/29 | 30 | 22 | 34 | 30 | 44/47b |
| “Kirsten murine sarcoma” | 0 | 0 | 0 | 0 | 0 | 0 |
| “Kirsten rat sarcoma” | 0 | 0 | 3 | 0 | 1 | 0 |
| Total | 28 | 45 | 57 | 56 | 160 | 77 |
| Total | 423 | |||||
| Total after deduplication | 25 | 31 | 28 | 36 | 113 | 50 |
| Total after deduplication | 283 | |||||
The ESMO conference proceedings search located 423 records, with 283 remaining after deduplication.
American Association for Cancer Research conference proceedings (Internet), 2007–13
The AACR website had multiple search options retrieving different sets of results. A combination of the following was used:
Search 1: Whole website
-
Searched 5 February 2013
-
Searched website for abstracts from 2007–10; search limited to KRAS terms in title only – retrieved 236 results
Search 2: Individual years
2009
-
Searched 6 February 2013
| Keywords | Title search (advanced search) | Boolean search in presentation title |
|---|---|---|
| KRAS or K-RAS or K RAS | 60 | |
| Kirsten AND rat AND sarcoma | 0 | |
| Kirsten AND murine AND sarcoma | 0 | |
| Total | 60 | |
2010
-
www.abstractsonline.com/plan/start.aspx?mkey={0591FA3B-AFEF-49D2-8E65-55F41EE8117E}
-
Searched 6 February 2013
| Keywords | Title search (advanced search) | Boolean search |
|---|---|---|
| KRAS or K-RAS or K RAS | 93 | |
| Kirsten AND rat AND sarcoma | 1 | |
| Kirsten AND murine AND sarcoma | 0 | |
| Total | 94 | |
2011
-
www.abstractsonline.com/plan/start.aspx?mkey={507D311A-B6EC-436A-BD67-6D14ED39622C}
-
Searched 6 February 2013
| Keywords | Title search (advanced search) | Boolean search |
|---|---|---|
| KRAS or K-RAS or K RAS | 82 | |
| Kirsten AND rat AND sarcoma | 1 | |
| Kirsten AND murine AND sarcoma | 0 | |
| Total | 83 |
2012
-
www.abstractsonline.com/plan/start.aspx?mkey={2D8C569E-B72C-4E7D-AB3B-070BEC7EB280}
-
Searched 6 February 2013
| Keywords | Title (advanced search) | Boolean search |
|---|---|---|
| KRAS or K-RAS or K RAS | 93 | |
| Kirsten AND rat AND sarcoma | 1 | |
| Kirsten AND murine AND sarcoma | 0 | |
| Total | 94 | |
The combined American Association for Cancer Research conference proceedings search located 567 records in total.
American Society of Clinical Oncology conference proceedings (Internet), 2007–13
www.asco.org/ASCOv2/Meetings/Abstracts
Searched 5 February 2013
Searched 2007–12 Annual Meetings
| Keywords | Searched in title | Searched in abstract | Total |
|---|---|---|---|
| KRAS | 204 | 204 | |
| K-RAS | 46 | 46 | |
| K RAS | 46 (same results as K-RAS) | ||
| Kirsten rat sarcoma | 0 | 1 | 1 |
| Kirsten murine sarcoma | 0 | 1 (same result as Kirsten rat sarcoma) | |
| Total | 251 |
The ASCO conference proceedings search located 251 records.
Association for Molecular Pathology conference proceedings, 2007–13
Searched 6 February 2013
2012 AMP Abstracts; Long Beach, CA, 25–27 October 2012 – http://download.journals.elsevierhealth.com/pdfs/journals/1525-1578/PIIS1525157812002115.pdf.
2011 AMP Abstracts; Grapevine, TX, 17–19 November 2011 – http://download.journals.elsevierhealth.com/pdfs/journals/1525-1578/PIIS1525157811002546.pdf.
2010 AMP Abstracts; San Jose, CA, 18–20 November 2010 – http://download.journals.elsevierhealth.com/pdfs/journals/1525-1578/PIIS1525157810601365.pdf.
2009 AMP Abstracts; Kissimmee, FL, 19–22 November 2009 – http://download.journals.elsevierhealth.com/pdfs/journals/1525-1578/PIIS1525157810602851.pdf.
2008 AMP Abstracts; Grapevine, TX, 29 October–2 November 2008 – http://download.journals.elsevierhealth.com/pdfs/journals/1525-1578/PIIS1525157810602000.pdf.
2007 AMP Abstracts; Los Angeles, CA, 7–10 November 2007 – http://download.journals.elsevierhealth.com/pdfs/journals/1525-1578/PIIS1525157810604424.pdf.
| Intervention | 2007 | 2008 | 2009 | 2010 | 2011 | 2012 |
|---|---|---|---|---|---|---|
| KRAS | 4 | 5 | 23 | 32 | 32 | 38 |
| K-RAS | 1/2 | 0 | 2/4 | 0/1 | 0 | 0 |
| K RAS | 0/2 | 0 | 0/3 | 0/1 | 0/1 | 0 |
| Kirsten murine sarcoma | 0 | 0 | 0 | 0 | 0 | 0 |
| Kirsten rat sarcoma | 0 | 0 | 0 | 0 | 0 | 0 |
| Total per year | 5 | 5 | 25 | 32 | 32 | 38 |
| Total | 137 | |||||
The AMP conference proceedings search located 137 records.
Cost-effectiveness search strategies
CRC + KRAS + Economics filter (limit: 2000-C)
EMBASE (OvidSP), 2000 to Week 4 2013
Searched 29 January 2013
-
exp colon cancer/ or exp rectum cancer/ or colorectal tumor/ (169,460)
-
((colorect$ or rectal$ or rectum$ or colon$ or sigma$ or sigmo$ or rectosigm$ or bowel$ or anal or anus) adj3 (cancer$ or neoplas$ or oncolog$ or malignan$ or tumo?r$ or carcinoma$ or adenocarcinoma$ or metasta$ or meta-sta$ or sarcoma$ or adenom$ or lesion$)).ti,ab,ot,hw. (246,253)
-
(m-CRC or CRC).ti,ab,ot. (14,068)
-
((cecum or cecal or caecum or caecal or il?eoc?ecal or il?eoc?ecum) adj3 (cancer$ or neoplas$ or oncolog$ or malignan$ or tumo?r$ or carcinoma$ or adenocarcinoma$ or metasta$ or meta-sta$ or sarcoma$ or adenom$ or lesion$)).ti,ab,ot. (2125)
-
(large intestin$ adj3 (cancer$ or neoplas$ or oncolog$ or malignan$ or tumo?r$ or carcinoma$ or adenocarcinoma$ or metasta$ or meta-sta$ or sarcoma$ or adenom$ or lesion$)).ti,ab,ot. (1871)
-
(lower intestin$ adj3 (cancer$ or neoplas$ or oncolog$ or malignan$ or tumo?r$ or carcinoma$ or adenocarcinoma$ or metasta$ or meta-sta$ or sarcoma$ or adenom$ or lesion$)).ti,ab,ot. (26)
-
or/1-6 (250,031)
-
k ras oncogene/ (4967)
-
(k ras or kras or k-ras or V-Ki-ras$ or V-K-ras or V-Ki-ras or v ki ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS).af. (17,128)
-
(Kirsten adj3 (murine or rat) adj3 sarcoma$).ti,ab,ot. (399)
-
(thera?screen$ or therascreen$).af. (67)
-
(Cobas adj3 (k ras or kras or k-ras or V-Ki-ras$ or V-K-ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS)).af. (8)
-
(sanger sequencing adj3 (k ras or kras or k-ras or V-Ki-ras$ or V-K-ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS)).af. (15)
-
(pyrosequencing adj3 (k ras or kras or k-ras or V-Ki-ras$ or V-K-ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS)).af. (25)
-
((HRM or HRMA or dHPLC) adj3 (k ras or kras or k-ras or V-Ki-ras$ or V-K-ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS)).af. (13)
-
(high resolution adj3 melt$ adj3 (k ras or kras or k-ras or V-Ki-ras$ or V-K-ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS)).af. (8)
-
(SNapShot adj3 (k ras or kras or k-ras or V-Ki-ras$ or V-K-ras or V-Ki-ras or v ki ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS)).af. (5)
-
(Next generation sequencing adj3 (k ras or kras or k-ras or V-Ki-ras$ or V-K-ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS)).af. (1)
-
high resolution melting analysis/ (701)
-
19 and (8 or 9 or 10) (62)
-
or/8-18,20 (17,382)
-
7 and 21 (5731)
-
health-economics/ (32,282)
-
exp economic-evaluation/ (194,421)
-
exp health-care-cost/ (186,276)
-
exp pharmacoeconomics/ (160,882)
-
or/23-26 (445,930)
-
(econom$ or cost or costs or costly or costing or price or prices or pricing or pharmacoeconomic$).ti,ab. (542,934)
-
(expenditure$ not energy).ti,ab. (21,678)
-
(value adj2 money).ti,ab. (1191)
-
budget$.ti,ab. (22,178)
-
or/28-31 (565,244)
-
27 or 32 (823,889)
-
letter.pt. (811,274)
-
editorial.pt. (424,059)
-
note.pt. (543,769)
-
or/34-36 (1,779,102)
-
33 not 37 (742,302)
-
(metabolic adj cost).ti,ab. (800)
-
((energy or oxygen) adj cost).ti,ab. (3005)
-
((energy or oxygen) adj expenditure).ti,ab. (18,652)
-
or/39-41 (21,682)
-
38 not 42 (737,540)
-
22 and 43 (310)
-
limit 44 to yr=“2000 -Current” (303)
-
limit 45 to embase (285)
Costs filter: Centre for Reviews and Dissemination. NHS EED Economics Filter: EMBASE (Ovid) Weekly Search. York: CRD; 2010. URL: www.york.ac.uk/inst/crd/intertasc/nhs_eed_strategies.html (accessed 17 March 2011).
