Notes
Article history
The contractual start date for this research was in April 2023. This article began editorial review in August 2023 and was accepted for publication in February 2024. The authors have been wholly responsible for all data collection, analysis and interpretation and for writing up their work. The Health and Social Care Delivery Research editors and publisher have tried to ensure the accuracy of the authors’ article and would like to thank the reviewers for their constructive comments on the draft document. However, they do not accept liability for damages or losses arising from material published in this article.
Permissions
Copyright statement
Copyright © 2024 Webster et al. This work was produced by Webster et al. under the terms of a commissioning contract issued by the Secretary of State for Health and Social Care. This is an Open Access publication distributed under the terms of the Creative Commons Attribution CC BY 4.0 licence, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. See: https://creativecommons.org/licenses/by/4.0/. For attribution the title, original author(s), the publication source – NIHR Journals Library, and the DOI of the publication must be cited.
2024 Webster et al.
Background
Rates of acute respiratory infection have increased since the COVID-19 pandemic. 1 In response, NHS England has established new acute respiratory infection (ARI) hubs and ARI virtual wards. 2,3 These are intended to reduce pressure on other parts of the health service by providing care for people with respiratory infections. This review was prepared to inform national guidance on the initial assessment and management of acute respiratory infection in people aged over 16 in England, published by the National Institute for Health and Care Excellence (NICE). 4 The NICE guideline (NG237) has a broader scope than the present review and the reader should refer to that document for recommendations from NICE.
Epidemiology and burden of acute respiratory infections
Acute respiratory infections comprise any infection of the upper or lower respiratory tract, including the nose (common cold), sinuses (sinusitis), middle ear (acute otitis media), larynx (laryngitis) and pharynx (pharyngitis/tonsillitis), as well as the lower airways (acute bronchitis) and lung (pneumonia). They can affect all individuals, but are particularly common in children, older adults and people with pre-existing lung disease. Acute respiratory infections represent a major cause of illness across the UK and worldwide. Estimates from the World Health Organization (WHO) suggest that there were 17.2 billion upper respiratory tract infections5 and 488.9 million cases of lower respiratory tract infection6 globally in 2019, accounting for approximately 2.4 million deaths worldwide. 6 Acute respiratory infections therefore have a high burden on the healthcare system, with significant associated healthcare and societal costs. One study estimated direct annual medical costs associated with acute respiratory infections in the UK at £86M,7 including costs of general practitioner (GP) consultations, prescribed medications and any required hospital admissions. The causes of ARI are varied, but predominantly involve viruses (such as influenza, respiratory syncytial virus, parainfluenza, rhinovirus, adenovirus, coronavirus and human metapneumovirus8) or bacteria (including Streptococcus pneumoniae, Haemophilus influenzae, Moraxella catarrhalis, Mycoplasma pneumoniae, Chlamydia pneumoniae, Staphylococcus aureus, Bordetella pertussis, Gram-negative rods and Legionella9).
Presentation of acute respiratory infections
The symptoms of respiratory infections can vary from relatively mild, self-limiting problems to more severe symptoms requiring urgent assessment and potentially hospital admission. They often include a combination of symptoms including sore throat, runny nose, cough, fever and shortness of breath. Many people with acute respiratory infections will manage their own symptoms without seeking advice from a healthcare professional. Among individuals who present to a professional, distinguishing between those in whom symptoms are likely to resolve without treatment and those in whom symptoms may deteriorate and require intervention is key. Ideally this distinction would be made rapidly, using information available at the time of the consultation, such as readily available symptoms and signs. This could also include a point-of-care diagnostic test.
Diagnostic pathway for suspected acute respiratory infections
Clinical diagnosis of an acute respiratory infection is the norm, based on the typical symptoms and signs of disease. Identification of a specific causative pathogen is frequently not required, especially if symptoms are mild and considered likely to resolve spontaneously. However, it is important to identify people whose symptoms may not resolve without intervention. This includes those with severe symptoms, who may require admission to hospital for escalation of care. It may also include those with a bacterial infection, where symptoms are less likely to be self-limiting and may require antibiotics.
In some instances, a clinical diagnosis may be supplemented with laboratory confirmation of a bacterial or viral infection. These tests can be used as an ‘add-on’ to clinical diagnosis, or to help triage people who may require additional care. Tests could be based on the measurement of substances that fluctuate with the presence of different types of infection. These biological markers (‘biomarkers’) may include proteins produced by the body in response to an infection (such as C-reactive protein, CRP) or levels of certain cell types (including white cell counts). Point-of-care tests measuring these biomarkers are known as ‘host-response’ point-of-care tests. Alternatively, diagnosis may be based on isolation of a specific bacterium or virus, known as ‘microbiological’ point-of-care tests. Identification of the causative pathogen can be challenging, however, because many of the species responsible for infections can be carried as commensal organisms. Consequently, identification of an organism does not definitively mean that this is the cause of the individual’s symptoms, so a false positive test result may be produced. Conversely, there may be low rates of shedding for some pathogens, or the sampling technique may be inadequate. This can lead to false-negative test results. Furthermore, standard microbiological diagnosis often takes too long to influence immediate management in primary care because samples may need to be transported to a central laboratory, and identification of an organism may require culture for several days. Decisions regarding initial treatment are therefore frequently taken without the benefit of a microbiological result. The lack of a ‘gold standard’ diagnostic test to distinguish between bacterial and viral infection means that it can be difficult to diagnose these conditions, and also makes it difficult to assess the accuracy of new tests.
Treatment pathway for suspected acute respiratory infections
The initial treatment of acute respiratory infections is determined by two key features. First, treatment depends on the severity of the symptoms at presentation – including an assessment of whether the individual is unwell enough to require hospital admission, or management in an intermediate care facility (such as a virtual ward). Second, treatment depends on the anticipated prognosis for the illness – with consideration of whether the infection is likely to resolve or deteriorate without intervention. The likely prognosis will depend on features specific to the individual (such as their age and the presence of comorbidities) as well as features of the infection itself (including whether a bacterial or viral cause is suspected).
Despite most acute respiratory infections being caused by viruses, antibiotics are frequently prescribed for these conditions. The reasons for this are multifactorial but may include patient expectations, time pressures, diagnostic uncertainty and concerns about medico-legal consequences of perceived undertreatment. 10,11
Relevant health inequalities
People on lower incomes and with poorer living situations are at higher risk of infectious diseases. 12 In the UK, the incidence of pneumonia in people over 65 is 70% higher in those living in the lowest socioeconomic quintile compared with the highest quintile. 13 These higher rates of ARI are linked to increased rates of domestic damp and mould,14 air pollution,15 functional impairment, unhealthy lifestyles and comorbidities. 16 Therefore, rapid and accurate diagnostic tests that enable early treatment could play a role in reducing the inequalities in morbidity and mortality from ARIs.
People living in deprived areas are also at increased risk of carrying resistant bacteria. 17 Antibiotic prescribing is higher in deprived areas and for people on low incomes. 18,19 This is partly due to the higher incidence of infections. However, clinician antibiotic prescribing is influenced by many factors including uncertainty, fear of negative outcomes and perceived and actual patient expectation. 20,21 This can lead to high antibiotic prescribing becoming the norm in some areas. 22 Tests that reduce diagnostic uncertainty are an important tool to reduce unnecessary prescription of antibiotics in deprived communities, which in turn could contribute to reduced carriage of resistant bacteria.
Aims and objectives
This systematic review aimed to determine the accuracy of the following tests in adults (>16 years) who present in an acute care setting:
-
symptoms and signs to diagnose bacterial respiratory infections
-
rapid, point-of-care tests to diagnose bacterial or viral respiratory infections
-
rapid, point-of-care tests to diagnose influenza and respiratory syncytial virus (RSV).
Methods
Details of the protocol for this systematic review were registered on PROSPERO and can be accessed at https://www.crd.york.ac.uk/prospero/display_record.php?ID=CRD42023427097. There were no changes to the protocol during the review process. The principal approach used was an overview of systematic reviews.
Search strategy
We undertook systematic literature searches to identify published clinical evidence relevant to the review question. Database searches used subject headings, free-text terms and, where appropriate, study design filters. We conducted two main sets of searches, the first to identify systematic reviews of diagnostic test accuracy studies (up to 22 May 2023) and the second to identify primary diagnostic test accuracy studies (up to 6 June 2023), where there were gaps in the available evidence. We searched for systematic reviews in the following databases: MEDLINE, EMBASE, Cochrane Database of Systematic Reviews, NIHR Journals Library and Epistemonikos. We searched for primary studies in MEDLINE and EMBASE. A pragmatic search of the International Trials Registers (ClinicalTrials.gov and the WHO International Clinical Trials Registry Platform) was also conducted but did not yield any relevant results.
The searches were iterative, with the initial search structured around broad, top-level terms for the index tests (rapid point-of-care tests or clinical prediction rules) combined with terms for the target condition or causative agents of respiratory tract infections. Later searches included the addition of relevant host-response biomarkers or named tests (devices), as the retrieval of relevant research evidence evolved. No date restrictions were placed on the searches.
Details of the search strategies (reviews and primary studies) can be found in Appendix 1. Searches for grey literature or unpublished literature were not undertaken.
Eligibility criteria
Systematic reviews that fulfilled the following criteria were eligible for inclusion. Where no systematic reviews were identified, primary studies that fulfilled the same criteria were included.
Population
We included reviews (or primary studies) of participants aged 16 years or over with suspected acute respiratory infection. We included remote settings (such as via telephone or video call) and face-to face settings (e.g. care homes, community pharmacies, primary care, emergency departments or outpatient settings). We excluded reviews or primary studies where more than one quarter of the participants had a diagnosis of COVID-19; were inpatients in hospital; had a respiratory infection during end-of-life care; had aspiration pneumonia, bronchiectasis, cystic fibrosis or known immunosuppression; or had symptoms of otitis media or sinusitis. We also excluded studies where more than one quarter of participants were children (aged <16 years).
Index tests
We assessed index tests that could be used at point of care to distinguish between bacterial and viral respiratory infections. We included index tests used specifically to identify bacterial infections and those used to specifically identify viral infections, as well as tests which may be able to distinguish between the two types of infection. We intended that any included point-of-care tests could be conducted and provide results within 45 minutes or less. However, it should be noted that the duration of the test was often not reported. If the test duration was not explicit, but after investigation it appeared sufficiently close to this time frame (e.g. likely to be feasible within approximately 1 hour) then we included the test in the review.
We included the following tests:
-
Symptoms and signs of acute respiratory infection, which were either assessed individually, or in combination, as part of a clinical decision tool.
-
Biomarker point-of-care tests including the following:
-
CRP
-
procalcitonin
-
a combination of CRP and myxovirus resistance protein A (MxA)
-
a combination of TNF-related apoptosis-induced ligand (TRAIL), interferon-γ-induced protein-10 (IP-10) and CRP
-
white cell differential count
We included other point-of-care tests that had been assessed in published systematic reviews. However, when it was necessary to expand the search to primary studies, we only included the specific tests listed above.
We also included:
-
-
Multiplex or single point-of-care tests for the following viral pathogens:
-
respiratory syncytial virus (RSV)
-
influenza (A or B)
-
We did not include point-of-care tests that aimed specifically to diagnose SARS-CoV-2 or group A streptococcus, because there is existing guidance from NICE on testing for these organisms. When assessing primary studies, we excluded those where the index test had been performed on frozen/stored samples, as we did not consider this to be conducted ‘at point of care’.
Reference standard
We accepted any reference standard that could be used to distinguish viral and bacterial infections, including confirmation of bacterial infection or viral infection through laboratory testing, radiological assessment, expert consensus or a clinical algorithm.
Study design
We primarily included existing systematic reviews. We defined ‘systematic’ reviews as reviews which (1) stated clear and unambiguous eligibility criteria, (2) undertook a comprehensive search (either stated as their aim or implied by use of two or more bibliographic databases), (3) provided details of the included studies (e.g. with a table of characteristics, and references for all included studies) and (4) used tools to assess the validity of primary studies [e.g. Quality Assessment of Diagnostic Accuracy Studies version 2 (QUADAS-2)]. 23
Where no applicable systematic reviews were identified, or where there were evidence gaps (e.g. no evidence on an index test) in the systematic reviews, we conducted searches for diagnostic test accuracy studies. We included diagnostic cross-sectional or diagnostic cohort studies (known as one-gate designs). We excluded diagnostic case-control studies (two-gate designs), which often overestimate accuracy. 24,25
We also excluded studies not published in English, preprints, dissertations and theses, registry entries for ongoing trials, editorials, letters, news items and commentaries, animal studies, conference abstracts and posters.
Screening and inclusion assessment
Titles and abstracts identified by the searches were independently screened by two reviewers [Katie Webster (KW) and Tom Parkhouse (TP)]. We obtained full copies of all reports considered potentially relevant and these were independently assessed for inclusion by two reviewers (KW, TP). Any disagreements were resolved by consensus, or discussion with additional reviewers (Deborah Caldwell, Julian Higgins and Hayley Jones) where necessary.
Assessment of identified systematic reviews
We selected the most robust and up-to-date evidence for each test, determined by consensus decision of two reviewers (TP, KW). Systematic reviews identified in the search were assessed for their applicability to the review question. Where multiple overlapping reviews were identified, we included the most relevant review, considering the comprehensiveness of the search, date of publication and relevance to the current review question. Reviews with largely overlapping scope were not assessed or extracted if the information had been superseded by a more recent publication. We extracted data from relevant analyses reported in systematic reviews that closely matched the review protocol.
Data extraction
Data were extracted using standardised data extraction forms developed in Microsoft Excel. Data extraction forms were piloted on a small sample of papers and adapted as necessary. Data were extracted by one reviewer and checked in detail by a second reviewer. Any disagreements were resolved by consensus.
We collected the following data, where reported: study design (systematic review or diagnostic accuracy study), funding sources (public, industry, mixed), study location and setting, presentation (symptoms), sex, age, inclusion criteria, rapid point-of-care test details (manufacturer, target condition/organism), reference standard test(s).
We collected data from systematic reviews on diagnostic accuracy measures, including sensitivity, specificity or area under the curve (AUC). For primary studies, we extracted data as 2 × 2 tables where possible, comparing the index test with the reference standard. When measures of accuracy (e.g. sensitivity, specificity, AUC) were reported without providing the information needed to calculate 2 × 2 tables, we extracted these data.
Risk-of-bias assessment
We assessed the risk of bias in results of systematic reviews using the Risk of Bias in Systematic Reviews (ROBIS) tool. 26 For additional primary test accuracy studies, we assessed risk of bias and applicability using QUADAS-2. 23 Quality assessment was undertaken by one reviewer and checked by a second reviewer. Any disagreements were resolved by consensus, or through discussion with a third reviewer (Penny Whiting).
Evidence synthesis
Having identified suitable systematic reviews for inclusion, we present an overview of reviews, according to methods reported in the Cochrane Handbook for Systematic Reviews of Interventions. 27 We summarised data reported within the included systematic reviews, including results of analyses presented by the original review authors.
Statistical analysis
For tests where no suitable systematic reviews were identified, we performed meta-analyses of sensitivity and specificity using data from primary diagnostic test accuracy studies. Where at least four studies were available, we fitted bivariate random effects models with binomial likelihoods, using the ‘metandi’ function in Stata version 17 (StataCorp LLC, College Station, Los Angeles, CA, United States). 28–30 Where fewer than four studies were available, univariate meta-analyses of sensitivity and specificity were conducted. Subgroup analyses were performed by device/manufacturer. We use coupled forest plots of sensitivity and specificity, allowing visual assessment of heterogeneity, and summary estimates with 95% confidence intervals (CIs). Study-level and summary results were also plotted in receiver operating characteristic (ROC) space, with 95% confidence ellipses around summary estimates representing the joint uncertainty in sensitivity and specificity. Heterogeneity across studies is quantified using τ2 statistics. These are estimates of the variance across studies of sensitivity and specificity on the log-odds scale. Ninety-five per cent prediction ellipses are also shown on the summary ROC plots.