MEDLINE (OvidSP), 2000 to January Week 3 2013
Searched 29 January 2013
-
exp Colorectal Neoplasms/ (134,899)
-
((colorect$ or rectal$ or rectum$ or colon$ or sigma$ or sigmo$ or rectosigm$ or bowel$ or anal or anus) adj3 (cancer$ or neoplas$ or oncolog$ or malignan$ or tumo?r$ or carcinoma$ or adenocarcinoma$ or metasta$ or meta-sta$ or sarcoma$ or adenom$ or lesion$)).ti,ab,ot,hw. (165,996)
-
(m-CRC or CRC).ti,ab,ot. (8243)
-
((cecum or cecal or caecum or caecal or il?eoc?ecal or il?eoc?ecum) adj3 (cancer$ or neoplas$ or oncolog$ or malignan$ or tumo?r$ or carcinoma$ or adenocarcinoma$ or metasta$ or meta-sta$ or sarcoma$ or adenom$ or lesion$)).ti,ab,ot. (1571)
-
(large intestin$ adj3 (cancer$ or neoplas$ or oncolog$ or malignan$ or tumo?r$ or carcinoma$ or adenocarcinoma$ or metasta$ or meta-sta$ or sarcoma$ or adenom$ or lesion$)).ti,ab,ot. (1542)
-
(lower intestin$ adj3 (cancer$ or neoplas$ or oncolog$ or malignan$ or tumo?r$ or carcinoma$ or adenocarcinoma$ or metasta$ or meta-sta$ or sarcoma$ or adenom$ or lesion$)).ti,ab,ot. (23)
-
or/1-6 (170,912)
-
Genes, ras/ (11,084)
-
(k ras or kras or k-ras or V-Ki-ras$ or V-K-ras or V-Ki-ras or v ki ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS).af. (9558)
-
(Kirsten adj3 (murine or rat) adj3 sarcoma$).ti,ab,ot. (346)
-
(thera?screen$ or therascreen$).af. (16)
-
(Cobas adj3 (k ras or kras or k-ras or V-Ki-ras$ or V-K-ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS)).af. (2)
-
(sanger sequencing adj3 (k ras or kras or k-ras or V-Ki-ras$ or V-K-ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS)).af. (5)
-
(pyrosequencing adj3 (k ras or kras or k-ras or V-Ki-ras$ or V-K-ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS)).af. (12)
-
((HRM or HRMA) adj3 (k ras or kras or k-ras or V-Ki-ras$ or V-K-ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS)).af. (5)
-
(high resolution adj3 melt$ adj3 (k ras or kras or k-ras or V-Ki-ras$ or V-K-ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS)).af. (4)
-
(SNapShot adj3 (k ras or kras or k-ras or V-Ki-ras$ or V-K-ras or V-Ki-ras or v ki ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS)).af. (3)
-
(Next generation sequencing adj3 (k ras or kras or k-ras or V-Ki-ras$ or V-K-ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS)).af. (0)
-
or/8-18 (16,720)
-
7 and 19 (3092)
-
economics/ (26,342)
-
exp “costs and cost analysis”/ (168,037)
-
economics, dental/ (1847)
-
exp “economics, hospital”/ (18,317)
-
economics, medical/ (8474)
-
economics, nursing/ (3868)
-
economics, pharmaceutical/ (2383)
-
(economic$ or cost or costs or costly or costing or price or prices or pricing or pharmacoeconomic$).ti,ab. (375,308)
-
(expenditure$ not energy).ti,ab. (15,563)
-
(value adj1 money).ti,ab. (18)
-
budget$.ti,ab. (15,762)
-
or/21-31 (493,254)
-
((energy or oxygen) adj cost).ti,ab. (2454)
-
(metabolic adj cost).ti,ab. (667)
-
((energy or oxygen) adj expenditure).ti,ab. (14,408)
-
or/33-35 (16,880)
-
32 not 36 (489,444)
-
letter.pt. (758,034)
-
editorial.pt. (307,072)
-
historical article.pt. (288,506)
-
or/38-40 (1,339,895)
-
37 not 41 (463,260)
-
20 and 42 (74)
-
limit 43 to yr=“2000 -Current” (69)
Costs filter: Centre for Reviews and Dissemination. NHS EED Economics Filter: MEDLINE (Ovid) Monthly Search. York: CRD; 2010. URL: www.york.ac.uk/inst/crd/intertasc/nhs_eed_strategies.html (accessed 28 September 2010).
MEDLINE In-Process & Other Non-Indexed Citations (OvidSP) and MEDLINE Daily Update (OvidSP), up to 28 January 2013
Searched 29 January 2013
-
exp Colorectal Neoplasms/ (132)
-
((colorect$ or rectal$ or rectum$ or colon$ or sigma$ or sigmo$ or rectosigm$ or bowel$ or anal or anus) adj3 (cancer$ or neoplas$ or oncolog$ or malignan$ or tumo?r$ or carcinoma$ or adenocarcinoma$ or metasta$ or meta-sta$ or sarcoma$ or adenom$ or lesion$)).ti,ab,ot,hw. (7699)
-
(m-CRC or CRC).ti,ab,ot. (1022)
-
((cecum or cecal or caecum or caecal or il?eoc?ecal or il?eoc?ecum) adj3 (cancer$ or neoplas$ or oncolog$ or malignan$ or tumo?r$ or carcinoma$ or adenocarcinoma$ or metasta$ or meta-sta$ or sarcoma$ or adenom$ or lesion$)).ti,ab,ot. (111)
-
(large intestin$ adj3 (cancer$ or neoplas$ or oncolog$ or malignan$ or tumo?r$ or carcinoma$ or adenocarcinoma$ or metasta$ or meta-sta$ or sarcoma$ or adenom$ or lesion$)).ti,ab,ot. (28)
-
(lower intestin$ adj3 (cancer$ or neoplas$ or oncolog$ or malignan$ or tumo?r$ or carcinoma$ or adenocarcinoma$ or metasta$ or meta-sta$ or sarcoma$ or adenom$ or lesion$)).ti,ab,ot. (0)
-
or/1-6 (7889)
-
Genes, ras/ (6)
-
(k ras or kras or k-ras or V-Ki-ras$ or V-K-ras or V-Ki-ras or v ki ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS).af. (666)
-
(Kirsten adj3 (murine or rat) adj3 sarcoma$).ti,ab,ot. (17)
-
(thera?screen$ or therascreen$).af. (5)
-
(Cobas adj3 (k ras or kras or k-ras or V-Ki-ras$ or V-K-ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS)).af. (0)
-
(sanger sequencing adj3 (k ras or kras or k-ras or V-Ki-ras$ or V-K-ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS)).af. (1)
-
(pyrosequencing adj3 (k ras or kras or k-ras or V-Ki-ras$ or V-K-ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS)).af. (0)
-
((HRM or HRMA) adj3 (k ras or kras or k-ras or V-Ki-ras$ or V-K-ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS)).af. (1)
-
(high resolution adj3 melt$ adj3 (k ras or kras or k-ras or V-Ki-ras$ or V-K-ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS)).af. (0)
-
(SNapShot adj3 (k ras or kras or k-ras or V-Ki-ras$ or V-K-ras or V-Ki-ras or v ki ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS)).af. (1)
-
(Next generation sequencing adj3 (k ras or kras or k-ras or V-Ki-ras$ or V-K-ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS)).af. (0)
-
or/8-18 (671)
-
7 and 19 (270)
-
economics/ (1)
-
exp “costs and cost analysis”/ (143)
-
economics, dental/ (0)
-
exp “economics, hospital”/ (8)
-
economics, medical/ (0)
-
economics, nursing/ (0)
-
economics, pharmaceutical/ (1)
-
(economic$ or cost or costs or costly or costing or price or prices or pricing or pharmacoeconomic$).ti,ab. (33,295)
-
(expenditure$ not energy).ti,ab. (992)
-
(value adj1 money).ti,ab. (3)
-
budget$.ti,ab. (1659)
-
or/21-31 (35,017)
-
((energy or oxygen) adj cost).ti,ab. (186)
-
(metabolic adj cost).ti,ab. (46)
-
((energy or oxygen) adj expenditure).ti,ab. (681)
-
or/33-35 (890)
-
32 not 36 (34,752)
-
letter.pt. (19,083)
-
editorial.pt. (12,353)
-
historical article.pt. (144)
-
or/38-40 (31,562)
-
37 not 41 (34,345)
-
20 and 42 (17)
Costs filter:
Costs filter: Centre for Reviews and Dissemination. NHS EED Economics Filter: MEDLINE (Ovid) Monthly Search. York: CRD; 2010. URL: www.york.ac.uk/inst/crd/intertasc/nhs_eed_strategies.html (accessed 28 September 2010).