Analysis of subgroups
We sought data pertaining to the following subgroups of interest: setting of study; age of patients; presence of chronic comorbidity; people who are pregnant/post-partum; and different reference standards.
Interpretation of test accuracy
To aid in the interpretation of results, we identified test accuracy thresholds for sensitivity and specificity that we considered to represent an accurate test (75%) and a very accurate test (90%). We recognise that these thresholds are arbitrary, but used them to assist in the Grading of Recommendations Assessment, Development and Evaluation (GRADE) assessment of the results, and interpretation of the findings.
Assessment of the certainty of the evidence
We performed GRADE31 assessments on all syntheses, both those extracted from systematic reviews and those we undertook ourselves. However, this approach was adapted slightly to accommodate the inclusion of data from systematic reviews. For example, we were unable to determine a rating of inconsistency for many of the analyses reported in a systematic review – as no information on heterogeneity was provided. Consequently, the reported GRADE ratings may overestimate the certainty of the evidence for some outcomes, as this domain was not assessed.
Where possible, we examined the risk-of-bias assessments for the specific studies included in each analysis. Where the majority of studies were rated at unclear or high risk of bias for at least one domain, we downgraded the certainty of evidence. If risk of bias assessments for individual studies were not provided by the review authors, we assessed the studies directly using the QUADAS-2 tool. In some instances it was not possible to determine exactly which studies were included in a specific analysis. Our judgement of risk of bias was then based on the overall set of studies, rather than the specific studies included in each analysis.
Patient and public involvement or community engagement, and involvement
Due to the limited time available, we did not directly involve patients, the public or the community in the review. However, the draft scope for this review was developed by NICE with the input of a guideline committee that included patient and public representatives. In addition, the guideline scope was subject to a consultation and engagement process (https://www.nice.org.uk/guidance/gid-ng10376/documents/draft-scope-comments-and-responses).
Equality, diversity and inclusion
The review team included a representative for equality, diversity and inclusion (Christie Cabral).
Ethics
Ethical approval was not required for this project, as it is a secondary analysis of data already in the public domain.
Results
Systematic reviews
Results of the search
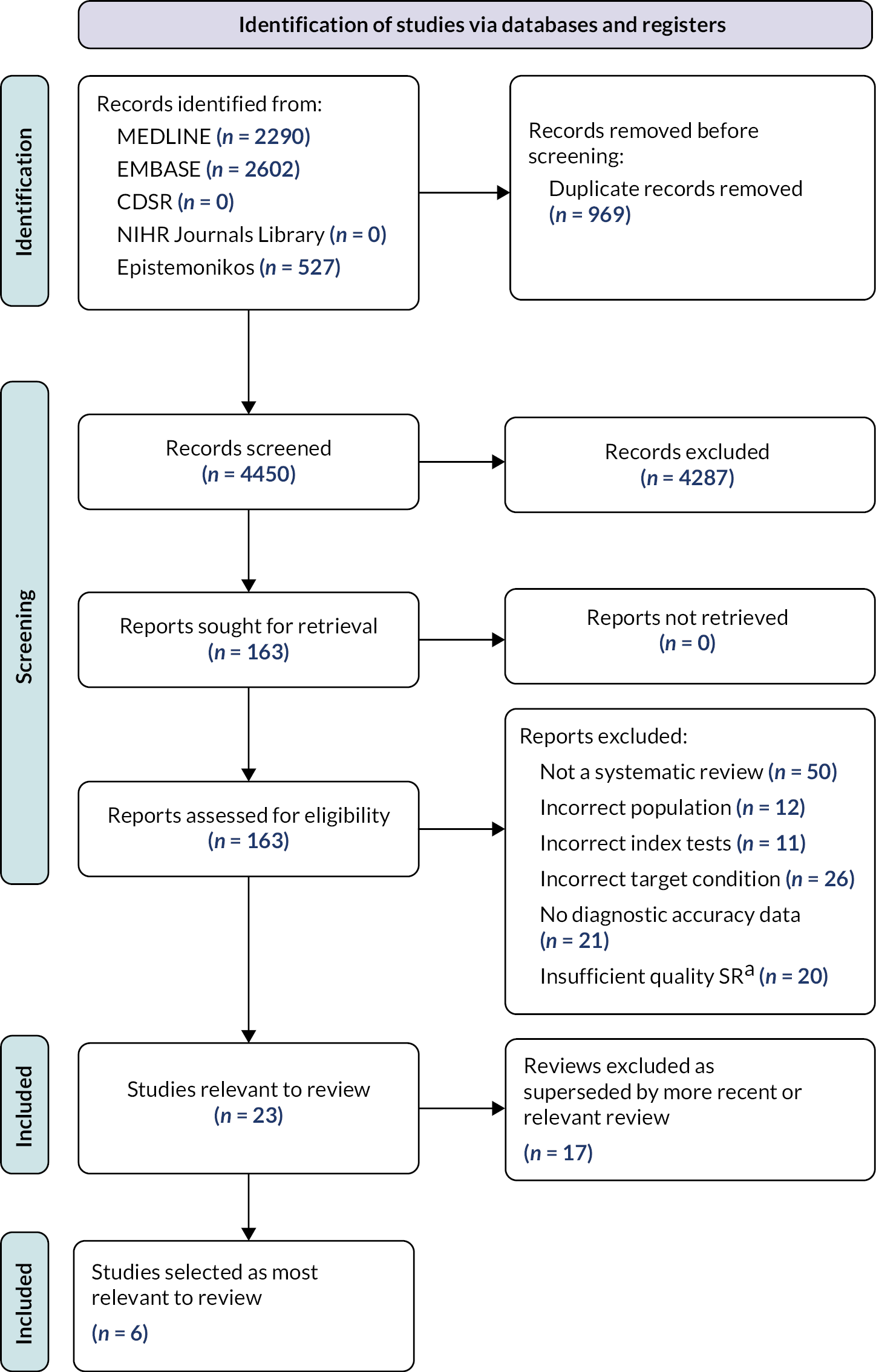
The systematic search for potentially relevant systematic reviews found 4450 references. The full texts of 163 articles were retrieved for closer inspection; 23 of these studies met the inclusion criteria for this review (see Appendix 2, Table 2 for a summary of these studies). The Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) flow diagram is shown in Figure 1.
FIGURE 1.
Identification of relevant systematic reviews. aSystematic review that searched only one database, or did not provide an assessment of methodological quality for included studies.
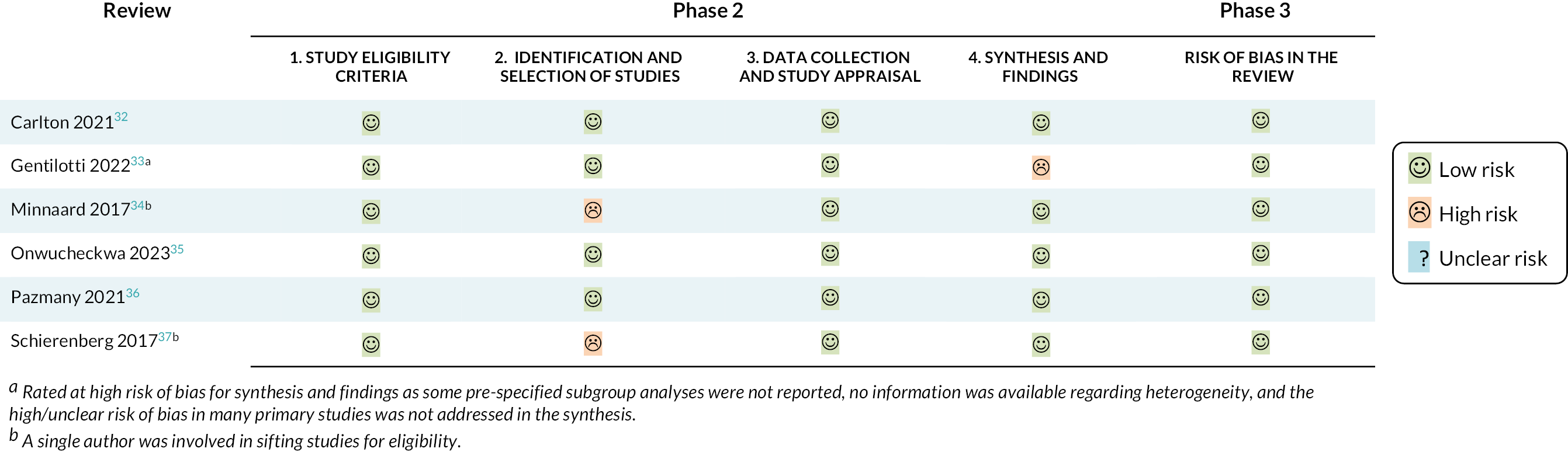
Six relevant systematic reviews were identified as being most aligned with the scope of this overview. Details of these six reviews are reported in Table 1, and the summary risk of bias assessment is shown in Figure 2. Details of all reviews and publications excluded after full text assessment – along with the main reason for exclusion – are given in Report Supplementary Material 1, Table S1.
| Reference | Population | Clinical features | Setting | Target condition assessed | Index tests | Reference standard |
|---|---|---|---|---|---|---|
| Carlton 202132 | Adults and children. Different analyses included different populations | People presenting with symptoms of acute respiratory tract infection | Primary, emergency or secondary care | Bacterial respiratory tract infection and viral respiratory tract infection | Combinations of biomarkers (at least two included). TRAIL, IP-10 and CRP (ImmunoXpert) CRP and MxA (FebriDx) CRP and neopterin |
Any reference standard, including expert consensus, clinical algorithms and microbiology |
| Gentilotti 202233 | Adults and children. Where possible, summary (subgroup) estimates were extracted which relate to adults only | Symptoms consistent with acute respiratory infection | All included studies relating to primary/emergency care settings, including primary care, emergency department, outpatient clinics and long-term care facilities. Where possible, summary (subgroup) estimates were extracted to show the effect in these different settings | Bacterial pneumonia and influenza | Symptoms and signs, host biomarkers (CRP and procalcitonin) and single pathogen tests for influenza | Any reference standard was permitted including X-ray, bacterial or viral culture, PCR, rapid antigen tests, lung ultrasound, composite analyses, expert opinion, microbiological diagnosis and rapid influenza tests |
| Minnaard 201734 | Adults | Suspected lower respiratory tract infection | Primary healthcare, ambulatory care or emergency department settings | Pneumonia | Combination of symptoms and signs plus CRP measurement | Chest X-ray |
| Onwuchekwa 202335 | Adults and children. Extracted data relate to adults only | No information provided | Primary care, emergency care and hospitalised participants | RSV | Direct immunofluorescence and rapid antigen tests | RT PCR |
| Pazmany 202136 | Adults with COPD | Presenting with an acute exacerbation of COPD | Primary care, emergency care and hospitalised participants | Bacterial acute exacerbation of COPD | Presence of purulent sputum | Microbiological culture |
| Schierenberg 201737 | Adults | Immunocompetent adults who self-referred with an acute or worsened cough or lower respiratory tract infection | Primary care, ambulatory care or emergency departments | Pneumonia | Combinations of symptoms and signs (clinical prediction models) | Chest X-ray, CT or MRI |
FIGURE 2.
ROBIS assessment for included systematic reviews. aRated at high risk of bias for synthesis and findings as some prespecified subgroup analyses were not reported, no information was available regarding heterogeneity, and the high/unclear risk of bias in many primary studies was not addressed in the synthesis. bA single author was involved in sifting studies for eligibility.
Symptoms and signs for the diagnosis of bacterial pneumonia
Four recent systematic reviews were identified which assessed the accuracy of individual symptoms and signs or combinations of symptoms and signs in diagnosing bacterial pneumonia. 33,34,36,37 The total number of studies included in each review ranged from 834,37 to 421. 33 The reviews were all considered to be at low risk of bias overall, although we had some concerns regarding the synthesis for one review33 (as the high risk of bias in the primary studies was not addressed in the synthesis, there was no information on heterogeneity and some subgroup analyses were not reported) and some concerns over the identification of studies for two other reviews34,37 (as a single author was involved in sifting studies). Of note, none of the reviews explicitly stated that case-control studies were excluded – the inclusion of such studies may result in overestimates of diagnostic test accuracy.
The reviews included primary studies of participants with symptoms of acute respiratory infection,33 cough or lower respiratory tract infection34,37 or an exacerbation of COPD. 36 One review included both adults and children33 but presented some subgroup data for adults and children. Where possible, we extracted data which related exclusively to adults. All of the studies specifically included participants in appropriate settings (primary, ambulatory or emergency care settings), although one review also included some hospitalised participants. 36 The target condition was pneumonia for two studies,34,37 bacterial pneumonia for one study33 and a bacterial exacerbation of COPD for the final study. 36
Individual symptoms and signs
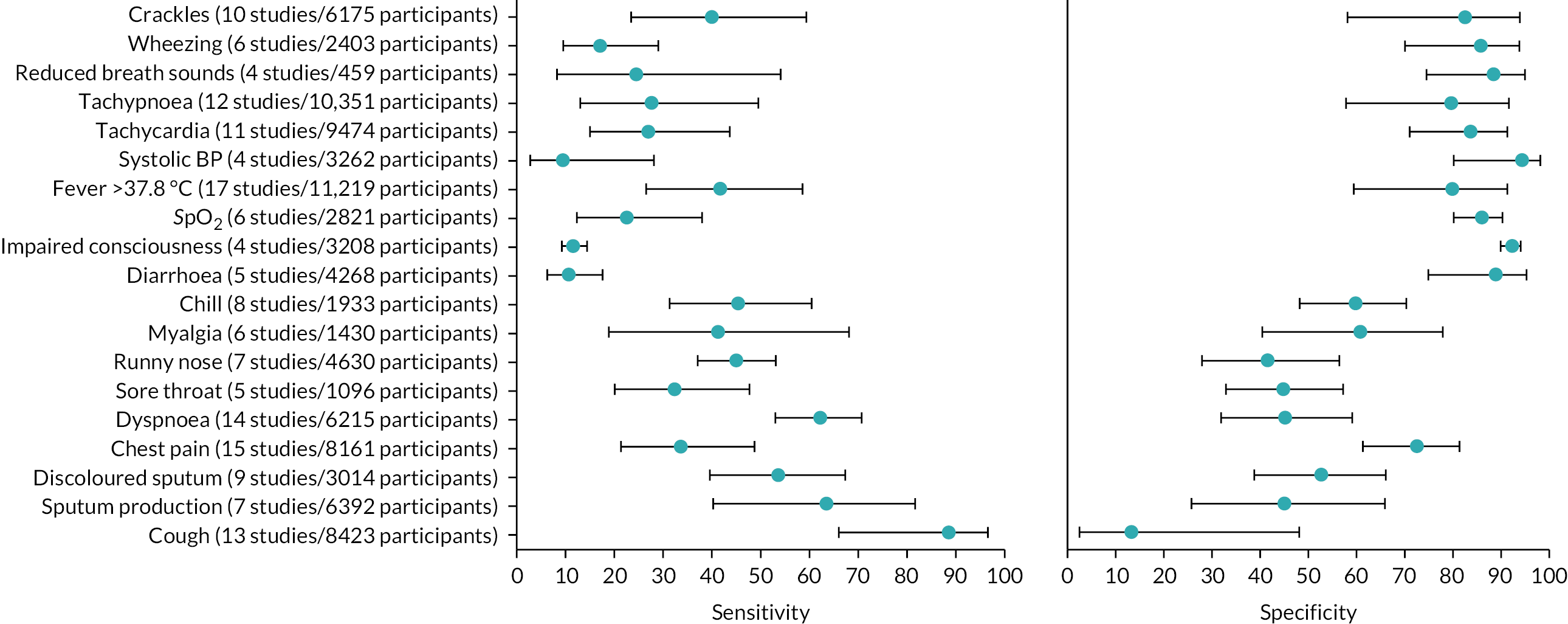
Overall, the estimated accuracy of symptoms and signs in the diagnosis of bacterial pneumonia was poor (Figure 3). Data were available from a single review by Gentilotti and colleagues, for symptoms closely associated with acute respiratory infection (such as cough, sore throat and a runny nose), generic symptoms (including myalgia and diarrhoea) and a variety of clinical signs (including tachycardia, hypotension and low oxygen saturation). 33 The certainty of the evidence ranged from very low to moderate certainty (see Appendix 3, Table 3). Concerns were predominantly due to the risk of bias in the primary studies, and wide confidence intervals that crossed our prespecified thresholds for ‘accurate’ or ‘very accurate’ tests.
FIGURE 3.
Sensitivity and specificity of individual symptoms and signs to diagnose bacterial pneumonia. Pooled estimates from random effects meta-analyses. 33
Subgroup analysis: People with chronic obstructive pulmonary disease
We identified one review which provided some data on the presence of purulent sputum to identify those with bacterial exacerbations of COPD. 36 In keeping with data for the general population, the estimated sensitivity and specificity were poor [71%, 95% CI 42 to 90.3 studies, 259 participants (very-low-certainty evidence due to risk of bias and wide confidence interval) and 51%, 95% CI 30 to 73.3 studies, 259 participants (moderate-certainty evidence due to a risk of bias), respectively, see Appendix 3, Table 3]. We did not identify any additional information on the subgroups of interest in this review.
Combinations of symptoms and signs
Schierenberg’s37 review included an analysis of clinical prediction models used to detect bacterial pneumonia – including combinations of symptoms and signs. Across the six models considered, the area under the curve (AUC) ranged from 0.53 to 0.79. This was considered very-low-certainty evidence due to heterogeneity between the individual estimates which ranged from not useful to useful, and a risk of publication bias (see Appendix 3, Table 3).
The same authors34 performed a separate review which investigated the addition of CRP to the models. When using a combination of CRP, symptoms and signs, the AUC was found to increase by 0.075 (95% CI 0.044 to 0.107). They also reported the accuracy of combinations of symptoms and signs plus CRP for diagnosing bacterial pneumonia at two risk thresholds: 2.5% and 20% (i.e. where individuals with a predicted risk of either ≥ 2.5% or ≥ 20% were classed as having bacterial pneumonia). Across the eight studies included in the review, the lower threshold was estimated to have high sensitivity (97%, 95% CI 95 to 98, moderate-certainty evidence) but the estimated specificity was poor (36%, 95% CI 34 to 37, moderate-certainty evidence). Raising the risk threshold to 20% resulted in much higher estimated specificity (90%, 95% CI 89 to 91, moderate-certainty evidence), but the estimated sensitivity dropped to 70% (95% CI 66 to 73, low-certainty evidence) (see Appendix 3, Table 3).
Biomarker point-of-care tests to detect bacterial or viral respiratory tract infection
Two recent systematic reviews were identified which investigated the use of host biomarkers. 32,33
C-reactive protein
Data were identified from one review. 33 Sensitivity and specificity of CRP to detect bacterial infection varied across the different thresholds assessed. These ranged from 10 mg/l [4 studies, 944 participants; estimated sensitivity 92% (95% CI 56 to 99) very-low-certainty evidence; estimated specificity 43% (95% CI 22 to 66) moderate-certainty evidence] to 100 mg/l [6 studies, 4418 participants; estimated sensitivity 52% (95% CI 31 to 72) moderate-certainty evidence; estimated specificity 91% (95% CI 79 to 97) low-certainty evidence] (see Appendix 3, Table 4).
Procalcitonin
Accuracy of procalcitonin to detect bacterial infection was assessed at three thresholds, by a single review. 33 Test accuracy varied across these thresholds. At > 0.1 mcg/ml the sensitivity and specificity were 74% (95% CI 38% to 93% and 36% to 94%) respectively; 4 studies; 1092 participants; very-low-certainty evidence). At > 0.25 mcg/ml the sensitivity was 44% (95% CI 14% to 79%) and specificity was 89% (95% CI 50% to 98%; 5 studies; 4019 participants; low- and very-low-certainty evidence). At > 0.5 mcg/ml the sensitivity was 44% (95% CI 19% to 73%) and specificity was 93% (95% CI 43% to 100%; 4 studies; 1195 participants; low- and very low-certainty evidence).
TNF-related apoptosis-induced ligand, IP-10 and CRP (ImmunoXpert)
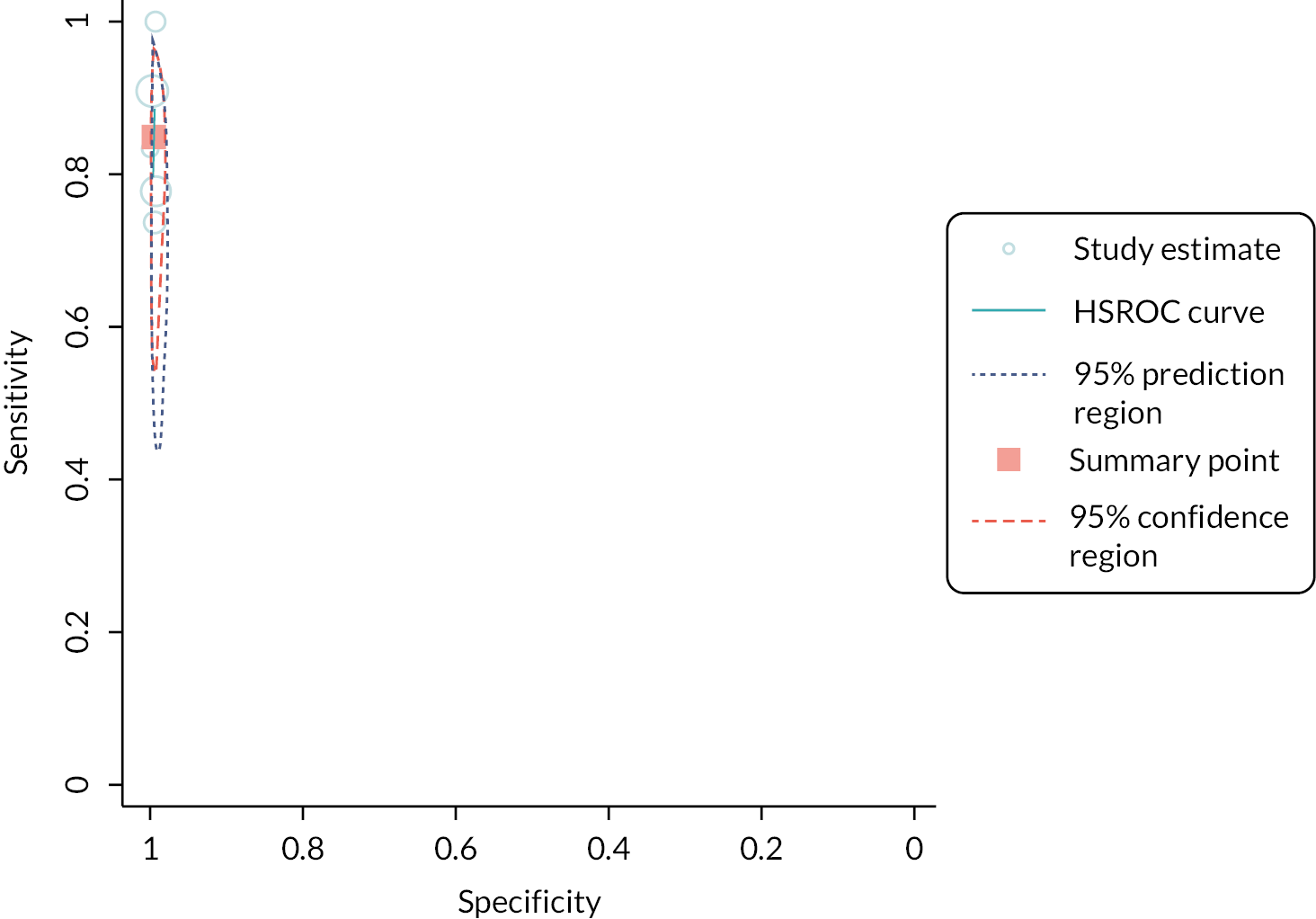
The diagnostic accuracy of ImmunoXpert for bacterial infections had an estimated sensitivity of 85% (95% CI 75% to 91%) and estimated specificity of 86% (95% CI 73% to 93%; 4 studies; 1291 participants). 32 However, the evidence was again considered very low certainty (see Appendix 3, Table 4).
C-reactive protein and MxA (FebriDx)
FebriDx had an estimated sensitivity of 84% (95% CI 75 to 90; low-certainty evidence) and estimated specificity of 93% (95% CI 90 to 95; moderate-certainty evidence; 4 studies; 598 participants)32 (see Appendix 3, Table 4).
Other host biomarkers
The Carlton review32 identified one study (198 participants) which examined the combination of CRP and neopterin to diagnose bacterial infection. This was shown to have an estimated sensitivity of 80% (95% CI 71 to 86) and estimated specificity of 82% (95% CI 71 to 89), though there was very low certainty in the evidence for both estimates (see Appendix 3, Table 4).
Single pathogen tests for influenza and RSV
Influenza
The Gentilotti33 review assessed the accuracy of various single pathogen tests for influenza. However, of the six single pathogen tests included, only immunochromatography had an estimate for an adult-specific population. Estimates for the remaining tests are taken from studies that included both adults and children, and we therefore had serious concerns about indirectness in the GRADE assessment. This should be taken into consideration when interpreting the findings.
Most of the tests that rely on direct antigen detection (immunochromatography, direct immunofluorescence, optical immunoassays and MariPOC) showed adequate sensitivity (ranging from 56% to 82%) and high specificity (range 88–99%), although the certainty of the evidence was considered to be very low or low. This was due to concerns over the potential for bias, indirectness for some estimates (where analyses included children) and wide confidence intervals. Tests using a chemiluminescent neuraminidase assay showed adequate sensitivity (81%, 95% CI 51% to 94%, 787 participants, 4 studies) and specificity (82%, 95% CI 65% to 91%, 787 participants, 4 studies), but the certainty of the evidence was also very low due to the risk of bias and the inclusion of children in the analysis (see Appendix 3, Table 5).
Overall, the diagnostic accuracy of tests based on nucleic acid amplification (both PCR-based and non-PCR-based) appeared higher than those based on antigen detection, with sensitivity ranging from 91% to 95.1% and specificity from 97.5% to 98%. However, the certainty of the evidence was again considered low or very low due to the risk of bias, inclusion of children in the analyses and confidence intervals that crossed our pre-specified threshold for a useful test (90%) (see Appendix 3, Table 5).
Respiratory syncytial virus
A recent systematic review was identified which investigated the use of single pathogen tests for diagnosing RSV. 35 The vast majority of included studies were focused on children. However, we did identify two studies38,39 within this review that considered an adult population, and assessed tests that could be used in a point-of-care setting (direct immunofluorescence and rapid antigen testing). Both of these tests showed high specificity, but poor sensitivity for the detection of RSV, and the certainty of the evidence was low and very low (see Appendix 3, Table 5).
Primary studies
White cell differential count
We identified no systematic reviews that considered white cell differential count for the diagnosis of bacterial respiratory infection. Therefore we undertook a search for primary studies. Four hundred and fifty-five references were identified by the search. We retrieved the full texts of 48 studies for closer inspection, and included four studies (see Appendix 4, Figure 4 for the PRISMA flow diagram, Appendix 4, Table 6 for details of the included studies and Appendix 4, Figure 5 for the QUADAS-2 assessments of included studies). Details of all primary studies excluded at full text, along with the main reason for exclusion are given in the Report Supplementary Material 1, Table S2.
Three studies assessed the accuracy of total white cell count in diagnosing pneumonia,40–42 while the remaining study43 looked at diagnostic accuracy for bacterial pharyngitis. Participants were heterogeneous, and included people with symptoms of lower respiratory tract infection41 and those who already had a diagnosis of community-acquired pneumonia. 42 It should be noted that the results of these studies may not be fully applicable to primary or emergency care settings, as white cell counts were not conducted at point of care and therefore would be unlikely to provide results within 45 minutes. We had additional concerns regarding one study40 that incorporated white cell counts as part of the reference standard, and another study41 that excluded people with more severe illness or malignancy.
Pneumonia
Two studies including a total of 864 participants41,42 reported sensitivity estimates ranging from 10.1% to 71.1%, and specificity estimates ranging from 31.3% to 94.6%, depending on the threshold used (see Appendix 4, Table 6 for further details). One study of 284 participants40 reported an area under the curve of 0.65. The evidence was considered very low certainty (see Appendix 5, Table 7).
Bacterial pharyngitis
A single study of 179 participants43 was identified that looked at the use of white cell count to diagnose bacterial pharyngitis. The study simply reported an AUC of 0.68 (no confidence intervals were reported). This was low-certainty evidence (see Appendix 5, Table 7).
Multiplex tests
We did not identify an existing systematic review addressing multiplex tests that could be used in the point-of-care setting, so we undertook a search for primary studies. Five hundred and eighty-seven references were identified by the search. We retrieved the full texts of 130 studies for closer inspection. Twelve of these studies met the criteria specified in the review protocol. See Appendix 6 for the PRISMA flow diagram (see Appendix 6, Figure 6), details of the included studies (see Appendix 6, Table 8) and QUADAS-2 assessments of the included studies (see Appendix 6, Figure 7). Most of the studies included were considered to be at low risk of bias for at least five of the seven QUADAS-2 domains. The main concerns were regarding the use of an inappropriate reference standard (such as the use of a rapid antigen test, or incorporation of index test results as part of the reference standard), concerns over participant flow and timing (high numbers of excluded participants), and poor applicability of the index test (if samples were not analysed in a point-of-care setting). Details of all primary studies excluded at full text, along with the main reason for exclusion are given in the supplementary information (see Report Supplementary Material 1, Table S3). Twelve diagnostic accuracy studies were identified. All considered the accuracy of the tests to diagnose at least two viruses, including influenza A, influenza B and RSV.
Influenza A
Eight studies, across seven papers (2212 participants), reported on the detection of influenza A. 44–50 The diagnostic accuracy of these tests was very high, with an estimated sensitivity of 98% (95% CI 91% to 100%) and specificity of 99% (95% CI 97% to 99%), respectively. However, the certainty of the evidence was low. Sufficient data were available to analyse two specific multiplex tests separately: Cobas Liat and Xpert Xpress. See Appendix 7, Table 9 for the overall results and GRADE assessment, Appendix 7, Figure 8 for the results of the meta-analysis, and Appendix 7, Figure 9 for the results of the individual studies shown in ROC space.
Influenza B
Six studies, across five papers (1823 participants), assessed detection of influenza B. 44,46,48–50 The pooled estimate for sensitivity was 95% (95% CI 89% to 98%) and for specificity was 99% (95% CI 98% to 99.6%). While potentially useful, the evidence was considered very low and low certainty, respectively. Again, separate analyses were conducted for Cobas Liat and Xpert Xpress. See Appendix 8, Table 10 for the overall results and GRADE assessment, Appendix 8, Figure 10 for the meta-analysis, and Appendix 8, Figure 11 for the results of the individual studies shown in ROC space.