NHS Economic Evaluation Database (NHS EED) (Wiley), 2000 to Issue 4, 2012
Searched 22 January 2013
#1 MeSH descriptor: [Colorectal Neoplasms] explode all trees (4380)
#2 ((colorect* or rectal* or rectum* or colon* or sigma* or sigmo* or rectosigm* or bowel* or anal or anus) near/3 (cancer* or neoplas* or oncolog* or malignan* or tumour* or tumor* or carcinoma* or adenocarcinoma* or metasta* or meta-sta* or sarcoma* or adenom* or lesion*)) (7773)
#3 (m-CRC or CRC) (715)
#4 ((cecum or cecal or caecum or caecal or ileocecal or ileocaecal or ileocaecum or ileocecum) near/3 (cancer* or neoplas* or oncolog* or malignan* or tumour* or tumor* or carcinoma* or adenocarcinoma* or metasta* or meta-sta* or sarcoma* or adenom* or lesion*)) (24)
#5 (large intestin* near/3 (cancer* or neoplas* or oncolog* or malignan* or tumour* or tumor*or carcinoma* or adenocarcinoma* or metasta* or meta-sta* or sarcoma* or adenom* or lesion*)) (86)
#6 (lower intestin* near/3 (cancer* or neoplas* or oncolog* or malignan* or tumour* or tumor* or carcinoma* or adenocarcinoma* or metasta* or meta-sta* or sarcoma* or adenom* or lesion*)) (114)
#7 #1 or #2 or #3 or #4 or #5 or #6 (8053)
#8 MeSH descriptor: [Genes, ras] this term only (46)
#9 (k ras or kras or K-ras or V-Ki-ras* or V-K-ras or V-Ki-ras or v ki ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS) (386)
#10 (Kirsten near/3 (murine or rat) near/3 sarcoma*) (7)
#11 (thera screen* or thera-screen* or therascreen*) (13)
#12 (Cobas) (115)
#13 (sanger sequencing) (7)
#14 (pyrosequencing) (18)
#15 (HRM or HRMA) (11)
#16 (high resolution near/3 melt*) (1)
#17 (SNapShot) (50)
#18 (“Next generation sequencing”) (2)
#19 #8 or #9 or #10 or #11 or #12 or #13 or #14 or #15 or #16 or #17 or #18 (605)
#20 #7 and #19 from 2000 (98)
The NHS EED search retrieved three references.
Science Citation Index (SCI-EXPANDED) (Web of Knowledge), 2000 to 25 January 2013
Searched 30 January 2013
Databases=SCI-EXPANDED Timespan=2000-01-01 - 2013-01-30.
#27 117 #6 and #17 and #26
#26 532,023 #21 not #25
#25 33,411 #22 or #23 or #24
#24 15,970 TS=((energy or oxygen) NEAR expenditure)
#23 2095 TS=(metabolic NEAR cost)
#22 17,130 TS=((energy or oxygen) NEAR cost)
#21 551,568 #18 or #19 or #20
#20 909 TS=(value NEAR money)
#19 10,944 TS=(expenditure* not energy)
#18 546,907 TS=(economic* or cost or costs or costly or costing or price or prices or pricing or pharmacoeconomic* or budget*)
#17 10,434 #16 OR #15 OR #14 OR #13 OR #12 OR #11 OR #10 OR #9 OR #8 OR #7
#16 27 TS=((Next SAME generation SAME sequencing) SAME (k ras or kras or V-Ki-ras* or V-K-ras or c-ki-ras or c-k-ras or ki-ras or k-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS))
#15 23 TS=(SNapShot SAME (k ras or kras or k-rasor V-Ki-ras* or V-K-ras or V-Ki-ras or v ki ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS))
#14 76 TS=((high SAME resolution SAME melt*) SAME (k ras or kras or k-ras or V-Ki-ras* or V-K-ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS))
#13 59 TS=((HRM or HRMA or dHPLC) SAME (k ras or kras or k-ras or V-Ki-ras* or V-K-ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS))
#12 95 TS=(pyrosequencing SAME (k ras or kras or k-ras or V-Ki-ras* or V-K-ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS))
#11 50 TS=((sanger SAME sequencing) SAME (k ras or kras or k-ras or V-Ki-ras* or V-K-ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS))
#10 3 TS=(Cobas SAME (k ras or kras or k-ras or V-Ki-ras* or V-K-ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS))
#9 17 TS=(thera$screen* or therascreen*)
#8 97 TS=(Kirsten NEAR/3 (murine or rat) NEAR/3 sarcoma*)
#7 10,424 TS=(k ras or kras or K-ras or V-Ki-ras* or V-K-ras or V-Ki-ras or v ki ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS)
#6 132,645 #1 or #2 or #3 or #4 or #5
#5 1321 TS=(lower SAME intestin* NEAR/3 (cancer* or neoplas* or oncolog* or malignan* or tumo$r* or carcinoma* or adenocarcinoma* or metasta* or meta-sta* or sarcoma* or adenom* or lesion*))
#4 1097 TS=(large SAME intestin* NEAR/3 (cancer* or neoplas* or oncolog* or malignan* or tumo$r* or carcinoma* or adenocarcinoma* or metasta* or meta-sta* or sarcoma* or adenom* or lesion*))
#3 481 TS=((cecum or cecal or caecum or caecal or il$eoc$ecal or il$eoc$ecum) NEAR/3 (cancer* or neoplas* or oncolog* or malignan* or tumo$r* or carcinoma* or adenocarcinoma* or metasta* or meta-sta* or sarcoma* or adenom* or lesion*))
#2 8999 TS=(m-CRC or CRC)
#1 129,783 TS=((colorect* or rectal* or rectum* or colon* or sigma* or sigmo* or rectosigm* or bowel* or anal or anus) NEAR/3 (cancer* or neoplas* or oncolog* or malignan* or tumo?r* or carcinoma* or adenocarcinoma* or metasta* or meta-sta* or sarcoma* or adenom* or lesion*))
EconLit (EBSCO), 2000 to 30 January 2013
Searched 30 January 2013
Search modes – Boolean/phrase
-
S6 s1 or s2 or s3 or s4 Limiters - Published Date from: 20000101- (104)
-
S5 s1 or s2 or s3 or s4 (135)
-
S4 TX ((colorect* or rectal* or rectum* or colon* or sigma* or sigmo* or rectosigm* or bowel* or anal or anus) N4 (cancer* or neoplas* or oncolog* or malignan* or tumour* or tumor* or carcinoma* or adenocarcinoma* or metasta* or meta-sta* or sarcoma* or adenom* or lesion*)) (90)
-
S3 TX(thera screen* or thera-screen* or therascreen*) (0)
-
S2 TX(Kirsten murine sarcoma* or Kirsten rat sarcoma*) (0)
-
S1 TX(k ras or kras or k-ras or V-Ki-ras* or V-K-ras or V-Ki-ras or v ki ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS) (45)
Health Economic Evaluations Database (HEED) (Internet), up to 30 January 2013
http://onlinelibrary.wiley.com/book/10.1002/9780470510933
Searched 30 January 2013
Compound search (all data), unable to limit by date
| Keywords | Results |
|---|---|
| k ras or kras or k-ras or V-Ki-ras$ or V-K-ras or V-Ki-ras or v ki ras or c-ki-ras or c-k-ras or ki-ras or ki ras or Kras1 or Kras2 or KRAS1P or RASK or RASK1 or RASK2 or Kirsten RAS | 11 |
| Kirsten murine sarcoma OR Kirsten rat sarcoma | 2 |
| thera screen OR thera-screen OR therascreen | 0 |
| Total | 13 |
The HEED search retrieved 13 records.