Influenza A and/or B
Seven papers46,48,50–54 reported on the detection of influenza A and/or B, as a combined measure (2162 participants). However, in two of these papers,53,54 the multiplex test of interest was used as the reference standard, not as the index test. As such, we did not include these studies in the analysis, but instead presented the results as the percentage positive agreement and percentage negative agreement between tests (see entries for these studies in Appendix 6, Table 8). We were able to include two further studies44,49 which assessed detection of influenza A and B separately. In total, the analysis included eight studies, across the seven included papers. The pooled estimate for sensitivity was 97% (95% CI 93% to 99%) and for specificity was 97% (95% CI 95% to 98%). Both estimates were considered to be low-certainty evidence. Separate analyses were also conducted for Cobas Liat and Xpert Xpress. See Appendix 9, Table 11 for the overall results and GRADE assessment, Appendix 9, Figure 12 for the meta-analysis, and Appendix 9, Figure 13 for the results of the individual studies shown in ROC space.
Respiratory syncytial virus
Five studies assessed RSV (2273 participants). 44,45,47,49,55 There was moderate-certainty evidence that the specificity of these tests was very high. The pooled estimate was 99.5% (95% CI 99% to 100%). Sensitivity was also relatively high, with a pooled estimate of 85% (95% CI 74% to 92%). However, in this case, the evidence was considered very low certainty, owing to a serious risk of bias in the studies and very serious imprecision. Separate analyses were also conducted for Cobas Liat and Xpert Xpress. See Appendix 10, Table 12 for the overall results and GRADE assessment, Appendix 10, Figure 14 for the meta-analysis, and Appendix 10, Figure 15 for the results of the individual studies shown in ROC space.
Discussion
The evidence identified in this review shows limited diagnostic accuracy for symptoms and signs of bacterial infection and for point-of-care tests that rely on a single biomarker (such as CRP or procalcitonin). Point-of-care tests that include multiple biomarkers may have slightly higher diagnostic accuracy. However, the evidence was predominantly assessed as low or very low certainty, due to limitations which include the risk of bias in primary studies, indirectness of the evidence and imprecision of the effect estimates.
We identified several tests used to diagnose influenza in an adult population, including tests that detect the presence of influenza antigens and those that detect nucleic acids. Diagnostic accuracy appeared highest for nucleic acid amplification tests – either those that test exclusively for influenza or multiplex tests (capable of diagnosing additional pathogens). The evidence was again considered to be predominantly low or very low certainty. The available data on RSV was very limited – the majority of primary studies were conducted in children, and therefore not applicable to this review. Consequently, we are unable to draw conclusions about the accuracy of direct antigen tests for RSV. The specificity of multiplex tests for RSV is probably high. However, the sensitivity may be lower, and the evidence was low certainty.
We used rigorous methods and extensive searches to ensure that all relevant evidence was identified for this review. Nonetheless, the majority of the evidence identified was considered to be low or very low certainty when assessed with the GRADE framework. In part, this was due to concerns over the potential for bias in the primary studies, and some concerns over indirectness in the included populations (where analyses included children, or some participants who were hospitalised). However, many of the concerns were due to imprecision in the effect estimates – as the confidence intervals crossed thresholds that we considered to represent an accurate or very accurate test (taken to be a sensitivity or specificity of 75% and 90%, respectively). We acknowledge that these thresholds are arbitrary and that readers, or different authors, may consider different thresholds to represent a useful test. This would impact on the certainty in the estimates. Furthermore, we noted that most systematic reviews did not report any information on heterogeneity in the primary studies included in their analyses. Consequently, we were unable to assess inconsistency when applying GRADE, and our assessment of evidence certainty may be considered optimistic.
In this review we primarily sought evidence about the accuracy of tests to distinguish between viral and bacterial causes of ARI that take no more than 45 minutes to yield a result. Symptoms and signs of infection are part of routine clinical assessment and therefore would add no time to the decision-making process. Many of the diagnostic tests identified give results within 10 to 15 minutes, making them suitable for use in a primary care setting or emergency department. However, multiplex tests typically require more time, with many taking up to 1 hour or longer. The extent to which such tests would fit into routine clinical practice needs careful consideration. However, the clear benefit of the multiplex platforms is the possibility of testing a single sample for multiple viral and bacterial pathogens, as well as the apparent increase in diagnostic accuracy.
It should be noted that most of the evidence identified in this review looked at the diagnostic accuracy of tests in isolation – that is the accuracy of a single test to determine the cause of a respiratory infection. In reality, clinical diagnosis involves assessment of a constellation of symptoms and signs, as well as the results of specific tests. We identified only one review that assessed the incremental benefit of assessing CRP in conjunction with symptoms and signs of infection. 34 The addition of CRP showed a small increase in diagnostic accuracy as compared to symptoms and signs alone. Due to a lack of published evidence, it is currently unclear whether this is also the case for tests that examine other biomarkers.
Symptoms and signs of respiratory infection are often used to determine eligibility for studies of test accuracy. For example, many studies will enrol individuals with a fever or cough for further testing. Consequently, it is possible that estimates of accuracy for individual symptoms and signs could be artificially high – as the prevalence of these symptoms is high in the study population. Nonetheless, this situation does reflect routine clinical practice, where healthcare professionals are using these features to determine who requires further testing.
We accepted any reference standard to diagnose bacterial infection. This was partly because there is no agreement on what constitutes an ideal reference test. Microbiological testing may be regarded as an essential component of determining a viral or bacterial cause of an infection. However, these tests are likely to detect the presence of commensal organisms and are known to produce false-negative results (due to inadequacy of sampling technique or culture methods). 56,57 Consequently, a variety of reference standards were used in the studies included in this review – ranging from radiological imaging, microbiological assessment (such as culture and/or PCR) and consensus opinion of an expert panel.
For some tests, there were similarities between the index test and the reference standard used. In particular, a number of pathogen-specific tests used PCR techniques as both the index test and the reference standard. Given the similar methods used, the results of these tests are likely to be correlated, and therefore the accuracy of the index tests may be overestimated. 58
Cost-effectiveness was not assessed as part of this review. However, the cost of different types of tests varies and some may be prohibitively expensive for use in a primary care setting. This will need careful consideration before implementing a new testing strategy.
This review focused only on the diagnostic accuracy of tests. For patients and clinicians, the most important questions are likely to be about the impact of using these tests on health outcomes. For example, does testing for bacterial infections result in better health than relying on clinical judgement alone? Will more people avoid side effects from the prescription of unnecessary antibiotics? Will hospital admissions be reduced, or people suffer fewer complications from severe bacterial infections? Assessing these outcomes requires studies that consider the implementation of these tests followed by clinical management based on the test results.
It is recognised that prescription and use of antibiotics may be affected by factors commonly associated with health inequalities, such as age and ethnicity. 59 Testing to help determine who needs antibiotics could help to reduce these inequities in health care, but only if the tests themselves are used appropriately. CRP is one of the only tests for which there is evidence in relation to equity of use. Despite their limited diagnostic accuracy, the use of CRP tests for people with ARI may reduce antibiotic prescribing without increasing negative health outcomes,60 in part because they may enable clinicians to communicate a ‘no antibiotic’ treatment decision more easily. 59,61 A study from Denmark (where CRP tests are widely used) found that clinicians were less likely to use a CRP test when prescribing antibiotics for those who were unemployed or receiving disability pension, immigrants or children of immigrants. 62 It is not clear why this happens, but consequently these groups may still be more likely to be prescribed unnecessary antibiotics. For these tests to help address (rather than reproduce) inequities, there needs to be clear guidance and monitoring of use with respect to underserved groups.
At present there is an absence of evidence regarding diagnostic accuracy to support current clinical practice (where symptoms and signs are used to diagnose bacterial infection) or to justify the introduction of microbiological or host-response point-of-care tests. Policy makers should resist seeing point-of-care tests as the ‘silver bullet’ to solve healthcare system pressures until there is adequate evidence to demonstrate they are safe, clinically effective and cost-effective. There are concerns that introduction of point-of-care tests may unintentionally increase healthcare demand, as patients’ illnesses become ‘medicalised’ with attendances for testing.
We recommend further research to define an adequate reference standard for respiratory infection diagnosis. This could be based on a better understanding of the natural history of the microbiology, and/or prognosis, of infections. In addition, it should be established whether point-of-care tests add diagnostic value over and above current practice – the use of symptoms and signs to identify individuals at risk of more severe illness, or who require additional treatment. There is a lack of high-quality evidence regarding the diagnostic accuracy of point-of-care tests for ARI in the community and emergency department setting. The diagnostic accuracy of such point-of-care tests should be assessed specifically in this setting where the population is different – with generally less severe infections and consequently different microbiology and immune responses. This means that data from inpatients cannot necessarily be extrapolated to the outpatient setting, as the diagnostic accuracy of the test may vary according to disease severity. Finally, it will be important to assess if the use of point-of-care tests will medicalise illness and lead to unintended increased demand for NHS care for ARIs.
Conclusion
The majority of the evidence identified in this review was considered to be low or very low certainty, highlighting that future studies may change the overall estimates of accuracy. Nonetheless, from the evidence identified in this review it appears that individual symptoms and signs, or existing clinical prediction models (incorporating multiple symptoms and signs) are unlikely to be sufficiently accurate to distinguish between bacterial and viral infections. Diagnostic accuracy of individual host biomarkers also appears to be insufficient, although certain combinations of biomarkers may have higher sensitivity and specificity. As may be expected, the accuracy of different types of rapid tests for influenza and RSV varied. The highest diagnostic accuracy was seen with tests that rely on amplification of viral nucleic acid (including PCR and non-PCR-based techniques).
Further work is required to determine the optimum reference standard, and whether the introduction of point-of-care tests may add value to current diagnostic pathways. It remains to be seen whether additional testing would improve health outcomes for patients, or simply lead to an increase in healthcare consultations and resource costs.
Additional information
CRediT contribution statement
Katie E Webster (https://orcid.org/0009-0002-7997-4133): Data curation, Formal analysis, Investigation, Methodology, Project administration, Visualisation, Writing – Original draft, Writing – reviewing and editing. Tom Parkhouse (https://orcid.org/0000-0001-6773-5376): Data curation, Formal analysis, Investigation, Methodology, Visualisation, Writing – Original draft, Writing – reviewing and editing. Sarah Dawson (https://orcid.org/0000-0002-6682-063X): Investigation, Methodology, Writing – reviewing and editing, Other contributions (searches). Hayley E Jones (https://orcid.org/0000-0002-4265-2854): Formal analysis, Investigation, Methodology, Visualisation, Writing – reviewing and editing. Emily L Brown (https://orcid.org/0000-0002-8232-1769): Investigation, Writing – reviewing and editing. Alastair D Hay (https://orcid.org/0000-0003-3012-375X): Investigation, Writing – reviewing and editing. Penny Whiting (https://orcid.org/0000-0003-1138-5682): Investigation, Methodology, Writing – reviewing and editing. Christie Cabral (https://orcid.org/0000-0002-9884-0555): Investigation, Writing – reviewing and editing. Deborah M Caldwell (https://orcid.org/0000-0001-8014-7480): Funding acquisition, Investigation, Methodology, Project administration, Supervision, Writing – reviewing and editing. Julian PT Higgins (https://orcid.org/0000-0002-8323-2514): Funding acquisition, Investigation, Methodology, Project administration, Supervision, Writing – reviewing and editing.
Acknowledgements
This review was based on a draft protocol developed by NICE.
The authors thank Elisa Gentilotti and Anna Gorska for providing additional data from their published systematic review.
Data-sharing statement
This is secondary research and therefore the data generated are not suitable for sharing beyond that contained within the manuscript. Further information can be obtained from the corresponding author.
Ethics statement
Ethical approval was not required for this secondary research project.
Information governance statement
No identifiable data were used as part of this review.
Disclosure of interests
Full disclosure of interests: Completed ICMJE forms for all authors, including all related interests, are available in the toolkit on the NIHR Journals Library report publication page at https://doi.org/10.3310/JLCP4570.
Primary conflicts of interest: Alastair D Hay is Chief Investigator, and Emily L Brown is a study team member and member of the Trial Management Group of the NIHR-funded RAPID-TEST RCT (https://fundingawards.nihr.ac.uk/award/NIHR131758) which is investigating bioMérieux’s BioFire® rapid microbiological point-of-care test.
Alastair D Hay is a committee member of the EME-Funding committee 2020–24, HTA CET committee.
No other authors have any competing interests to declare.
Department of Health and Social Care disclaimer
This publication presents independent research commissioned by the National Institute for Health and Care Research (NIHR). The views and opinions expressed by authors in this publication are those of the authors and do not necessarily reflect those of the NHS, the NIHR, MRC, NIHR Coordinating Centre, the Health Technology Assessment programme or the Department of Health and Social Care.
This research article was published based on current knowledge at the time and date of publication. NIHR is committed to being inclusive and will continually monitor best practice and guidance in relation to terminology and language to ensure that we remain relevant to our stakeholders.
Study registration
The main study protocol was registered as PROSPERO CRD42023427097. https://www.crd.york.ac.uk/prospero/display_record.php?ID=CRD42023427097
Funding
This article presents independent research funded by the National Institute for Health and Care Research (NIHR) Health Technology Assessment (HTA) programme as award number NIHR159948.
This article reports on one component of the research award Diagnostic accuracy of point-of-care tests for acute respiratory infection: a systematic review of reviews. For more information about this research please view the award page [https://fundingawards.nihr.ac.uk/award/NIHR159948].
About this article
The contractual start date for this research was in April 2023. This article began editorial review in August 2023 and was accepted for publication in February 2024. The authors have been wholly responsible for all data collection, analysis and interpretation and for writing up their work. The Health and Social Care Delivery Research editors and publisher have tried to ensure the accuracy of the authors’ article and would like to thank the reviewers for their constructive comments on the draft document. However, they do not accept liability for damages or losses arising from material published in this article.
Copyright
Copyright © 2024 Webster et al. This work was produced by Webster et al. under the terms of a commissioning contract issued by the Secretary of State for Health and Social Care. This is an Open Access publication distributed under the terms of the Creative Commons Attribution CC BY 4.0 licence, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. See: https://creativecommons.org/licenses/by/4.0/. For attribution the title, original author(s), the publication source – NIHR Journals Library, and the DOI of the publication must be cited.
List of abbreviations
- AECOPD
- acute exacerbation of chronic obstructive pulmonary disease
- ARI
- acute respiratory infection
- AUC
- area under the curve
- BP
- blood pressure
- CDSR
- Cochrane Database of Systematic Reviews
- CI
- confidence interval
- COPD
- chronic obstructive pulmonary disease
- CRP
- C-reactive protein
- CT
- computed tomography
- DFA
- direct fluorescence antigen
- DTA
- diagnostic test accuracy
- EU
- European Union
- GRADE
- Grading of Recommendations Assessment, Development and Evaluation
- HSROC
- hierarchical summary receiver operating characteristic
- ICU
- intensive care unit
- IP-10
- interferon-γ-induced protein-10
- IQR
- interquartile range
- LRTI
- lower respiratory tract infection
- mPCR
- multiplex polymerase chain reaction
- MRI
- magnetic resonance imaging
- MxA
- myxovirus resistance protein A
- NAAT
- nucleic acid amplification test
- NICE
- National Institute for Health and Care Excellence
- PCR
- polymerase chain reaction
- POCT
- point-of-care test
- PRISMA
- Preferred Reporting Items for Systematic Reviews and Meta-Analyses
- QUADAS-2
- Quality Assessment of Diagnostic Accuracy Studies version 2
- RADT
- rapid antigen detection tests
- RFT
- rapid flu test
- RIDT
- rapid influenza diagnostic test
- ROBIS
- Risk of Bias in Systematic Reviews
- ROC
- receiver operating characteristic
- RSV
- respiratory syncytial virus
- RTI
- respiratory tract infection
- RT PCR
- real-time polymerase chain reaction
- SD
- standard deviation
- SpO2
- oxygen saturations
- TRAIL
- TNF-related apoptosis-induced ligand
- URTI
- upper respiratory tract infection
- WHO
- World Health Organization
Notes
Supplementary material can be found on the NIHR Journals Library report page (https://doi.org/10.3310/JLCP4570).