Appendix 2 Data extraction tables
| Study details | Selection | Population | Intervention details | KRAS test details | Other | ||
|---|---|---|---|---|---|---|---|
| Detail | EGFR inhibitor | Standard chemotherapy | |||||
| Bokemeyer 201128,53,56 (OPUS) Country: Germany, Spain, Italy, Ukraine, Russian Federation, Romania and Poland Study design: RCT Funding: Merck Serono Recruitment: August 2006 Total no. randomised: 337 No. of KRAS wild-type patients randomised: 179 No. with liver-limited metastases randomised: 48 |
Inclusion criteria: Adults (aged ≥ 18 years); histologically confirmed first occurrence of non-resectable, EGFR-expressing mCRC with at least one radiologically measurable lesion; life expectancy of at least 12 weeks; maximum ECOG performance status 2; adequate hepatic, renal and bone function. KRAS wild-type population data only extracted Exclusion criteria: Pregnancy; previous EGFR-targeted therapy or previous chemotherapy (excluding adjuvant treatment) for mCRC; uncontrolled severe organ or metabolic dysfunction |
Age (years), median (range): 61 (24–82) Male, n: 97 Criteria for unresectability: NR Performance status: ECOG 0: 70; 1: 93; 2: 16 Previous treatments: 153 had previous surgery, 34 had previous adjuvant chemotherapy and 24 had radiotherapy |
Intervention | FOLFOX-4 + cetuximab | FOLFOX-4 | KRAS test: one-step LightCycler PCR reaction (LightMix k-ras Gly12 assay) Manufacturer: TIB MOLBIOL Analysis software: LightCycler 2.0 system Mutations targeted: KRAS codons 12 and 13 missense mutations |
Data for KRAS wild-type population and liver metastases-only subgroup are post-hoc analyses. Baseline data are for entire wild-type population |
| Dose | As standard + cetuximab, initial dose 400 mg/m2, subsequent doses 250 mg/m2 | Oxaliplatin 85 mg/m2; folinic acid 200 mg/m2 followed by 5-FU as a 400 mg/m2 bolus then a 600 mg/m2 infusion over 22 hours | |||||
| As standard + weekly | Days 1 and 2 of a 14-day cycle | ||||||
| Median (IQR) duration (weeks) | Cetuximab 24 (13–38); oxaliplatin 23 (14–31); 5-FU 24 (14–36) | Oxaliplatin 24 (16–30); 5-FU 24 (16–32) | |||||
| No. with KRAS wild type treated | 82 | 97 | |||||
| No. with liver metastases treated | 25 | 23 | |||||
| Folprecht 201052 (CELIM) Country: Germany and Austria Study design: RCT Funding: Merck Serono, Sanofi-Aventis and Pfizer Recruitment: December 2004–March 2008 No. randomised: 111 No. of KRAS wild-type patients randomised: 70 No. with liver-limited metastases randomised: 111 |
Inclusion criteria: Unresectable, histologically confirmed colorectal liver metastases; no extrahepatic metastases (patients with synchronous liver metastases were eligible if the primary tumour had been resected before chemotherapy); Karnofsky perfomance score ≥ 80%; adequate hepatic, renal and bone marrow function Exclusion criteria: Previous chemotherapy (except adjuvant chemotherapy with an interval of ≥ 6 months); previous EGFR-targeted therapy; concurrent antitumour therapy; clinically relevant coronary artery disease; inflammatory bowel disease; previous malignancy; age < 18 years |
Age (years), median (range): 63 (56–71) Male, n: 71 Liver metastases: < 5 metastases: 30; 5–10 metastases: 58; > 10 metastases: 19; NR: 4 No. with previous liver resection: 14 Criteria for unresectability: Five or more liver metastases or metastases that were viewed as technically non-resectable by the local liver surgeon and radiologist on the basis of inadequate future liver remnant, or one of the following critera: infiltration of all hepatic liver veins, infiltration of both hepatic arteries or both portal vein branches Performance status: NR Previous treatments: nine patients had adjuvant radiotherapy, 18 had adjuvant chemotherapy |
Intervention | Cetuximab + FOLFOX6 | Cetuximab + FOLFIRI | KRAS test: Direct sequencing and DxS KRAS Mutation Test Kit Manufacturer: DxS, Manchester, UK Analysis software: NR Mutations targeted: KRAS mutations in codons 12 and 13 |
|
| Dose | (Cetuximab 400 mg/m2 initially then 250 mg/m2 subsequently) + (100 mg/m2 oxaliplatin, 400 mg/m2 folinic acid and 400 mg/m2 bolus 5-FU followed by 2400 mg/m2 over 46 hours) | (Cetuximab 400 mg/m2 initially then 250 mg/m2 subsequently) + (180 mg/m2 irinotecan, 400 mg/m2 folinic acid and 400 mg/m2 bolus 5-FU followed by 2400 mg/m2 over 46 hours) | |||||
| (Day 1 of a weekly cycle) + (day 1 of a 2-weekly cycle) | (Day 1 of a weekly cycle) + (day 1 of a 2-weekly cycle) | ||||||
| No. with KRAS wild type treated | 56 | 55 | |||||
| No. with liver metastases treated | 56 | 55 | |||||
| Kohne 201127,53 (CRYSTAL)a Country: Germany, Russian Federation, Poland, Singapore, South Korea, South Africa, France and Belgium (201 centres) Study design: RCT Funding: Meck Serono Recruitment: July 2004–November 2005 No. randomised: 1202 No. of KRAS wild-type patients randomised: 666 No. with liver-limited metastases randomised: 140 |
Inclusion criteria: Adults (aged ≥ 18 years); histologically confirmed adenocarcinoma of the colon or rectum; first occurrence of metastatic disease that could not be resected for curative purposes; EGFR expressing; maximum ECOG performance status 2; adequate hepatic, renal and haematological function Exclusion criteria: Previous anti-EGFR therapy or irinotecan-based therapy; previous chemotherapy for mCRC; adjuvant treatment within 6 months of the start of the trial; radiotherapy, surgery (excluding previous diagnostic biopsy) or any investigational drug with 30 days of the start of the trial |
Age (years), median (range): 61 (22–79) Male, n: 201 Criteria for unresectability: NR Performance status: ECOG 0: 203; ECOG 1: 131; ECOG 2: 14 Previous treatments: NR |
Intervention | Cetuximab + FOLFORI | FOLFORI | KRAS test: PCR clamping and melting curve method (LightMix k-ras Gly12 assay) Manufacturer: TIB MOLBIOL Analysis software: LightCycler 2.0 system Mutations targeted: KRAS codons 12 and 13 missense mutations |
Data for KRAS wild-type population and liver metastases-only subgroup are post-hoc analyses. Treatment continued until disease progression, toxic effects or withdrawal of consent. No definition of unresectable liver metastases was reported |
| Dose | As standard + initial 120-minute infusion of cetuximab 400 mg/m2 on day 1 followed by weekly 60-minute infusions of cetuximab 250 mg/m2 | 30- to 90-minute infusion of irinotecan 180 mg/m2, 120-minute infusion of racemic leucovorin 400 mg/m2 or L-leucovorin 200 mg/m2, 5-FU 400 mg/m2 bolus then 46-hour infusion of 2400 mg/m2 | |||||
| Weekly | Day 1 of a 14-day cycle | ||||||
| Median (IQR) duration (weeks) | Cetuximab 25.0 (12.9–40.4); irinotecan 26.0 (14.0–40.3); 5-FU 26.0 (13.8–40.4) | Irinotecan 25.7 (15.1–39.5); 5-FU 25.7 (14.9–36.0) | |||||
| No. treated | 172 | 176 | |||||
| No. with liver metastases treated | 68 | 72 | |||||
| Maughan 201154 (COIN) Country: UK and Republic of Ireland Study design: RCT Funding: UK MRC Recruitment: March 2005–May 2008 No. randomised: 1630 No. of KRAS wild-type patients randomised: 729 No. with liver-limited metastases randomised: 178 |
Inclusion criteria: Adults (aged ≥ 18 years); histologically confirmed adenocarcinoma of the colon or rectum; inoperable metastatic or locoregional disease; no previous chemotherapy for metastatic disease; WHO performance status 0–2; adequate hepatic, renal and haematological function; no adjuvant chemotherapy or rectal chemoradiotherapy within 1 month of the start of the trial Exclusion criteria: Unfit for chemotherapy; severe uncontrolled medical illness; psychiatric illness inhibiting informed consent; partial or complete bowel obstruction; pre-existing neuropathy of grade 1 or higher; requirement for treatment with contraindicated medication; another previous or current malignant disease that may affect treatment response; known hypersensitivity to any study treatment; brain metastases |
Age (years), median (range): 64 (56–70) Male, n: 498 Criteria for unresectability: NR Performance status: WHO 0: 348; WHO 1: 337; WHO 2: 44; WHO 3: 0 Previous treatments: NR |
Intervention | Cetuximab + standard chemotherapy (oxaliplatin + 5-FU and folinic acid, or oxaliplatin + capecitabine) | Standard chemotherapy (oxaliplatin + 5-FU and folinic acid, or oxaliplatin + capecitabine) | KRAS test: Pyrosequencing and MALDI-TOF mass array, with Sanger sequencing for discordant samples (< 1%) Manufacturer: In house Analysis software: Sequenom, San Diego, CA Mutations targeted: KRAS mutations in codons 12, 13 and 61 |
Data for KRAS wild-type population and liver metastases-only subgroup are post-hoc analyses. These participants were also BRAF and NRAS wild type. Unavailable tumour blocks = 141; insufficient tumour material for processing = 163; not successfully genotyped = 22 Standard chemotherapy was FOLFOX for some patients (approximately one-third of the whole trial) and XELOX for some patients (approximately two-thirds of the whole trial). Proportions unknown for the subgroup with liver metastases 178 KRAS wild-type patients had liver metastases only; 153 were included in the analysis (25 patients missing) |
| Dose | As standard + initial 2-hour infusion of 400 mg/m2 cetuximab and 250 mg/m2 over 1 hour subsequently | 2-hour infusion of L-folinic acid 175 mg or racemic folinic acid 350 mg, 2-hour infusion of oxaliplatin 85 mg/m2, followed by 400 mg/m2 bolus 5-FU and then 46-hour infusion of 2400 mg/m2 5-FU, or 2-hour infusion of 130 mg/m2 oxaliplatin followed by oral capecitabine (1000 mg/m2) twice a day for 2 weeks, reduced to 850 mg/m2 in later patients | |||||
| Weekly | 2-weekly for folonic acid, 3-weekly for capecitabine | ||||||
| Median (IQR) duration (weeks) | 28.1 (15.4–42.0) | 29.3 (15.7–40.1) | |||||
| No. treated | 357 | 358 | |||||
| No. with liver metastases treated | 87 | 91 | |||||
| Xu 201255 Country: China Study design: RCT Funding: NR Recruitment: June 2008–December 2011 No. eligible: NR No. randomised: 116 No. of KRAS wild-type patients randomised: 116 No. with liver-limited metastases randomised: 116 |
Inclusion criteria: Resected primary colorectal tumour; non-resectable synchronous liver-limited metastases; KRAS wild type Exclusion criteria: None reported |
Age: NR Male, n: NR Criteria for unresectability: NR Performance status: NR Previous treatments: NR |
Intervention | Cetuximab + FOLFIRI or FOLFOX6 | FOLFIRI or FOLFOX6 | KRAS test: Pyrosequencing Analysis software: NR Manufacturer: NR Mutations targeted: NR |
|
| Dose | NR | NR | |||||
| Frequency | NR | NR | |||||
| Mean number of cycles | NR | NR | |||||
| Duration | NR | NR | |||||
| No. with KRAS wild type treated | 59 | 57 | |||||
| No. with liver metastases treated | 59 | 57 | |||||
Appendix 3 Risk of bias assessments
QUADAS-2 assessments
Folprecht et al.52 (CELIM)
| Domain 1: Patient selection | |
|---|---|
| A. Risk of bias | |
| Describe methods of patient selection: Phase II RCT comparing FOLFOX6 + cetuximab with FOLFIRI + cetuximab in patients with unresectable liver metastases from CRC. In total, 111 participants were included, of whom 94 received KRAS testing and were included in this assessment. There were no inappropriate exclusions from the trial | |
| Was a consecutive or random sample of patients enrolled? | Yes |
| Was a case–control design avoided? | Yes |
| Did the study avoid inappropriate exclusions? | Yes |
| Could the selection of patients have introduced bias? | Risk: low |
| B. Concerns regarding applicability | |
| Describe included patients (prior testing, presentation, intended use of index test and setting): Study participants were described as having technically non-resectable or five or more liver metastases from CRC; it was unclear whether some participants may have had potentially resectable metastases at baseline | |
| Is there concern that the included patients do not match the review question? | Concern: unclear |
| Domain 2: Index test(s) | |
| A. Risk of bias | |
| Describe the index test and how it was conducted and interpreted: Tumour KRAS mutation status (index test) was determined before the clinical outcome (reference standard) was known | |
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes |
| Could the conduct or interpretation of the index test have introduced bias? | Risk: low |
| B. Concerns regarding applicability | |
| Is there concern that the target condition as defined by the reference standard does not match the review question? Tumour KRAS mutation status was determined using the Therascreen KRAS PCR kit | Concern: low |
| Domain 3: Reference standard | |
| A. Risk of bias | |
| Describe the reference standard and how it was conducted and interpreted: Clinical outcome (objective response) was used as the reference standard; data on resection rates were not reported by tumour KRAS mutation status. Analysis of objective response by tumour KRAS mutation status was retrospective | |
| Is the reference standard likely to correctly classify the target condition? | Yes |
| Were the reference standard results interpreted without knowledge of the results of the index test? | Unclear |
| Could the reference standard, its conduct or its interpretation have introduced bias? | Risk: low |
| B. Concerns regarding applicability | |
| Is there concern that the included patients do not match the review question? | Concern: high |
| Domain 4: Flow and timing | |
| A. Risk of bias | |
| Describe any patients who did not receive the index test(s) and/or reference standard or who were excluded from the 2 × 2 table: 17 (15%) participants were not included in the analysis. It was not clear whether this was because tumour KRAS mutation status was unknown or because follow-up data were not available | |
| Describe the time interval and any interventions between the index test(s) and the reference standard: Tumour response was assessed every four cycles (8 weeks) for a maximum of 2 years | |
| Was there an appropriate interval between the index test(s) and the reference standard? | Yes |
| Did all patients receive a reference standard? | Unclear |
| Did patients receive the same reference standard? | Yes |
| Were all patients included in the analysis? | No |
| Could the patient flow have introduced bias? | Risk: high |
Maughan et al.54 (COIN)
| Domain 1: Patient selection | |
|---|---|
| A. Risk of bias | |
| Describe methods of patient selection: RCT comparing cetuximab + FOLFOX or XELOX with FOLFOX or XELOX. All patients received KRAS mutation testing but only the subgroup of patients with unresectable liver metastases were included in this assessment. There were no inappropriate exclusions from the trial | |
| Was a consecutive or random sample of patients enrolled? | Yes |
| Was a case–control design avoided? | Yes |
| Did the study avoid inappropriate exclusions? | Yes |
| Could the selection of patients have introduced bias? | Risk: low |
| B. Concerns regarding applicability | |
| Describe included patients (prior testing, presentation, intended use of index test and setting): All study participants included in this assessment had inoperable liver metastases from CRC and no extrahepatic metastases or previous chemotherapy. Patients receiving combination chemotherapy before resection of operable liver metastases were explicitly excluded | |
| Is there concern that the included patients do not match the review question? | Concern: low |
| Domain 2: Index test(s) | |
| A. Risk of bias | |
| Describe the index test and how it was conducted and interpreted: Tumour KRAS mutation status (index test) was determined before the clinical outcome (reference standard) was known | |
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes |
| Could the conduct or interpretation of the index test have introduced bias? | Risk: low |
| B. Concerns regarding applicability | |
| Is there concern that the target condition as defined by the reference standard does not match the review question? Tumour KRAS mutation status was determined using pyrosequencing and MALDI-TOF, targeting mutations in codons 12, 13 and 61 | Concern: low |
| Domain 3: Reference standard | |
| A. Risk of bias | |