Supplementary material has been provided by the authors to support the report and any files provided at submission will have been seen by peer reviewers, but not extensively reviewed. Any supplementary material provided at a later stage in the process may not have been peer reviewed.
References
- UK Health Security Agency . Surveillance of Influenza and Other Seasonal Respiratory Viruses in the UK, Winter 2022 to 2023 2023. Surveillance of influenza and other seasonal respiratory viruses in winter 2021 to 2022 - GOV.UK (www.gov.uk) (accessed 27 July 2023).
- NHS England . Combined Adult and Paediatric Acute Respiratory Infection (ARI) Hubs (Previously RCAS Hubs) 2022. BW2064-combined-adult-paediatric-ari-hubs-october-22.pdf (england.nhs.uk) (accessed 27 July 2023).
- NHS England . Guidance Note: Acute Respiratory Infection Virtual Ward 2022. NHS England » Guidance note: Acute respiratory infection virtual ward (accessed 27 July 2023).
- National Institute for Health and Care Excellence . Suspected Acute Respiratory Infection in Over 16s: Assessment at First Presentation and Initial Management. NICE 2023. Suspected acute respiratory infection in over 16s: assessment at first presentation and initial management (nice.org.uk) (accessed 12 December 2023).
- Jin X, Ren J, Li R, Gao Y, Zhang H, Li J, et al. Global burden of upper respiratory infections in 204 countries and territories, from 1990 to 2019. EClinicalMedicine 2021;37. https://doi.org/10.1016/j.eclinm.2021.100986.
- Safiri S, Mahmoodpoor A, Kolahi AA, Nejadghaderi SA, Sullman MJM, Mansournia MA, et al. Global burden of lower respiratory infections during the last three decades. Front Public Health 2022;10. https://doi.org/10.3389/fpubh.2022.1028525.
- Meier GC, Watkins J, McEwan P, Pockett RD. Resource use and direct medical costs of acute respiratory illness in the UK based on linked primary and secondary care records from 2001 to 2009. PLOS ONE 2020;15. https://doi.org/10.1371/journal.pone.0236472.
- Creer DD, Dilworth JP, Gillespie SH, Johnston AR, Johnston SL, Ling C, et al. Aetiological role of viral and bacterial infections in acute adult lower respiratory tract infection (LRTI) in primary care. Thorax 2006;61:75-9. https://doi.org/10.1136/thx.2004.027441.
- Musher DM, Abers MS, Bartlett JG. Evolving understanding of the causes of pneumonia in adults, with special attention to the role of pneumococcus. Clin Infect Dis 2017;65:1736-44. https://doi.org/10.1093/cid/cix549.
- Fletcher-Lartey S, Yee M, Gaarslev C, Khan R. Why do general practitioners prescribe antibiotics for upper respiratory tract infections to meet patient expectations: a mixed methods study. BMJ Open 2016;6. https://doi.org/10.1136/bmjopen-2016-012244.
- O’Connor R, O’Doherty J, O’Regan A, Dunne C. Antibiotic use for acute respiratory tract infections (ARTI) in primary care; what factors affect prescribing and why is it important? A narrative review. Ir J Med Sci 2018;187:969-86. https://doi.org/10.1007/s11845-018-1774-5.
- Ayorinde A, Ghosh I, Ali I, Zahair I, Olarewaju O, Singh M, et al. Health inequalities in infectious diseases: a systematic overview of reviews. BMJ Open 2023;13. https://doi.org/10.1136/bmjopen-2022-067429.
- Millett ER, Quint JK, Smeeth L, Daniel RM, Thomas SL. Incidence of community-acquired lower respiratory tract infections and pneumonia among older adults in the United Kingdom: a population-based study. PLOS ONE 2013;8. https://doi.org/10.1371/journal.pone.0075131.
- Fisk WJ, Eliseeva EA, Mendell MJ. Association of residential dampness and mold with respiratory tract infections and bronchitis: a meta-analysis. Environ Health 2010;9. https://doi.org/10.1186/1476-069X-9-72.
- MacIntyre EA, Gehring U, Molter A, Fuertes E, Klumper C, Kramer U, et al. Air pollution and respiratory infections during early childhood: an analysis of 10 European birth cohorts within the ESCAPE Project. Environ Health Perspect 2014;122:107-13. https://doi.org/10.1289/ehp.1306755.
- Almirall J, Serra-Prat M, Bolíbar I, Balasso V. Risk factors for community-acquired pneumonia in adults: a systematic review of observational studies. Respiration 2017;94:299-311. https://doi.org/10.1159/000479089.
- Nomamiukor BO, Horner C, Kirby A, Hughes GJ. Living conditions are associated with increased antibiotic resistance in community isolates of Escherichia coli. J Antimicrob Chemother 2015;70:3154-8. https://doi.org/10.1093/jac/dkv229.
- Covvey JR, Johnson BF, Elliott V, Malcolm W, Mullen AB. An association between socioeconomic deprivation and primary care antibiotic prescribing in Scotland. J Antimicrob Chemother 2014;69:835-41. https://doi.org/10.1093/jac/dkt439.
- Thomson K, Berry R, Robinson T, Brown H, Bambra C, Todd A. An examination of trends in antibiotic prescribing in primary care and the association with area-level deprivation in England. BMC Public Health 2020;20. https://doi.org/10.1186/s12889-020-09227-x.
- Rose J, Crosbie M, Stewart A. A qualitative literature review exploring the drivers influencing antibiotic over-prescribing by GPs in primary care and recommendations to reduce unnecessary prescribing. Perspect Public Health 2021;141:19-27. https://doi.org/10.1177/1757913919879183.
- Cabral C, Lucas PJ, Ingram J, Hay AD, Horwood J. It’s safer to …’ parent consulting and clinician antibiotic prescribing decisions for children with respiratory tract infections: an analysis across four qualitative studies. Soc Sci Med 2015;136-137:156-64. https://doi.org/10.1016/j.socscimed.2015.05.027.
- Wang KY, Seed P, Schofield P, Ibrahim S, Ashworth M. Which practices are high antibiotic prescribers? A cross-sectional analysis. Br J Gen Pract 2009;59:e315-20. https://doi.org/10.3399/bjgp09X472593.
- Whiting P, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med 2011;155:529-36. https://doi.org/10.7326/0003-4819-155-8-201110180-00009.
- Whiting PF, Rutjes AW, Westwood ME, Mallett S, Group Q-S. A systematic review classifies sources of bias and variation in diagnostic test accuracy studies. J Clin Epidemiol 2013;66:1093-104. https://doi.org/10.1016/j.jclinepi.2013.05.014.
- Lijmer JG, Mol BW, Heisterkamp S, Bonsel GJ, Prins MH, van der Meulen JHP, et al. Empirical evidence of design-related bias in studies of diagnostic tests. JAMA 1999;282:1061-6. https://doi.org/10.1001/jama.282.11.1061.
- Whiting P, Savovic J, Higgins JPT, Caldwell DM, Reeves BC, Shea B, et al. ROBIS Group . ROBIS: a new tool to assess risk of bias in systematic reviews was developed. J Clin Epidemiol 2016;69:225-34. https://doi.org/10.1016/j.jclinepi.2015.06.005.
- Pollock M, Fernandes RM, Becker LA, Pieper D, Hartling L, . Cochrane Handbook for Systematic Reviews of Interventions. Version 6.3 (updated February 2022). Cochrane; 2022.
- Chu H, Cole SR. Bivariate meta-analysis of sensitivity and specificity with sparse data: a generalized linear mixed model approach. J Clin Epidemiol 2006;59:1331-2. https://doi.org/10.1016/j.jclinepi.2006.06.011.
- Reitsma JB, Glas AS, Rutjes AW, Scholten RJ, Bossuyt PM, Zwinderman AH. Bivariate analysis of sensitivity and specificity produces informative summary measures in diagnostic reviews. J Clin Epidemiol 2005;58:982-90. https://doi.org/10.1016/j.jclinepi.2005.02.022.
- Harbord RM, Whiting P. Metandi: meta-analysis of diagnostic accuracy using hierarchical logistic regression. Stata J 2009;9:211-29. https://doi.org/10.1177/1536867X0900900203.
- Guyatt GH, Oxman AD, Vist GE, Kunz R, Falck-Ytter Y, Alonso-Coello P, et al. GRADE Working Group . GRADE: an emerging consensus on rating quality of evidence and strength of recommendations. BMJ 2008;336:924-6. https://doi.org/10.1136/bmj.39489.470347.AD.
- Carlton HC, Savovic J, Dawson S, Mitchelmore PJ, Elwenspoek MMC. Novel point-of-care biomarker combination tests to differentiate acute bacterial from viral respiratory tract infections to guide antibiotic prescribing: a systematic review. Clin Microbiol Infect 2021;27:1096-108. https://doi.org/10.1016/j.cmi.2021.05.018.
- Gentilotti E, De Nardo P, Cremonini E, Gorska A, Mazzaferri F, Canziani LM, et al. Diagnostic accuracy of point-of-care tests in acute community-acquired lower respiratory tract infections. A systematic review and meta-analysis. Clin Microbiol Infect 2022;28:13-22. https://doi.org/10.1016/j.cmi.2021.09.025.
- Minnaard MC, de Groot JAH, Hopstaken RM, Schierenberg A, de Wit NJ, Reitsma JB, et al. The added value of C-reactive protein measurement in diagnosing pneumonia in primary care: a meta-analysis of individual patient data. CMAJ 2017;189:E56-63. https://doi.org/10.1503/cmaj.151163.
- Onwuchekwa C, Moreo LM, Menon S, Machado B, Curcio D, Kalina W, et al. Under-ascertainment of Respiratory Syncytial Virus infection in adults due to diagnostic testing limitations: a systematic literature review and meta-analysis. J Infect Dis 2023;228:173-84. https://doi.org/10.1093/infdis/jiad012.
- Pazmany P, Soos A, Hegyi P, Dohos D, Kiss S, Szakacs Z, et al. Inflammatory biomarkers are inaccurate indicators of bacterial infection on admission in patients with acute exacerbation of chronic obstructive pulmonary disease-A systematic review and diagnostic accuracy network meta-analysis. Front Med (Lausanne) 2021;8. https://doi.org/10.3389/fmed.2021.639794.
- Schierenberg A, Minnaard MC, Hopstaken RM, van de Pol AC, Broekhuizen BD, de Wit NJ, et al. External validation of prediction models for pneumonia in primary care patients with lower respiratory tract infection: an individual patient data meta-analysis. PLOS ONE 2016;11. https://doi.org/10.1371/journal.pone.0149895.
- Franck KT, Schneider UV, Ma CMG, Knudsen D, Lisby G. Evaluation of immuview RSV antigen test (SSI siagnostica) and BinaxNOW RSV card (alere) for rapid detection of respiratory syncytial virus in retrospectively and prospectively collected respiratory samples. J Med Virol 2020;92:2992-8. https://doi.org/10.1002/jmv.26369.
- Simpson JL, Moric I, Wark PA, Johnston SL, Gibson PG. Use of induced sputum for the diagnosis of influenza and infections in asthma: a comparison of diagnostic techniques. J Clin Virol 2003;26:339-46. https://doi.org/10.1016/s1386-6532(02)00084-7.
- Castro-Guardiola A, Armengou-Arxé A, Viejo-Rodríguez A, Peñarroja-Matutano G, Garcia-Bragado F. Differential diagnosis between community-acquired pneumonia and non-pneumonia diseases of the chest in the emergency ward. Eur J Intern Med 2000;11:334-9. https://doi.org/10.1016/S0953-6205(00)00118-7.
- Holm A, Nexoe J, Bistrup LA, Pedersen SS, Obel N, Nielsen LP, et al. Aetiology and prediction of pneumonia in lower respiratory tract infection in primary care. Br J Gen Pract 2007;57:547-54.
- Liu YF, Gao Y, Chen MF, Cao B, Yang XH, Wei L. Etiological analysis and predictive diagnostic model building of community-acquired pneumonia in adult outpatients in Beijing, China. BMC Infect Dis 2013;13:1-10. https://doi.org/10.1186/1471-2334-13-309.
- Gulich MS, Matschiner A, Glück R, Zeitler HP. Improving diagnostic accuracy of bacterial pharyngitis by near patient measurement of C-reactive protein (CRP). Br J Gen Pract 1999;49:119-21.
- Escarate E, Jones CG, Clarke E, Clark P, Norton S, Bag S, et al. Rapid on-site molecular Point of Care Testing during influenza outbreaks in aged care facilities improves antiviral use and reduces hospitalisation. Aust N Z J Public Health 2022;46:884-8. https://doi.org/10.1111/1753-6405.13307.
- Farfour E, Yung T, Baudoin R, Vasse M. Evaluation of four fully integrated molecular assays for the detection of respiratory viruses during the co-circulation of SARS-CoV-2, influenza and RSV. J Clin Med 2022;11. https://doi.org/10.3390/jcm11143942.
- Maignan M, Viglino D, Hablot M, Termoz Masson N, Lebeugle A, Collomb Muret R, et al. Diagnostic accuracy of a rapid RT-PCR assay for point-of-care detection of influenza A/B virus at emergency department admission: a prospective evaluation during the 2017/2018 influenza season. PLOS ONE 2019;14. https://doi.org/10.1371/journal.pone.0216308.
- Morris TC, Bird PW, Horvath-Papp E, Dhillon JK, May S, Tang JW. Xpert Xpress Flu/RSV: validation and impact evaluation at a large UK hospital trust. J Med Virol 2021;93:5146-51. https://doi.org/10.1002/jmv.26860.
- Valentin T, Kieslinger P, Stelzl E, Santner BI, Groselj-Strele A, Kessler HH, et al. Prospective evaluation of three rapid molecular tests for seasonal influenza in patients presenting at an emergency unit. J Clin Virol 2019;111:29-32. https://doi.org/10.1016/j.jcv.2019.01.003.
- Yin N, Van Nuffelen M, Bartiaux M, Preseau T, Roggen I, Delaunoy S, et al. Clinical impact of the rapid molecular detection of RSV and influenza A and B viruses in the emergency department. PLOS ONE 2022;17. https://doi.org/10.1371/journal.pone.0274222.
- Youngs J, Iqbal Y, Glass S, Riley P, Pope C, Planche T, et al. Implementation of the cobas Liat influenza point-of-care test into an emergency department during a high-incidence season: a retrospective evaluation following real-world implementation. J Hosp Infect 2019;101:285-8. https://doi.org/10.1016/j.jhin.2018.12.008.
- Boku S, Naito T, Murai K, Tanei M, Inui A, Nisimura H, et al. Near point-of-care administration by the attending physician of the rapid influenza antigen detection immunochromatography test and the fully automated respiratory virus nucleic acid test: contribution to patient management. Diagn Microbiol Infect Dis 2013;76:445-9. https://doi.org/10.1016/j.diagmicrobio.2013.04.029.
- Hansen GT, Moore J, Herding E, Gooch T, Hirigoyen D, Hanson K, et al. Clinical decision making in the emergency department setting using rapid PCR: results of the CLADE study group. J Clin Virol 2018;102:42-9. https://doi.org/10.1016/j.jcv.2018.02.013.
- Peretz A, Zadok BS, Azrad M. Performance of the Influ a + B K-SeT(R) assay as compared to two RT-PCR assays for detection of influenza virus. Diagn Microbiol Infect Dis 2020;98. https://doi.org/10.1016/j.diagmicrobio.2020.115097.
- Tanei M, Yokokawa H, Murai K, Sakamoto R, Amari Y, Boku S, et al. Factors influencing the diagnostic accuracy of the rapid influenza antigen detection test (RIADT): a cross-sectional study. BMJ Open 2014;4. https://doi.org/10.1136/bmjopen-2013-003885.
- Zuurbier RP, Korsten K, Verheij TJM, Butler C, Adriaenssens N, Coenen S, et al. REspiratory Syncytial Virus Consortium in EUrope (RESCEU) Investigators . Performance assessment of a rapid molecular respiratory syncytial virus point-of-care test: a prospective community study in older adults. J Infect Dis 2022;226:S63-70. https://doi.org/10.1093/infdis/jiab600.
- Theerthakarai R, El-Halees W, Ismail M, Solis RA, Khan MA. Nonvalue of the initial microbiological studies in the management of nonsevere community-acquired pneumonia. Chest 2001;119:181-4. https://doi.org/10.1378/chest.119.1.181.
- Ewig S, Schlochtermeier M, Goïke N, Niederman MS. Applying sputum as a diagnostic tool in pneumonia: limited yield, minimal impact on treatment decisions. Chest 2002;121:1486-92. https://doi.org/10.1378/chest.121.5.1486.
- Trikalinos TA, Balion CM. Chapter 9: options for summarizing medical test performance in the absence of a ‘gold standard’. J Gen Intern Med 2012;27:S67-75. https://doi.org/10.1007/s11606-012-2031-7.
- Harvey EJ, De Brun C, Casale E, Finistrella V, Ashiru-Oredope D. Influence of factors commonly known to be associated with health inequalities on antibiotic use in high-income countries: a systematic scoping review. J Antimicrob Chemother 2023;78:861-70. https://doi.org/10.1093/jac/dkad034.
- Smedemark SA, Aabenhus R, Llor C, Fournaise A, Olsen O, Jørgensen KJ. Biomarkers as point‐of‐care tests to guide prescription of antibiotics in people with acute respiratory infections in primary care. Cochrane Database Syst Rev 2022;2022. https://doi.org/10.1002/14651858.CD010130.pub3.
- Tonkin-Crine S, Anthierens S, Hood K, Yardley L, Cals JW, Francis NA, et al. GRACE INTRO/CHAMP consortium . Discrepancies between qualitative and quantitative evaluation of randomised controlled trial results: achieving clarity through mixed methods triangulation. Implement Sci 2016;11. https://doi.org/10.1186/s13012-016-0436-0.
- Sydenham RV, Hansen MP, Justesen US, Pedersen LB, Aabenhus RM, Wehberg S, et al. Factors associated with C-reactive protein testing when prescribing antibiotics in general practice: a register-based study. BMC Prim Care 2022;23. https://doi.org/10.1186/s12875-021-01614-6.
- Bruning AHL, Leeflang MMG, Vos J, Spijker R, de Jong MD, Wolthers KC, et al. Rapid tests for influenza, respiratory syncytial virus, and other respiratory viruses: a systematic review and meta-analysis. Clin Infect Dis 2017;65:1026-32. https://doi.org/10.1093/cid/cix461.
- Chartrand C, Leeflang MM, Minion J, Brewer T, Pai M. Accuracy of rapid influenza diagnostic tests: a meta-analysis. Ann Intern Med 2012;156:500-11. https://doi.org/10.7326/0003-4819-156-7-201204030-00403.
- Chartrand C, Tremblay N, Renaud C, Papenburg J. Diagnostic accuracy of rapid antigen detection tests for respiratory syncytial virus infection: systematic review and meta-analysis. J Clin Microbiol 2015;53:3738-49. https://doi.org/10.1128/JCM.01816-15.
- Engel MF, Paling FP, Hoepelman AI, van der Meer V, Oosterheert JJ. Evaluating the evidence for the implementation of C-reactive protein measurement in adult patients with suspected lower respiratory tract infection in primary care: a systematic review. Fam Pract 2012;29:383-93. https://doi.org/10.1093/fampra/cmr119.
- Falk G, Fahey T. C-reactive protein and community-acquired pneumonia in ambulatory care: systematic review of diagnostic accuracy studies. Fam Pract 2009;26:10-21. https://doi.org/10.1093/fampra/cmn095.
- Hill AT, Gold PM, El Solh AA, Metlay JP, Ireland B, Irwin RS. CHEST Expert Cough Panel . Adult outpatients with acute cough due to suspected pneumonia or influenza: CHEST guideline and expert panel report. Chest 2019;155:155-67. https://doi.org/10.1016/j.chest.2018.09.016.
- Han MY, Xie TA, Li JX, Chen HJ, Yang XH, Guo XG. Evaluation of lateral-flow assay for rapid detection of influenza virus. Biomed Res Int 2020;2020. https://doi.org/10.1155/2020/3969868.
- Hoult G, Gillespie D, Wilkinson TMA, Thomas M, Francis NA. Biomarkers to guide the use of antibiotics for acute exacerbations of COPD (AECOPD): a systematic review and meta-analysis. BMC Pulm Med 2022;22. https://doi.org/10.1186/s12890-022-01958-4.
- Htun TP, Sun Y, Chua HL, Pang J. Clinical features for diagnosis of pneumonia among adults in primary care setting: a systematic and meta-review. Sci Rep 2019;9. https://doi.org/10.1038/s41598-019-44145-y.
- Huang HS, Tsai CL, Chang J, Hsu TC, Lin S, Lee CC. Multiplex PCR system for the rapid diagnosis of respiratory virus infection: systematic review and meta-analysis. Clin Microbiol Infect 2018;24:1055-63. https://doi.org/10.1016/j.cmi.2017.11.018.
- Lee J, Song JU, Kim YH. Diagnostic accuracy of the Quidel Sofia rapid influenza fluorescent immunoassay in patients with influenza-like illness: a systematic review and meta-analysis. Tuberc Respir Dis (Seoul) 2021;84:226-36. https://doi.org/10.4046/trd.2021.0033.
- Merckx J, Wali R, Schiller I, Caya C, Gore GC, Chartrand C, et al. Diagnostic accuracy of novel and traditional rapid tests for influenza infection compared with reverse transcriptase polymerase chain reaction: a systematic review and meta-analysis. Ann Intern Med 2017;167:394-409. https://doi.org/10.7326/M17-0848.
- Nicholson KG, Abrams KR, Batham S, Medina MJ, Warren FC, Barer M, et al. Randomised controlled trial and health economic evaluation of the impact of diagnostic testing for influenza, respiratory syncytial virus and Streptococcus pneumoniae infection on the management of acute admissions in the elderly and high-risk 18- to 64-year-olds. Health Technol Assess 2014;18:1-274. https://doi.org/10.3310/hta18360.
- Petrozzino JJ, Smith C, Atkinson MJ. Rapid diagnostic testing for seasonal influenza: an evidence-based review and comparison with unaided clinical diagnosis. J Emerg Med 2010;39:476-490.e1. https://doi.org/10.1016/j.jemermed.2009.11.031.
- van der Meer V, Neven AK, van den Broek PJ, Assendelft WJ. Diagnostic value of C reactive protein in infections of the lower respiratory tract: systematic review. BMJ 2005;331. https://doi.org/10.1136/bmj.38483.478183.EB.
- Vos LM, Bruning AHL, Reitsma JB, Schuurman R, Riezebos-Brilman A, Hoepelman AIM, et al. Rapid molecular tests for influenza, respiratory syncytial virus, and other respiratory viruses: a systematic review of diagnostic accuracy and clinical impact studies. Clin Infect Dis 2019;69:1243-53. https://doi.org/10.1093/cid/ciz056.
- Wu MH, Lin CC, Huang SL, Shih HM, Wang CC, Lee CC, et al. Can procalcitonin tests aid in identifying bacterial infections associated with influenza pneumonia? A systematic review and meta-analysis. Influenza Other Respir Viruses 2013;7:349-55. https://doi.org/10.1111/j.1750-2659.2012.00386.x.
Appendix 1
Searches for systematic reviews
Database: Ovid MEDLINE(R) ALL <1946 to May 22, 2023> Final search strategy
| 1 | [Respiratory Tract Infection (RTI)] |
| 2 | exp Respiratory Tract Infections/ |
| 3 | exp Otorhinolaryngologic Diseases/ |
| 4 | ((airway* or bronchopulmonar* or broncho-pulmonar* or tracheobronch* or tracheo-bronch* or pulmonar* tract or pulmonary or respirat* tract or respiratory or (ear adj3 nose adj3 throat) or ENT or otorhinolaryng*) adj3 (infect* or coinfect* or inflamm*)).tw,kf. |
| 5 | ((chest or lung? or lobar or pleura?) adj3 (absces* or infect* or coinfect* or inflamm*)).tw,kf. |
| 6 | (bronchit* or bronchiolit* or allergic bronchopulmon* or bronchopneumon* or common cold* or coryza or croup or empyem* or epipharyngit* or epiglottit* or epiglotit* or flu or influenza or laryngit* or laryngotracheobronchit* or laryngo tracheo bronchit* or laryngo tracheobronchit* or laryngotracheit* or nasopharyngit* or otitis media or parainfluenza or pharyngit* or pleurisy or pneumoni* or pleuropneumoni* or rhinit* or rhinopharyngit* or rhinosinusit* or severe acute respiratory syndrome or SARS or sinusit* or sore throat* or throat infection* or supraglottit* or supraglotit* or tonsillit* or tonsilit* or tracheit* or whooping cough or pertussis or pertusis).mp. |
| 7 | ((acute* or exacerbat* or flare*) adj3 (asthma* or copd or coad or chronic obstructive pulmonary disease or chronic obstructive airway* disease or chronic obstructive lung disease)).mp. |
| 8 | ((acute* or subacute* or exacerbat* or prolonged) adj3 cough*).mp. |
| 9 | (RTI or LRTI or URTI or ARTI or AURI or ALRI).tw,kf. |
| 10 | or/2-9 |
| 11 | [RTI Viral Infection] |
| 12 | exp Respiratory System/ and (exp Viruses/ or exp Virus Diseases/) |
| 13 | exp Pneumonia, Viral/ or *Orthomyxoviridae Infections/ or Influenza, Human/ |
| 14 | ((airway* or respiratory or pulmonary or bronchopulmonar* or broncho-pulmonar* or tracheobronch* or tracheo-bronch* or (ear adj3 nose adj3 throat) or ENT or otorhinolaryng*) adj3 (nonbacter* or viral* or virus* or adenovir*)).tw,kf. |
| 15 | (rhinovir* or rhino* vir* or coryzavir* or coryza* vir* or influenzavir* or influenza* vir* or (H1N1 or H3N2) or parainfluenzavir* or parainfluenza* vir* or pneumovir* or pneumo* vir* or human metapneumovir* or human meta-pneumovir* or HMPV or respiratory syncytial vir*).mp. or RSV.tw,kf. |
| 16 | or/12-15 |
| 17 | [RTI Bacterial Infection] |
| 18 | exp Respiratory System/ and (exp Bacteria/ or exp Bacterial Infections/) |
| 19 | Pneumonia, Bacterial/ or Chlamydial Pneumonia/ or Pneumonia, Mycoplasma/ or Pneumonia, Pneumococcal/ or Pneumonia, Staphylococcal/ |
| 20 | ((airway* or respiratory or pulmonary or bronchopulmonar* or broncho-pulmonar* or tracheobronch* or tracheo-bronch* or (ear adj3 nose adj3 throat) or ENT or otorhinolaryng*) adj3 (bacter* or bacilli* or bacili* or corynebac* or mycobac* or nonvir* or pathogen*)).tw,kf. |
| 21 | (strep* pneumon* or diplococ* pneumon* or pneumococ* or staph* pneumon* or chlamyd* pneumon* or myco* pneumon* or influenza bacil* or bacteri* influenza* or h?emophil* influenza*).mp. |
| 22 | ((strep* adj3 (throat* or pharyn* or tonsil*)) or (strep* and (airway* or pulmonary or brochopulmonar* or brocho-pulmonar* or respiratory* or (ear adj3 nose adj3 throat) or ENT or Otorhinolaryng*))).mp. |
| 23 | (GABHS or (“group a” adj3 strep*)).tw,kf. |
| 24 | strep* pyogen*.mp. |
| 25 | or/18-24 |
| 26 | [Rapid Tests] |
| 27 | Point-of-Care Systems/ |
| 28 | (POCT or POCTs or (((point adj2 care) or poc) adj3 (analys* or antigen? or assay* or device? or immunoassay* or classif* or detect* or determin* or diagnos* or differenti* or identif* or method* or kit or kits or panel? or platform? or predict* or rapid or routine* or screen* or system* or technique* or test* or (cassette? or dipstick? or film* or stick or strip or fluorescent anti*)))).tw,kf. |
| 29 | (point adj2 care).ti,kf. |
| 30 | (((near adj2 patient) or nearpatient or rapid* or bedside? or bed-side? or extra-laboratory or extralaboratory) adj3 (analys* or antigen? or assay* or immunoassay* or classif* or detect* or determin* or diagnos* or differenti* or identif* or method* or kit or kits or panel? or predict* or screen* or system* or technique* or test* or fluorescent anti*)).tw,kf. |
| 31 | (((near adj2 patient) or nearpatient or bedside? or bed-side? or extra-laboratory or extralaboratory) adj3 rapid*).tw,kf. |
| 32 | Rapid Diagnostic Tests/ |
| 33 | (rapid* adj3 (detect* or diagnos* or screen*)).tw,kf. |
| 34 | (time-to-result? or ((quick* or rapid* or short* or time*) adj3 (turnaround or turn-around))).tw,kf. |
| 35 | (antigen? adj3 (analys* or assay* or immunoassay* or classif* or detect* or determin* or diagnos* or differenti* or identif* or method* or kit or kits or panel? or predict* or rapid or routine* or screen* or system* or technique* or test*)).tw,kf. |
| 36 | (RADT or RADTs or RDT or RDTs).tw,kf. |
| 37 | (biomarker* or bio* marker* or ((biologic* or bacteri* or viral or virus or immuno* or inflammat* or molecular or protein or serum) adj marker*)).tw,kf. |
| 38 | ((rapid adj3 (molecular or PCR or polymerase chain reaction)) or singleplex* or single-plex* or multiplex* or multi-plex*).mp. |
| 39 | lab-on-a-chip.tw,kf. |
| 40 | ((lateral flow adj (assay* or immunoassay* or test*)) or LFA or LFIA).tw,kf. |
| 41 | (immunochromatograph* or immuno-chromatograph* or immuno-chromato-graph* or direct immunofluorescence or direct immuno-fluorescence or enzym* immunoassay* or enzym* immuno-assay* or fluorescence immunoassay* or fluorescence immuno-assay* or optical immunoassay* or optical immuno-assay*).mp. or (ICA or EIA or FIA or OIA).tw,kf. |
| 42 | ((chemiluminescen* or chemi-luminescen*) adj (immunoassay* or immuno-assay* or assay*)).mp. |
| 43 | (((mobile or portable or handheld or hand-held) adj3 (analy#er? or device? or meters or metres)) and (blood? or plasma or saliva or sputum or spit or mucus or urine or urea or urinalys* or fluids or gas or gases)).mp. |
| 44 | or/27-43 |
| 45 | (10 or 16 or 25) and 44 |
| 46 | [Systematic Review Filter] |
| 47 | (systematic review or meta-analysis).pt. |
| 48 | systematic review/ or meta-analysis/ or network meta-analysis/ |