| Describe the reference standard and how it was conducted and interpreted: Clinical outcome (resection) was used as the reference standard. It was not clear whether investigators assessing resectability were aware of tumour KRAS mutation status. Treatment arms included FOLFOX or XELOX as standard chemotherapy and XELOX was not specified as standard chemotherapy for this review | |
| Is the reference standard likely to correctly classify the target condition? | Yes |
| Were the reference standard results interpreted without knowledge of the results of the index test? | Unclear |
| Could the reference standard, its conduct or its interpretation have introduced bias? | Risk: low |
| B. Concerns regarding applicability | |
| Is there concern that the included patients do not match the review question? | Concern: high |
| Domain 4: Flow and timing | |
| A. Risk of bias | |
| Describe any patients who did not receive the index test(s) and/or reference standard or who were excluded from the 2 × 2 table: Tumour response was assessed every 12 weeks. In total, 19% of the original participants did not receive KRAS mutation testing because no sample was available; testing failed in a further 1%. However, it was not clear how many, if any, participants in the unresectable liver metastases subgroup did not receive testing. A total of 153 participants with KRAS wild-type tumours were included in the analysis [25 (14%) missing] | |
| Was there an appropriate interval between the index test(s) and the reference standard? | Yes |
| Did all patients receive a reference standard? | Unclear |
| Did patients receive the same reference standard? | Yes |
| Were all patients included in the analysis? | No |
| Could the patient flow have introduced bias? | Risk: high |
Cochrane risk of bias assessments
Bokemeyer et al.28,53,56 (OPUS)
| Support for judgement | Risk of bias | |
|---|---|---|
| Random sequence generation | 1 : 1 randomisation was carried out using a stratified permuted-block procedure, with ECOG performance status as a stratification factor | Unclear |
| Allocation concealment | No details reported | Unclear |
| Participant/personnel blinding | Open-label design | High |
| Outcome assessor blinding | Outcomes assessed by a blinded independent review committee | Low |
| Incomplete outcome data | Outcomes for the whole study polulation were analysed using ITT analysis and outcome data appeared to be reported for all patients with liver-limited metasteses, however, details were limited | Low |
| Selective outcome reporting | All specified outcomes appear to be reported | Low |
Van Cutsem et al.27,53 (CRYSTAL)
| Support for judgement | Risk of bias | |
|---|---|---|
| Random sequence generation | 1 : 1 randomisation was carried out using a stratified permuted-block procedure, with ECOG performance status as a stratification factor | Unclear |
| Allocation concealment | No details reported | Unclear |
| Participant/personnel blinding | Open-label design | High |
| Outcome assessor blinding | No details reported | Unclear |
| Incomplete outcome data | Outcomes for the whole study polulation were analysed using ITT analysis and outcome data appeared to be reported for all patients with liver-limited metasteses; however, data for these patients were reported only in an abstract and details were limited | Low |
| Selective outcome reporting | All specified outcomes appear to be reported | Low |
Maughan et al.54 (COIN)
| Support for judgement | Risk of bias | |
|---|---|---|
| Random sequence generation | Patients were randomly assigned with minimisation by the MRC Clinical Trials Unit by telephone | Low |
| Allocation concealment | Treatment allocation was not masked | High |
| Participant/personnel blinding | Open-label design | High |
| Outcome assessor blinding | No details reported | Unclear |
| Incomplete outcome data | Outcomes for the whole study polulation were analysed using ITT analysis and outcome data appeared to be reported for all patients with liver-limited metasteses; however, data for these patients were reported only in an abstract and details were limited | Low |
| Selective outcome reporting | All specified outcomes appear to be reported | Low |
Xu et al.55
| Support for judgement | Risk of bias | |
|---|---|---|
| Random sequence generation | No details reported | Unclear |
| Allocation concealment | No details reported | Unclear |
| Participant/personnel blinding | Open-label design | High |
| Outcome assessor blinding | No details reported | Unclear |
| Incomplete outcome data | No details reported | Unclear |
| Selective outcome reporting | No details reported | Unclear |
Appendix 4 Survey of NHS laboratories participating in the UK National External Quality Assurance Scheme pilot for KRAS mutation testing
Appendix 5 Table of excluded studies with rationale
| Study | Reason for exclusion (once a study had failed on one criterion it was not assessed further) | |||
|---|---|---|---|---|
| 1. Not a primary study | 2. Did not include patients with mCRC and a resected or resectable primary tumour whose metastases are confined to the liver and are unresectable | 3. KRAS mutation test not performed and/or test and mutation not specified or deducible | 4. Study did not report on response to treatment, survival or PFS | |
| Adams 200981 | ✓ | |||
| Adams 201082 | ✓ | |||
| Adams 201083 | ✓ | |||
| Alberts 201084 | ✓ | |||
| Assenat 201185 | ✓ | |||
| Baker 200886 | ✓ | |||
| Baloglu 201287 | ✓ | |||
| Bokemeyer 200888 | ✓ | |||
| Bokemeyer 200956 | ✓ | |||
| Bokemeyer 200989 | ✓ | |||
| Bokemeyer 201090 | ✓ | |||
| Bokemeyer 201291 | ✓ | |||
| Chuko 201092 | ✓ | |||
| Cohen 200893 | ✓ | |||
| Colucci 201094 | ✓ | |||
| Di Salvatore 201095 | ✓ | |||
| Dubus 200996 | ✓ | |||
| Folprecht 200897 | ✓ | |||
| Folprecht 200998 | ✓ | |||
| Folprecht 201099 | ✓ | |||
| Gajate 2012100 | ✓ | |||
| Gao 2011101 | ✓ | |||
| Garufi 2009102 | ✓ | |||
| Goldberg 2010103 | ✓ | |||
| Griebsch 2011104 | ✓ | |||
| Harbison 2012105 | ✓ | |||
| Huang 2011106 | ✓ | |||
| Ibrahim 2010107 | ✓ | |||
| Jones 2013108 | ✓ | |||
| Jonker 2009109 | ✓ | |||
| Kimura 2012110 | ✓ | |||
| Kohne 2009111 | ✓ | |||
| Ku 2012112 | ✓ | |||
| Lang 2009113 | ✓ | |||
| Lievre 2006114 | ✓ | |||
| Lin 2010115 | ✓ | |||
| Lin 2011116 | ✓ | |||
| Linardou 2008117 | ✓ | |||
| Loupakis 2012118 | ✓ | |||
| Malapelle 2012119 | ✓ | |||
| Malapelle 2012120 | ✓ | |||
| Mancuso 2008121 | ✓ | |||
| Maughan 2009122 | ✓ | |||
| Maughan 2010123 | ✓ | |||
| Maughan 2010124 | ✓ | |||
| Mayer 2010125 | ✓ | |||
| Merck KGaA 2011126 | ✓ | |||
| Modest 2012127 | ✓ | |||
| Molinari 2010128 | ✓ | |||
| Moosmann 2011129 | ✓ | |||
| Ocvirk 2009130 | ✓ | |||
| Ocvirk 201057 | ✓ | |||
| Passardi 2011131 | ✓ | |||
| Petrelli 2011132 | ✓ | |||
| Petrelli 2012133 | ✓ | |||
| Piessevaux 2010134 | ✓ | |||
| Piessevaux 2011135 | ✓ | |||
| Piessevaux 2011136 | ✓ | |||
| Qiu 2010137 | ✓ | |||
| Raoul 2009138 | ✓ | |||
| Rivera 2009139 | ✓ | |||
| Rose 2012140 | ✓ | |||
| Salazar 2012141 | ✓ | |||
| Schuch 2008142 | ✓ | |||
| Serna 2011143 | ✓ | |||
| Shinozaki 2012144 | ✓ | |||
| Simon 2011145 | ✓ | |||
| Stenger 2012146 | ✓ | |||
| Stintzing 2010147 | ✓ | |||
| Taieb 2012148 | ✓ | |||
| Tejpar 2011149 | ✓ | |||
| Tejpar 2011150 | ✓ | |||
| Tejpar 2011151 | ✓ | |||
| Tejpar 2012152 | ✓ | |||
| Tsoukalas 2010153 | ✓ | |||
| Tsoukalas 2011154 | ✓ | |||
| Tsoukalas 2012155 | ✓ | |||
| Tveit 2010156 | ✓ | |||
| Tveit 2011157 | ✓ | |||
| Tveit 201258 | ✓ | |||
| Ubago 2011158 | ✓ | |||
| Vale 2009159 | ✓ | |||
| Van Cutsem 2008160 | ✓ | |||
| Van Cutsem 200927 | ✓ | |||
| Van Cutsem 2009161 | ✓ | |||
| Van Cutsem 2010162 | ✓ | |||
| Van Cutsem 2010163 | ✓ | |||
| Van Cutsem 2011164 | ✓ | |||
| Wasan 2011165 | ✓ | |||
| Whitehall 2009166 | ✓ | |||
| Yen 2010167 | ✓ | |||
| Zhang 2011168 | ✓ | |||
Appendix 6 Cost-effectiveness acceptability curves for sensitivity analyses
Cost-effectiveness acceptability curve for the linked evidence analysis sensitivity analysis: mortality second line based on average of first- and third-line mortality.
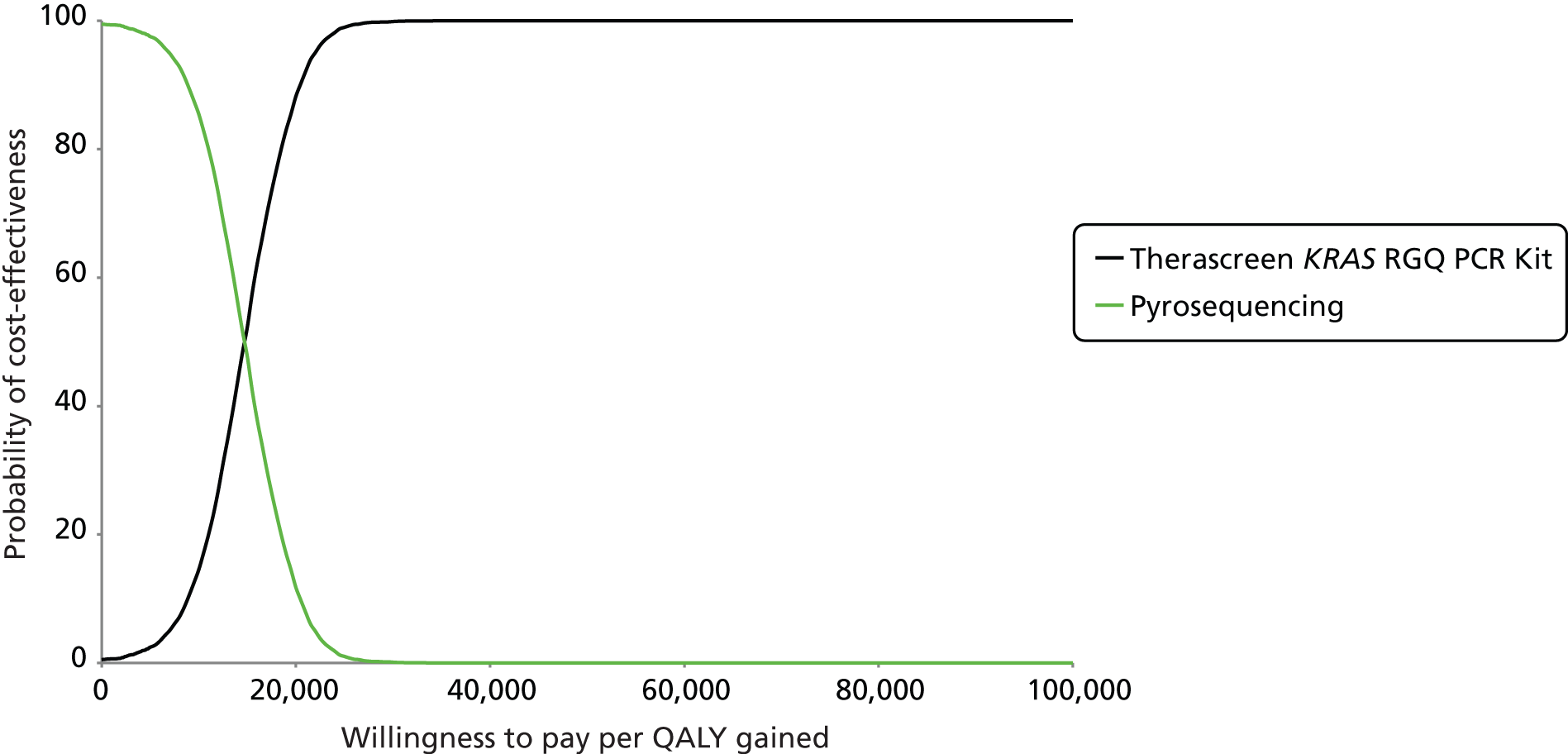
Cost-effectiveness acceptability curve for linked evidence analysis sensitivity analysis: proportion of patients with unknown mutation status based on the survey.