| 49 | (meta-analys* or metaanalys* or meta-synth* or metasynth*).tw,kf. |
| 50 | (((systematic* or quantitativ* or methodologic*) adj5 (review* or overview*)) or (systematic* adj3 analys*)).tw,kf. |
| 51 | (systematic or structured or evidence or diagnostic or predicti* or trials or studies).ti. and ((review or overview or look or examination or update* or summary).ti. or review.pt.) |
| 52 | (quantitativ$ adj5 synthes*).tw,kf. |
| 53 | ((research adj3 (integrati* or overview*)) or (integrative adj2 review*) or research integration).tw,kf. |
| 54 | scoping review?.ti,kf. or (review.ti,kf,pt. and (trials as topic or studies as topic).hw.) |
| 55 | ((diagnostic or evidence) adj3 review*).tw,kf. |
| 56 | review.pt. and (medline or medlars or embase or pubmed or scisearch or psychinfo or psycinfo or psychlit or psyclit or cinahl or electronic database* or bibliographic database* or computeri#ed database* or online database* or pooling or pooled or mantel haenszel or peto or dersimonian or der simonian or fixed effect or ((hand adj2 search*) or (manual* adj2 search*))).tw,kf,hw. |
| 57 | exp technology assessment, biomedical/ |
| 58 | (technology assessment* or HTA or HTAs or technology overview* or technology appraisal*).tw,kf. |
| 59 | (0266-4623 or 1469-493X or 1366-5278 or 1530-440X or 2046-4053).is. |
| 60 | or/47-59 |
| 61 | [DTA Filter] |
| 62 | Diagnosis/ |
| 63 | “Diagnostic Techniques and Procedures”/ |
| 64 | Diagnostic Test Approval/ |
| 65 | Diagnostic Tests, Routine/ |
| 66 | Molecular Diagnostic Techniques/ |
| 67 | exp Reagent Kits, Diagnostic/ |
| 68 | (diagnos* adj3 (analys* or assay* or immunoassay* or classif* or differenti* or method* or kit or kits or panel? or predict* or screen* or system* or technique* or test*)).ab. |
| 69 | diagnos*.ti,kf,hw. |
| 70 | “sensitivity and specificity”/ or “predictive value of tests”/ or roc curve/ or signal-to-noise ratio/ or “limit of detection”/ |
| 71 | false negative reactions/ or false positive reactions/ |
| 72 | (sensitivity or specificity).tw,kf. |
| 73 | likelihood ratio.tw,kf. |
| 74 | (predict* adj4 val*).tw,kf. or predict*.ti. |
| 75 | ((accura* or reliab* or valid*) and (point-of-care or POC or (rapid adj2 (analys* or assay* or immunoassay* or classif* or detect* or diagnos* or differenti* or predict* or technique* or test*)))).tw,kf. |
| 76 | ((accura* or reliab* or valid*) and (bacteri* and (viral or virus*) and (analys* or assay* or immunoassay* or classif* or detect* or diagnos* or differenti* or predict* or technique* or test*))).tw,kf. |
| 77 | area under curve/ |
| 78 | (observer adj variation*).tw,kf. |
| 79 | (roc adj curve*).tw,kf. |
| 80 | likelihood functions/ |
| 81 | (false adj (positiv* or negativ*)).tw,kf. |
| 82 | QUADAS*.mp. |
| 83 | Diagnosis, Differential/ |
| 84 | (codetect* or co-detect* or codiagnos* or co-diagnos*).tw,kf. |
| 85 | ((discriminat* or differenti* or dual*) adj (detect* or diagnos*)).mp. |
| 86 | (bacteri* adj5 (viral or virus*) adj5 (analys* or assay* or immunoassay* or classif* or detect* or codetect* or determin* or diagnos* or codiagnos* or differenti* or discriminat* or distinguish* or identif* or method* or misdiagnos* or predict* or kit or kits or panel? or predict* or rapid or routine* or screen* or system* or technique* or test*)).tw,kf,hw. |
| 87 | or/62-86 |
| 88 | 45 and 60 and 87 |
| 89 | [Other] |
| 90 | (bacteri* adj5 (viral or virus*) adj5 (detect* or diagnos* or differenti* or predict* or screen* or test*)).tw,kf. |
| 91 | (bacteri* and (viral or virus*) and (codetect* or co-detect* or codiagnos* or co-diagnos*)).tw,kf. |
| 92 | (10 or 16 or 25) and 60 and (90 or 91) |
| 93 | (((prescribing or prescription?) adj guideline?) or ((antibiotic? or antimicrobial) adj stewardship?)).mp. |
| 94 | ((guide or guiding or predict* or ration* or reduc* or steward*) adj3 (antibiotic* or antivir* or anti-vir* or antimicrob* or anti-microb*)).tw,kf. |
| 95 | 45 and 60 and (93 or 94) |
| 96 | 88 or 92 or 95 |
| 97 | remove duplicates from 96 |
| 98 | [Symptoms & Signs] |
| 99 | Symptom Assessment/ |
| 100 | Patient Acuity/ |
| 101 | ((sign? adj3 symptom*) or ((sign? or symptom*) adj2 (score* or scoring))).tw,kf. |
| 102 | ((patient* or sign? or symptom* or illness* or disease* or disorder* or infection*) adj3 acuity).tw,kf. |
| 103 | exp Vital Signs/ |
| 104 | (peak flow or oxygen saturation or sats).mp. |
| 105 | Clinical Decision Rules/ |
| 106 | (clinic* predicti* or (clinic* adj5 (decision* or predicti*) adj5 (aid? or algorithm? or characteristic? or criteri* or evaluation? or index or indices or marker? or method* or model* or panel? or parameter? or rule or rules or score? or scoring or screen* or signs or symptoms or system? or technique? or test* or tool? or value? or variable*))).mp. |
| 107 | (clinical* adj (predicti* or predictor*)).tw,kf. |
| 108 | (rule in or ruled in or rule out or ruled out).tw,kf. |
| 109 | ((predict* or prognos* or cluster*) adj3 (sign? or symptom*)).tw,kf. |
| 110 | ((detect* or diagnos*) adj5 (sign? or symptom*)).tw,kf. |
| 111 | or/99-110 |
| 112 | (10 or 16 or 25) and 111 and 60 and 87 |
| 113 | [Host-response biomarkers] |
| 114 | Procalcitonin/ |
| 115 | (procalcitonin or pro-calcitonin or calcitonin precursor polyprotein or calcitonin related polypeptide alpha or calcitonin-1).mp. or PCT.tw,kf. |
| 116 | C-Reactive Protein/ |
| 117 | C-reactive protein.mp. or (CRP or HSCRP).tw,kf. |
| 118 | Myxovirus Resistance Proteins/ |
| 119 | (myxovirus resistance protein* or mx-protein* or MxA or (interferon adj2 induc* protein) or IP-10).mp. |
| 120 | (myxovirus resistance protein* or mx-protein* or MxA or (interferon adj2 induc* protein)).mp. |
| 121 | (FebriDx* or Febri-Dx*).mp. |
| 122 | TNF-Related Apoptosis-Inducing Ligand/ |
| 123 | ((tumor necrosis factor or TNF) adj2 related apoptosis adj2 ligand).tw,kf. |
| 124 | TRAIL.tw,kf. |
| 125 | Chemokine CXCL10/ |
| 126 | (ImmunoXpert* or Immuno-Xpert*).tw,kf. |
| 127 | (Interferon gamma inducible protein-10 or IFN-gamma-inducible protein-10 or IP-10 or IP10 or CXCL10 or CXCL-10).tw,kf. |
| 128 | (ImmunoXpert* or Immuno-Xpert* or MeMedBV* or MeMed-BV*).mp. |
| 129 | leukocyte count/ or lymphocyte count/ or cd4 lymphocyte count/ or cd4-cd8 ratio/ |
| 130 | ((WBC or white blood cell? or lymphocyte? or leukocyte? or CD4 or eosinophil? or neutrophil?) adj3 (count? or number? or ratio?)).tw,kf. |
| 131 | *leukocytes/ or exp *granulocytes/ or exp *leukocytes, mononuclear/ |
| 132 | *interleukins/ or interleukin-5/ or interleukin-6/ or interleukin-10/ |
| 133 | (il-5 or interleukin 5 or b-cell-growth-factor-ii or bcgf-ii or eosinophil differentiation factor or t-cell replacing factor).tw,kf. |
| 134 | (il-6 or interleukin-6 or b-cell differentiation factor or b-cell stimulatory factor-2 or bsf-2 or (differentiation-inducing protein adj1 myeloid) or hybridoma growth factor or plasmacytoma growth factor or hepatocyte stimulating factor or interferon beta-2 or ifn-beta-2 or mgi-2).tw,kf. |
| 135 | (il-10 or interleukin-10 or cytokine synthesis inhibitory factor or csif-10).tw,kf. |
| 136 | (interleukin*.tw,kf. or exp Interleukins/) and ((diagnos* or detect*).ti,kf,hw. or diagnosis.fs.) |
| 137 | or/114-136 |
| 138 | (10 or 16 or 25) and 137 and 60 and 87 |
| 139 | HEMATOLOGIC TESTS/ |
| 140 | ((h?em* or blood or plasma or serum) adj2 (test* or marker?)).tw,kf. |
| 141 | exp Cell Count/ |
| 142 | ((blood or RBC or red cell? or erythrocyt* or normocyt* or platelet* or thrombocyt*) adj3 (count* or distribution? or number* or paramet* or ratio?)).tw,kf. |
| 143 | Blood Sedimentation/ |
| 144 | (((blood or RBC or red cell? or erythrocyt*) adj2 sedimentation) or ESR).tw,kf. |
| 145 | exp BLOOD GAS ANALYSIS/ |
| 146 | blood gas*.tw,kf. |
| 147 | Oxygen/an, bl [Analysis, Blood] |
| 148 | Carbon Dioxide/an, bl [Analysis, Blood] |
| 149 | Sodium Bicarbonate/an, bl [Analysis, Blood] |
| 150 | (ABG or O2sat* or O2-sat* or O2CT or PaO2 or PaCO2 or HCO3 or (blood adj3 pH)).tw,kf. |
| 151 | (partial pressure and oxygen).hw. |
| 152 | (partial pressure adj3 (oxygen or O2)).tw,kf. |
| 153 | Sodium/bl [Blood] |
| 154 | ((blood or plasma or serum) adj2 (sodium or Na)).tw,kf. |
| 155 | ((blood or plasma or serum) adj2 marker?).tw,kf. |
| 156 | Fibrin Fibrinogen Degradation Products/ |
| 157 | (fibrin* adj2 degradation).tw,kf. |
| 158 | fibrinogen.tw,kf. or *fibrinogen/ or Fibrinogen/an, bl, ur [Analysis, Blood, Urine] |
| 159 | (d-dimer? or ddimer?).tw,kf. |
| 160 | Urine/an [Analysis] |
| 161 | (((urin* or urea) adj2 (analys* or test* or marker?)) or UAT).tw,kf. |
| 162 | Nitrogen/ur [Urine] |
| 163 | ((nitrogen or nitrate? or nitrite? or “N” or N2) adj3 (urea or urin*)).tw,kf. |
| 164 | Adrenomedullin/ |
| 165 | (adrenomedullin or proadrenomedullin or ADM or proADM).tw,kf. |
| 166 | exp Aspartate Aminotransferases/ |
| 167 | ((aspartat* adj3 (aminotrans* or amino-trans* or apoaminotrans* or apo-aminotrans* or apo-amino-trans* or apoamino-trans* or transaminas* or trans-aminas*)) or ((glutam* aspart* or glutam* oxaloacet*) adj3 (transaminas* or trans-aminas*)) or sgot).tw,kf. |
| 168 | Alanine Transaminase/ |
| 169 | ((alanine adj3 (aminotrans* or amino-trans* or transamin* or trans-amin*)) or (glutam* adj3 pyruvic adj3 trans*) or sgpt).tw,kf. |
| 170 | ((lipopolysac* or lipo-polysac* or lipo-poly-sac* or lipopoly-sac* or LPS) adj3 (bind* or bound*)).tw,kf. |
| 171 | Chitinases/ or Chitinase-3-like protein 1/ |
| 172 | (kitinase-3-like-1 or chitinase-3-like-1 or chitinase-3-like-protein-1 or CHI3L1).tw,kf. |
| 173 | Antibodies, Bacterial/an, bl [Analysis, Blood] |
| 174 | Antibodies, Viral/an, bl [Analysis, Blood] |
| 175 | Blood Proteins/an |
| 176 | Immunoglobulins/an |
| 177 | (“immunoglobulin M” or IgM or “immunoglobulin G” or IgG).tw,kf,hw. |
| 178 | *Serologic Tests/ |
| 179 | (((point adj2 care) or poc or (near adj2 patient) or nearpatient or rapid* or bedside? or bed-side? or extra-laboratory or extralaboratory) adj3 (serolog* or antibody or antibodies or immunoglobulin* or immune globulin*)).tw,kf. |
| 180 | ((serolog* or antibody or antibodies or immunoglobulin* or immune globulin*) and (analys* or assay* or immunoassay* or classif* or detect* or determin* or diagnos* or differenti* or identif* or method* or kit or kits or panel? or predict* or rapid or routine* or screen* or system* or technique* or test*)).ti,kf. |
| 181 | or/139-180 |
| 182 | (10 or 16 or 25) and 181 and 60 and 87 |
| 183 | 97 or 112 or 138 or 182 |
Database: Ovid EMBASE <1974 to 2023 May 24>
| 1 | Respiratory Tract Infection/ or exp Influenza/ or Laryngotracheobronchitis/ or Parainfluenza Virus Infection/ or Respiratory Syncytial Virus Infection/ or Viral Respiratory Tract Infection/ or Lower Respiratory Tract Infection/ or Chest Infection/ or Pertussis/ or Lung Infection/ or exp Infectious Pneumonia/ or Lung Abscess/ or exp Lung Mycosis/ or exp Viral Bronchiolitis/ or Upper Respiratory Tract Infection/ or exp Nose Infection/ or Oropharynx Candidiasis/ or Peritonsillar Abscess/ or Viral Upper Respiratory Tract Infection/ |
| 2 | Ear Nose Throat Disease/di or Otorhinolaryngology/ or exp Ear Infection/ or exp Otitis/ |
| 3 | ((airway* or bronchopulmonar* or broncho-pulmonar* or tracheobronch* or tracheo-bronch* or pulmonar* tract or pulmonary or respirat* tract or respiratory or (ear adj3 nose adj3 throat) or ENT or otorhinolaryng*) adj3 (infect* or coinfect* or inflamm*)).tw,kf. |
| 4 | ((chest or lung? or lobar or pleura?) adj3 (absces* or infect* or coinfect* or inflamm*)).tw,kf. |
| 5 | (bronchit* or bronchiolit* or allergic bronchopulmon* or bronchopneumon* or common cold* or coryza or croup or empyem* or epipharyngit* or epiglottit* or epiglotit* or flu or influenza or laryngit* or laryngotracheobronchit* or laryngo tracheo bronchit* or laryngo tracheobronchit* or laryngotracheit* or nasopharyngit* or otitis media or parainfluenza or pharyngit* or pleurisy or pneumoni* or pleuropneumoni* or rhinit* or rhinopharyngit* or rhinosinusit* or severe acute respiratory syndrome or SARS or sinusit* or sore throat* or throat infection* or supraglottit* or supraglotit* or tonsillit* or tonsilit* or tracheit* or whooping cough or pertussis or pertusis).mp. |
| 6 | ((acute* or exacerbat* or flare*) adj3 (asthma* or copd or coad or chronic obstructive pulmonary disease or chronic obstructive airway* disease or chronic obstructive lung disease)).mp. |
| 7 | ((acute* or subacute* or exacerbat* or prolonged) adj3 cough*).mp. |
| 8 | (RTI or LRTI or URTI or ARTI or AURI or ALRI).tw,kf. |
| 9 | or/1-8 |
| 10 | exp Respiratory System/ and exp Virus Infection/ |
| 11 | ((airway* or respiratory or pulmonary or bronchopulmonar* or broncho-pulmonar* or tracheobronch* or tracheo-bronch* or (ear adj3 nose adj3 throat) or ENT or otorhinolaryng*) adj3 (nonbacter* or viral* or virus* or adenovir*)).tw,kf. |
| 12 | Rhinovirus/ or exp Human Rhinovirus/ or exp Rhinovirus Infection/ |
| 13 | exp Influenza Virus/ or Orthomyxovirus Infection/ |