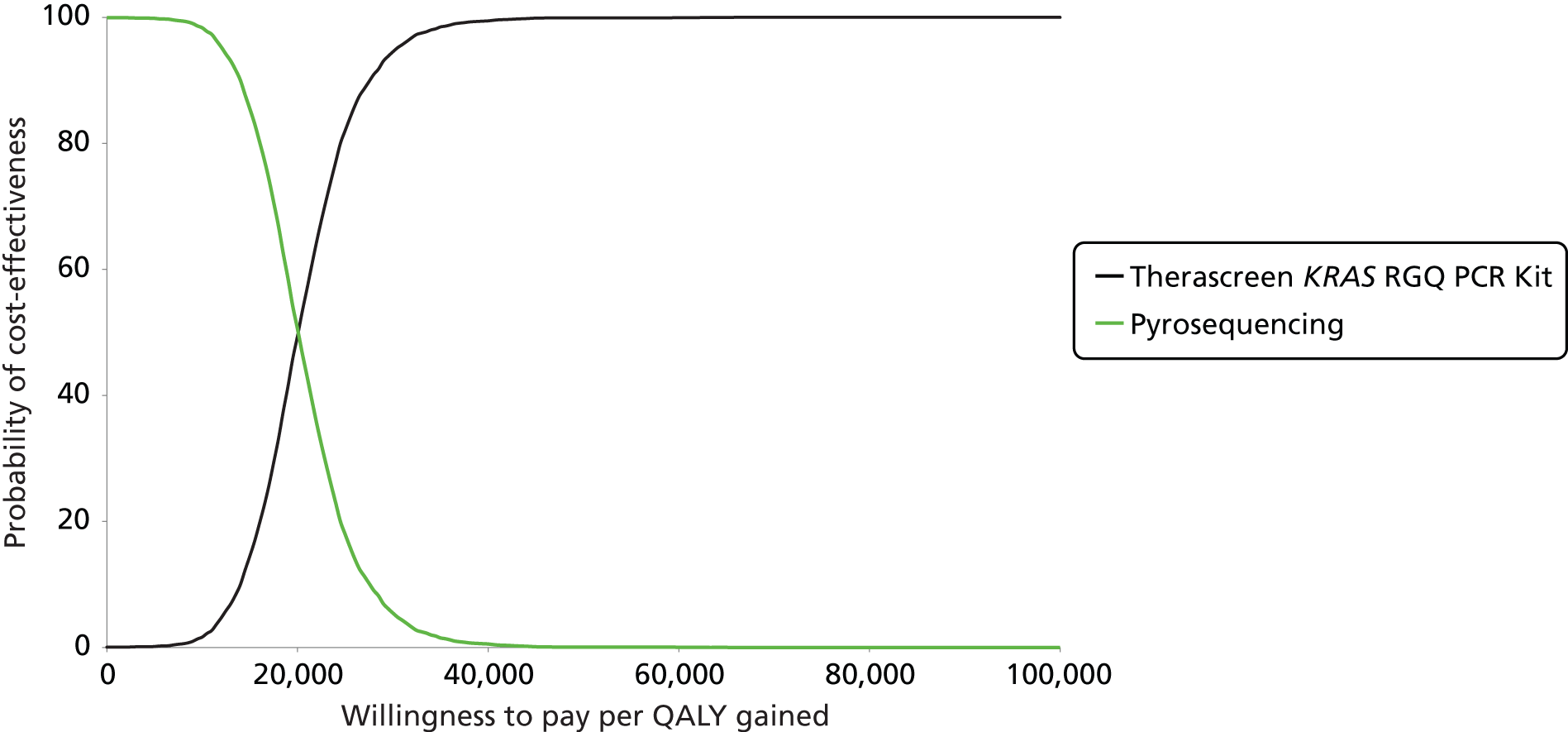
Appendix 7 National Institute for Health and Care Excellence guidance relevant to the management of metastatic colorectal cancer
Cancer service guidance
National Institute for Clinical Excellence. Improving Outcomes in Colorectal Cancer: Manual Update. Cancer service guidance. London: NICE; June 2004. URL: http://guidance.nice.org.uk/CSGCC (accessed 14 May 2013).
Clinical guidelines
National Institute for Health and Clinical Excellence. Colorectal Cancer – Capecitabine and Tegafur Uracil. NICE technology appraisal 61. London: NICE; May 2005. URL: http://guidance.nice.org.uk/TA61 (accessed 14 May 2013).
National Institute for Health and Clinical Excellence. Colorectal Cancer (Advanced) – Irinotecan, Oxaliplatin and Raltitrexed. NICE technology appraisal 93. London: NICE; August 2005. URL: http://guidance.nice.org.uk/TA93 (accessed 14 May 2013).
National Institute for Health and Clinical Excellence. Colon Cancer (Adjuvant) – Capecitabine and Oxaliplatin. NICE technology appraisal 100. London: NICE; April 2006. URL: http://guidance.nice.org.uk/TA100 (accessed 14 May 2013).
National Institute for Health and Clinical Excellence. Colorectal Cancer – Laparoscopic Surgery. NICE technology appraisal 105. London: NICE; August 2006. URL: http://guidance.nice.org.uk/TA105 (accessed 14 May 2013).
National Institute for Health and Clinical Excellence. Colorectal Cancer: the Diagnosis and Management of Colorectal Cancer. Clinical guideline CG131. London: NICE; November 2011. URL: http://guidance.nice.org.uk/CG131 (accessed 14 May 2013). [Date of review: to be confirmed.] [Clinical guideline CG131 updates and replaces TA93 and incorporates TA61, TA100 and TA105.]
Technology appraisals
National Institute for Health and Clinical Excellence. Colorectal Cancer (Metastatic) – Bevacizumab and Cetuximab. NICE technology appraisal 118. London: NICE; January 2007. URL: http://guidance.nice.org.uk/TA118 (accessed 14 May 2013).
National Institute for Health and Clinical Excellence. Colorectal Cancer (Metastatic) – Cetuximab (Terminated Appraisal). NICE technology appraisal 150. London: NICE; June 2008. URL: http://guidance.nice.org.uk/TA150 (accessed 14 May 2013).
National Institute for Health and Clinical Excellence. Cetuximab for the First-Line Treatment of Metastatic Colorectal Cancer. NICE technology appraisal 176. London: NICE; August 2009. URL: http://guidance.nice.org.uk/TA176 (accessed 14 May 2013). [Date of review: August 2012. The last review decision was in June 2011, when it was agreed that TA176 would be cross-referenced with CG131. The reason given for not incorporating TA176 into CG131 was ‘as the results of the further subgroup analyses of the COIN study could potentially lead to the need to update the recommendations in the future’.]
National Institute for Health and Clinical Excellence. Bevacizumab in Combination with Oxaliplatin and either Fluorouracil Plus Folinic Acid or Capecitabine for the Treatment of Metastatic Colorectal Cancer. NICE technology appraisal 212. London: NICE; December 2010. URL: http://guidance.nice.org.uk/TA212 (accessed 13 May 2013). [Date of review: to be confirmed.]
National Institute for Health and Clinical Excellence. Colorectal Cancer (Metastatic) 2nd Line: Cetuximab, Bevacizumab and Panitumumab. NICE technology appraisal 242. London: NICE; January 2012. URL: http://guidance.nice.org.uk/TA242 (accessed 14 May 2013). [Date of review: January 2015.] [Technology appraisal 242 replaces TA150 and partially updated TA118.]
National Institute for Health and Care Excellence. Aflibercept in Combination with Irinotecan and Fluorouracil-Based Therapy for Treating Metastatic Colorectal Cancer that has Progressed Following Prior Oxaliplatin-Based Chemotherapy. NICE technology appraisal guidance 307. London: NICE; 2014. URL: http://publications.nice.org.uk/aflibercept-in-combination-with-irinotecan-and-fluorouracil-based-therapy-for-treating-metastatic-ta307 (accessed 10 June 2014). [Date for review: August 2016.]
National Institute for Health and Clinical Excellence pathways
National Institute for Health and Clinical Excellence. NICE Pathways: Colorectal Cancer Overview. London: NICE; November 2011. URL: http://pathways.nice.org.uk/pathways/colorectal-cancer (accessed 14 May 2013).
Quality standards
National Institute for Health and Clinical Excellence. Colorectal Cancer (QS20). London: NICE; August 2012. URL: http://guidance.nice.org.uk/QS20 (accessed 14 May 2013).
Terminated
National Institute for Health and Clinical Excellence. Panitumumab in Combination with Chemotherapy for the Treatment of Metastatic Colorectal Cancer (Terminated Appraisal). NICE technology appraisal 240. London: NICE; December 2011. URL: http://guidance.nice.org.uk/TA240 (accessed 14 May 2013). [‘NICE is unable to recommend the use in the NHS of panitumumab in combination with chemotherapy for the treatment of metastatic colorectal cancer because no evidence submission was received from the manufacturer or sponsor of the technology’.]
Appendix 8 Preferred Reporting Items for Systematic Reviews and Meta-Analyses checklist
| Section/topic | No. | Checklist item | Reported on page no. |
|---|---|---|---|
| Title | |||
| Title | 1 | Identify the report as a systematic review, meta-analysis or both | p. |
| Abstract | |||
| Structured summary | 2 | Provide a structured summary including, as applicable: background; objectives; data sources; study eligibility criteria, participants and interventions; study appraisal and synthesis methods; results; limitations; conclusions and implications of key findings; systematic review registration number | Executive summary – pp. ; PROSPERO registration – p. |
| Introduction | |||
| Rationale | 3 | Describe the rationale for the review in the context of what is already known | Chapter 2, background – pp. |
| Objectives | 4 | Provide an explicit statement of questions being addressed with reference to participants, interventions, comparisons, outcomes and study design (PICOS) | Objectives – p. ; inclusion criteria – Table 2, p. |
| Methods | |||
| Protocol and registration | 5 | Indicate if a review protocol exists, if and where it can be accessed (e.g. web address) and, if available, provide registration information including registration number | Protocol (see www.metaxis.com/prospero/full_doc.asp?RecordID=3663); PROSPERO registration number: CRD42013003663 |
| Eligibility criteria | 6 | Specify study characteristics (e.g. PICOS, length of follow-up) and report characteristics (e.g. years considered, language, publication status) used as criteria for eligibility, giving rationale | Table 2 – p. |
| Information sources | 7 | Describe all information sources (e.g. databases with dates of coverage, contact with study authors to identify additional studies) in the search and date last searched | Chapter 3, Search strategy – pp. |
| Search | 8 | Present full electronic search strategy for at least one database, including any limits used, such that it could be repeated | Appendix 1 – pp. |
| Study selection | 9 | State the process for selecting studies (i.e. screening, eligibility, included in systematic review and, if applicable, included in the meta-analysis) | Chapter 3, Inclusion screening and data extraction – p. |
| Data collection process | 10 | Describe method of data extraction from reports (e.g. piloted forms, independently, in duplicate) and any processes for obtaining and confirming data from investigators | Chapter 3, Inclusion screening and data extraction – p. |
| Data items | 11 | List and define all variables for which data were sought (e.g. PICOS, funding sources) and any assumptions and simplifications made | Chapter 3, Inclusion screening and data extraction – p. ; Table 2 – p. |
| Risk of bias in individual studies | 12 | Describe methods used for assessing risk of bias of individual studies (including specification of whether this was carried out at the study or outcome level) and how this information is to be used in any data synthesis | Chapter 3, Quality assessment – p. |
| Summary measures | 13 | State the principal summary measures (e.g. risk ratio, difference in means) | Chapter 3, Methods of analysis/synthesis – pp. |
| Synthesis of results | 14 | Describe the methods of handling data and combining results of studies, if carried out, including measures of consistency (e.g. I2) for each meta-analysis | Chapter 3, Methods of analysis/synthesis – pp. |
| Risk of bias across studies | 15 | Specify any assessment of risk of bias that may affect the cumulative evidence (e.g. publication bias, selective reporting within studies) | NA |
| Additional analyses | 16 | Describe methods of additional analyses (e.g. sensitivity or subgroup analyses, meta-regression), if carried out, indicating which were prespecified | NA |
| Results | |||
| Study selection | 17 | Give numbers of studies screened, assessed for eligibility and included in the review, with reasons for exclusions at each stage, ideally with a flow diagram | Chapter 3, Results of the assessment of clinical effectiveness – pp. ; Figure 1 – p. |
| Study characteristics | 18 | For each study, present characteristics for which data were extracted (e.g. study size, PICOS, follow-up period) and provide the citations | Chapter 3, What is the accuracy of KRAS mutation testing for predicting response to treatment with cetuximab plus standard chemotherapy and subsequent resection rates? – pp. ; Chapter 3, How do outcomes from treatment with cetuximab plus standard chemotherapy vary according to which test is used to select patients for treatment? – pp. ; Appendix 2 – pp. |
| Risk of bias within studies | 19 | Present data on risk of bias of each study and, if available, any outcome-level assessment (see item 12) | Chapter 3, What is the accuracy of KRAS mutation testing for predicting response to treatment with cetuximab plus standard chemotherapy and subsequent resection rates? – p. ; Table 9 – p. ; Chapter 3, How do outcomes from treatment with cetuximab plus standard chemotherapy vary according to which test is used to select patients for treatment? – p. ; Table 12 – p. ; Appendix 3 – pp. |
| Results of individual studies | 20 | For all outcomes considered (benefits or harms), present, for each study: (a) simple summary data for each intervention group and (b) effect estimates and confidence intervals, ideally with a forest plot | Table 8 – p. ; Figure 4 – p. ; Table 11 – p. ; Figures 5–7 – pp. |
| Synthesis of results | 21 | Present results of each meta-analysis carried out, including confidence intervals and measures of consistency | NA |
| Risk of bias across studies | 22 | Present results of any assessment of risk of bias across studies (see item 15) | NA |
| Additional analysis | 23 | Give results of additional analyses, if carried out (e.g. sensitivity or subgroup analyses, meta-regression; see item 16) | NA |
| Discussion | |||
| Summary of evidence | 24 | Summarise the main findings including the strength of evidence for each main outcome; consider their relevance to key groups (e.g. health-care providers, users and policy-makers) | Chapter 5, Statement of principal findings – pp. |
| Limitations | 25 | Discuss limitations at study and outcome level (e.g. risk of bias) and at review level (e.g. incomplete retrieval of identified research, reporting bias) | Chapter 5, Strengths and limitations of the assessment and Uncertainties – pp. |
| Conclusions | 26 | Provide a general interpretation of the results in the context of other evidence, and implications for future research | Chapter 6 – pp. |
| Funding | |||
| Funding | 27 | Describe sources of funding for the systematic review and other support (e.g. supply of data) and the role of funders for the systematic review | NIHR HTA programme, project number 12/75/01 – p. |
Glossary
- Cost-effectiveness analysis
- An economic analysis that converts effects into health terms and describes the costs for additional health gain.
- Decision modelling
- A theoretical construct that allows the comparison of the relationship between costs and outcomes of alternative health-care interventions.
- False negative
- Incorrect negative test result – number of diseased persons with a negative test result.
- False positive
- Incorrect positive test result – number of non-diseased persons with a positive test result.
- Incremental cost-effectiveness ratio
- The difference in the mean costs of two interventions in the population of interest divided by the difference in the mean outcomes in the population of interest.
- Index test
- The test thats performance is being evaluated.
- Markov model
- An analytic method particularly suited to modelling repeated events or the progression of a chronic disease over time.
- Meta-analysis
- Statistical techniques used to combine the results of two or more studies and obtain a combined estimate of effect.
- Meta-regression
- Statistical technique used to explore the relationship between study characteristics and study results.
- Metastasis
- The spread of a disease from one organ or part to another, non-adjacent organ or part.
- Opportunity costs
- The cost of forgone outcomes that could have been achieved through alternative investments.
- Publication bias
- Bias arising from the preferential publication of studies with statistically significant results.
- Quality-adjusted life-year
- A measure of health gain, used in economic evaluations, in which survival duration is weighted or adjusted by the patient’s quality of life during the survival period.
- Quality of life
- An individual’s emotional, social and physical well-being and his or her ability to perform the ordinary tasks of living.
- Receiver operating characteristic curve
- A graph which illustrates the trade-offs between sensitivity and specificity that result from varying the diagnostic threshold.
- Reference standard
- The best currently available diagnostic test against which the index test is compared.
- Sensitivity
- Proportion of people with the target disorder who have a positive test result.
- Specificity
- Proportion of people without the target disorder who have a negative test result.
- True negative
- Correct negative test result – number of non-diseased persons with a negative test result.
- True positive
- Correct positive test result – number of diseased persons with a positive test result.
List of abbreviations
- ARMS
- amplification-refractory mutation system
- ASCO
- American Society of Clinical Oncology
- BRAF
- v-raf murine sarcoma viral oncogene homolog B
- BSC
- best supportive care
- CEAC
- cost-effectiveness acceptability curve
- CI
- confidence interval
- CR
- complete response
- CRC
- colorectal cancer
- CRD
- Centre for Reviews and Dissemination
- CT
- computed tomography
- DNA
- deoxyribonucleic acid
- ECOG
- Eastern Cooperative Oncology Group
- EGFR
- epidermal growth factor receptor
- ESMO
- European Society for Medical Oncology
- FDA
- Food and Drug Administration
- FFPE
- formalin fixed paraffin embedded
- FN
- false negative
- FOLFIRI
- irinotecan in combination with infusional fluorouracil plus folinic acid
- FOLFOX
- oxaliplatin in combination with infusional fluorouracil plus folinic acid
- FP
- false positive
- GERCOR
- Groupe Coopérateur Multidisciplinaire en Oncologie
- HR
- hazard ratio
- HRM
- high-resolution melt (analysis)
- HTA
- Health Technology Assessment
- ICER
- incremental cost-effectiveness ratio
- IQR
- interquartile range
- ITT
- intention to treat
- KRAS
- Kirsten rat sarcoma viral oncogene
- LY
- life-year
- MALDI-TOF
- matrix-assisted laser desorption ionisation time-of-flight
- mCRC
- metastatic colorectal cancer
- MDT
- multidisciplinary team
- MRC
- Medical Research Council
- MRI
- magnetic resonance imaging
- NCIC CO.17
- National Cancer Institute of Canada Clinical Trials Group CO.17
- NEQAS
- National External Quality Assurance Scheme
- NICE
- National Institute for Health and Care Excellence
- OR
- odds ratio
- ORR
- objective response rate
- OS
- overall survival
- PCO
- Provisional Clinical Opinion
- PCR
- polymerase chain reaction
- PD
- progressive disease
- PFS
- progression-free survival
- PR
- partial response
- PRESS-EBC
- Peer Review of Electronic Search Strategies Evidence-Based Checklist
- PSA
- probabilistic sensitivity analysis
- QALY
- quality-adjusted life-year
- RCT
- randomised controlled trial
- RECIST
- Response Evaluation Criteria in Solid Tumours
- SD
- stable disease
- TN
- true negative
- TP
- true positive
- WHO
- World Health Organization
- XELOX
- oxaliplatin and capecitabine
This monograph is based on the Technology Assessment Report produced for the National Institute for Health and Care Excellence (NICE). The full report contained a considerable number of data that were deemed commercial-in-confidence. The full report was used by the Appraisal Committee at NICE in their deliberations. The full report with each piece of commercial-in-confidence data removed and replaced by the statement ‘commercial-in-confidence information (or data) removed’ is available on the NICE website: www.nice.org.uk.
The present monograph presents as full a version of the report as is possible while retaining readability, but some sections, sentences, tables and figures have been removed. Readers should bear in mind that the discussion, conclusions and implications for practice and research are based on all the data considered in the original full NICE report.