| 14 | Respirovirus/ or Human Parainfluenza virus 1/ or Human Parainfluenza Virus 3/ or Respirovirus Infection/ |
| 15 | exp Virus Pneumonia/ |
| 16 | Pneumovirus/ or Pneumovirus Infection/ or exp Human Respiratory Syncytial Virus/ or Respiratory Syncytial Virus Infection/ |
| 17 | Metapneumovirus/ or Metapneumovirus Infection/ or Human Metapneumovirus/ or Human Metapneumovirus Infection/ |
| 18 | (rhinovir* or rhino* vir* or coryzavir* or coryza* vir* or influenzavir* or influenza* vir* or (H1N1 or H3N2) or parainfluenzavir* or parainfluenza* vir* or pneumovir* or pneumo* vir* or human metapneumovir* or human meta-pneumovir* or HMPV or respiratory syncytial vir*).mp. or RSV.tw,kf. |
| 19 | or/10-18 |
| 20 | exp Respiratory System/ and (exp Bacterium/ or exp Bacterial Infection/) |
| 21 | ((airway* or respiratory or pulmonary or bronchopulmonar* or broncho-pulmonar* or tracheobronch* or tracheo-bronch* or (ear adj3 nose adj3 throat) or ENT or otorhinolaryng*) adj3 (bacter* or bacilli* or bacili* or corynebac* or mycobac* or nonvir* or pathogen*)).tw,kf. |
| 22 | Bacterial Pneumonia/ or Chlamydial Pneumonia/ or Mycoplasma Pneumonia/ or Staphylococcal Pneumonia/ or exp Streptococcus Pneumonia/ |
| 23 | (strep* pneumon* or diplococ* pneumon* or pneumococ* or staph* pneumon* or chlamyd* pneumon* or myco* pneumon* or influenza bacil* or bacteri* influenza* or h?emophil* influenza*).mp. |
| 24 | ((strep* adj3 (throat* or pharyn* or tonsil*)) or (strep* and (airway* or pulmonary or brochopulmonar* or brocho-pulmonar* or respiratory* or (ear adj3 nose adj3 throat) or ENT or Otorhinolaryng*))).mp. |
| 25 | Streptococcus Infection/ or Streptococcus Group A/ or exp Group A Streptococcal Infection/ or Streptococcal Pharyngitis/ |
| 26 | (GABHS or (“group a” adj3 strep*)).tw,kf. |
| 27 | strep* pyogen*.mp. |
| 28 | or/20-27 |
| 29 | “systematic review”/ or meta analysis/ or network meta-analysis/ |
| 30 | review.pt. and (evidence based adj (medicine or practice)).mp. |
| 31 | (systematic or structured or evidence or diagnostic or predicti* or trials or studies).ti. and ((review or overview or look or examination or update* or summary).ti. or review.pt.) |
| 32 | (0266-4623 or 1469-493X or 1366-5278 or 1530-440X or 2046-4053).is. |
| 33 | (systematic review? or evidence report* or technology assessment?).jw. |
| 34 | (meta-analys* or metaanalys* or meta-synth* or metasynth*).ti,ab,kf,hw. |
| 35 | (((systematic* or methodologic*) adj3 (analys* or review* or overview*)) or (quantitativ* adj3 (review* or synthes*))).tw,kf. |
| 36 | (diagnostic test accuracy study or validation study or cohort analysis or cross-sectional study or case control study).hw. and review.ti,kf,pt. |
| 37 | ((integrative adj2 review*) or research integration).tw,kf. or scoping review?.ti,kf. |
| 38 | ((diagnostic or evidence) adj3 review*).tw,kf. |
| 39 | review.pt. and (medline or medlars or embase or pubmed or scisearch or psychinfo or psycinfo or psychlit or psyclit or cinahl or electronic database* or bibliographic database* or computeri#ed database* or online database* or pooling or pooled or mantel haenszel or peto or dersimonian or der simonian or fixed effect or ((hand adj2 search*) or (manual* adj2 search*))).ti,ab,kf,hw. |
| 40 | biomedical technology assessment/ |
| 41 | (technology assessment* or HTA or HTAs or technology overview* or technology appraisal*).tw,kf. |
| 42 | or/29-41 |
| 43 | Gold Standard/ |
| 44 | (reference standard? or gold standard?).tw,kf. |
| 45 | clinical diagnosis.mp. |
| 46 | Diagnostic Test Accuracy Study/ |
| 47 | Diagnostic Accuracy/ |
| 48 | (DTA or (diagnos* adj2 accura*)).tw,kf. |
| 49 | Validation Study/ |
| 50 | “Sensitivity and Specificity”/ |
| 51 | specificity.tw,kf. |
| 52 | Receiver Operating Characteristic/ |
| 53 | Reliability/ |
| 54 | Internal Validity/ |
| 55 | Internal Consistency/ |
| 56 | (validat* or validity).tw,kf. |
| 57 | likelihood ratio*.tw,kf. |
| 58 | Predictive Value/ |
| 59 | (predict* adj4 val*).tw,kf. or predict*.ti. |
| 60 | ((re-test or retest or test-retest) adj reliability).tw,kf. |
| 61 | Diagnostic Error/ or False Negative Result/ or False Positive Result/ or Missed Diagnosis/ |
| 62 | (false adj (positiv* or negativ*)).tw,kf. |
| 63 | receiver operating characteristic*.tw,kf. |
| 64 | ROC.tw,kf. |
| 65 | Area Under the Curve/ |
| 66 | Observer Variation/ |
| 67 | (observer adj variation*).tw,kf. |
| 68 | ((degree? or rate* or rating) adj3 agreement?).tw,kf. |
| 69 | Diagnosis/ |
| 70 | diagnos*.ti,kf. |
| 71 | (diagnos* adj3 (analys* or assay* or immunoassay* or classif* or differenti* or method* or kit or kits or panel? or predict* or screen* or system* or technique* or test*)).ab. |
| 72 | Diagnostic Procedure/ or Diagnostic Test/ or Diagnostic Test Approval/ or exp Diagnostic Kit/ or Diagnosis Time/ |
| 73 | Laboratory Diagnosis/ |
| 74 | Molecular Diagnosis/ |
| 75 | ((accura* or reliab* or valid*) and (point-of-care or POC or (rapid adj2 (analys* or assay* or immunoassay* or classif* or detect* or diagnos* or differenti* or predict* or technique* or test*)))).tw,kf. |
| 76 | ((accura* or reliab* or valid*) and (bacteri* and (viral or virus*) and (analys* or assay* or immunoassay* or classif* or detect* or diagnos* or differenti* or predict* or technique* or test*))).tw,kf. |
| 77 | “quality assessment of diagnostic accuracy studies”/ |
| 78 | QUADAS*.mp. |
| 79 | Differential Diagnosis/ |
| 80 | (codetect* or co-detect* or codiagnos* or co-diagnos*).tw,kf. |
| 81 | ((discriminat* or differenti* or dual*) adj (detect* or diagnos*)).mp. |
| 82 | (bacteri* adj5 (viral or virus*) adj5 (analys* or assay* or immunoassay* or classif* or detect* or codetect* or determin* or diagnos* or codiagnos* or differenti* or discriminat* or distinguish* or identif* or method* or misdiagnos* or predict* or kit or kits or panel? or predict* or rapid or routine* or screen* or system* or technique* or test*)).tw,kf,hw. |
| 83 | or/43-82 |
| 84 | 42 and 83 |
| 85 | Diagnostic Accuracy/ and Review/ |
| 86 | 84 or 85 |
| 87 | (9 or 19 or 28) and 86 |
| 88 | (COVID19 or COVID-19 or COVID2019 or COVID-2019 or 2019 nCoV or 2019nCoV or 2019-novel CoV or “SARS-CoV-2” or “SARS-CoV2” or SARSCoV2 or “SARSCoV-2” or 2019 nCoV or 2019nCoV or 2019-novel CoV or “SARS coronavirus 2” or “Severe Acute Respiratory Syndrome Coronavirus-2”).ti. |
| 89 | 87 not 88 |
| 90 | ((neonat* or infant* or child* or p?ediatri*) not adult*).ti. |
| 91 | 89 not 90 |
| 92 | “Point of Care System”/ |
| 93 | (POCT or POCTs or (((point adj2 care) or poc) adj3 (analys* or antigen? or assay* or device? or immunoassay* or classif* or detect* or determin* or diagnos* or differenti* or identif* or method* or kit or kits or panel? or platform? or predict* or rapid or routine* or screen* or system* or technique* or test* or (cassette? or dipstick? or film* or stick or strip or fluorescent anti*)))).tw,kf. |
| 94 | (point adj2 care).ti,kf. |
| 95 | (((near adj2 patient) or nearpatient or rapid* or bedside? or bed-side? or extra-laboratory or extralaboratory) adj3 (analys* or antigen? or assay* or immunoassay* or classif* or detect* or determin* or diagnos* or differenti* or identif* or method* or kit or kits or panel? or predict* or screen* or system* or technique* or test* or fluorescent anti*)).tw,kf. |
| 96 | (((near adj2 patient) or nearpatient or bedside? or bed-side? or extra-laboratory or extralaboratory) adj3 rapid*).tw,kf. |
| 97 | Rapid Test/ or Influenza A Rapid Test/ or Streptococcus Group A Rapid Test/ |
| 98 | (rapid test* or (rapid* adj3 (detect* or diagnos* or screen*))).tw,kf. |
| 99 | (time-to-result? or ((quick* or rapid* or short* or time*) adj3 (turnaround or turn-around))).tw,kf. |
| 100 | (antigen? adj3 (analys* or assay* or immunoassay* or classif* or detect* or determin* or diagnos* or differenti* or identif* or method* or kit or kits or panel? or predict* or rapid or routine* or screen* or system* or technique* or test*)).tw,kf. |
| 101 | (RADT or RADTs or RDT or RDTs).tw,kf. |
| 102 | (biomarker or bio* marker* or ((biologic* or bacteri* or viral or virus or immuno* or inflammat* or molecular or protein or serum) adj marker*)).tw,kf. |
| 103 | Multiplex Analyzer/ |
| 104 | exp Multiplex Polymerase Chain Reaction/ |
| 105 | Singleplex Polymerase Chain Reaction/ |
| 106 | ((rapid adj3 (molecular or PCR or polymerase chain reaction)) or singleplex* or single-plex* or multiplex* or multi-plex*).mp. |
| 107 | lab-on-a-chip.tw,kf. |
| 108 | ((lateral flow adj (assay* or immunoassay* or test*)) or LFA or LFIA).tw,kf. |
| 109 | (immunochromatograph* or immuno-chromatograph* or immuno-chromato-graph* or direct immunofluorescence or direct immuno-fluorescence or enzym* immunoassay* or enzym* immuno-assay* or fluorescence immunoassay* or fluorescence immuno-assay* or optical immunoassay* or optical immuno-assay*).mp. or (ICA or EIA or FIA or OIA).tw,kf. |
| 110 | ((chemiluminescen* or chemi-luminescen*) adj (immunoassay* or immuno-assay* or assay*)).mp. |
| 111 | (((mobile or portable or handheld or hand-held) adj3 (analy#er? or device? or meters or metres)) and (blood? or plasma or saliva or sputum or spit or mucus or urine or urea or urinalys* or fluids or gas or gases)).mp. |
| 112 | or/92-111 |
| 113 | 91 and 112 |
| 114 | (bacteri* adj5 (viral or virus*) adj5 (detect* or diagnos* or differenti* or predict* or screen* or test*)).tw,kf. |
| 115 | (bacteri* and (viral or virus*) and (codetect* or co-detect* or codiagnos* or co-diagnos*)).tw,kf. |
| 116 | (9 or 19 or 28) and 42 and (114 or 115) |
| 117 | 116 not (88 or 90) |
| 118 | 113 or 117 |
| 119 | limit 118 to conference abstract status |
| 120 | 118 not 119 |
| 121 | Health Status Indicator/ or Patient Acuity/ |
| 122 | Symptom Assessment/ |
| 123 | Symptomatology/ |
| 124 | *Symptom/ |
| 125 | ((sign? adj2 symptom*) and (score* or scoring)).tw,kf. |
| 126 | ((patient* or sign? or symptom* or illness* or disease* or disorder* or infection*) adj3 acuity).tw,kf. |
| 127 | Vital Sign/ |
| 128 | Decision Support System/ or Clinical Decision Rule/ |
| 129 | (clinic* predicti* or (clinic* adj5 (decision* or predicti*) adj5 (aid? or algorithm? or characteristic? or criteri* or evaluation? or index or indices or marker? or method* or model* or panel? or parameter? or rule or rules or score? or scoring or screen* or signs or symptoms or system? or technique? or test* or tool? or value? or variable*))).tw,kf. |
| 130 | (clinical* adj (predicti* or predictor*)).tw,kf. |
| 131 | (“rule in” or “ruled in” or “rule out” or “ruled out”).tw,kf. |
| 132 | ((predict* or prognos* or cluster*) adj3 (sign? or symptom*)).tw,kf. |
| 133 | ((detect* or diagnos*) and (sign? or symptom*)).ti,kf. |
| 134 | or/121-133 |
| 135 | 91 and 134 |
| 136 | limit 135 to conference abstract status |
| 137 | 135 not 136 |
| 138 | Procalcitonin Test Kit/ |
| 139 | *Procalcitonin/ or Procalcitonin/ec [Endogenous Compound] |
| 140 | (procalcitonin or pro-calcitonin or calcitonin precursor polyprotein or calcitonin related polypeptide alpha or calcitonin-1 or PCT).tw,kf. |
| 141 | *C reactive protein/ or C reactive protein/ec [Endogenous Compound] |
| 142 | (c-reactive protein or CRP or HSCRP).tw,kf. |
| 143 | Myxovirus Resistance Protein/ |
| 144 | (myxovirus resistance protein* or mx-protein* or MxA or (interferon adj2 induc* protein) or IP-10).tw,kf. |
| 145 | (FebriDx* or Febri-Dx*).af. |
| 146 | Tumor Necrosis Factor Related Apoptosis Inducing Ligand/ |
| 147 | ((tumor necrosis factor or TNF) adj2 related apoptosis adj2 ligand).tw,kf. |
| 148 | TRAIL.tw,kf. |
| 149 | C Reactive Protein/ and Endogenous Compound/ |
| 150 | Procalcitonin/ and Endogenous Compound/ |
| 151 | Gamma Interferon Inducible Protein 10/ |
| 152 | (Interferon gamma inducible protein-10 or IFN-gamma-inducible protein-10 or IP-10 or IP10 or CXCL10 or CXCL-10).tw,kf. |
| 153 | (ImmunoXpert* or Immuno-Xpert* or MeMedBV* or MeMed-BV*).af. |
| 154 | or/138-153 |
| 155 | 91 and 154 |
| 156 | limit 155 to conference abstract status |
| 157 | 155 not 156 |
| 158 | exp *Blood Cell Count/ |
| 159 | ((WBC or white blood cell? or white cell? or lymphocyte? or leukocyte? or monocyte? or CD4* or eosinophil? or neutrophil?) adj3 (count* or distribution? or number* or paramet* or ratio?)).tw,kf. |
| 160 | ((whole blood or blood cell or RBC or red cell? or erythrocyt* or normocyt* or platelet* or thrombocyt*) adj3 (count* or distribution? or number* or paramet* or ratio?)).tw,kf. |
| 161 | ((h?em* or blood or plasma or serum) adj2 (test* or marker?)).tw,kf. |
| 162 | *erythrocyte sedimentation rate/ |
| 163 | (((blood or RBC or red cell? or erythrocyt*) adj2 sedimentation) or ESR).tw,kf. |
| 164 | or/158-163 |
| 165 | 91 and 164 |
| 166 | limit 165 to conference abstract status |
| 167 | 165 not 166 |
| 168 | Blood Gas Analysis/ |
| 169 | blood gas*.tw,kf. |
| 170 | Oxygen Saturation/ |
| 171 | (ABG or O2sat* or O2-sat* or O2CT or PaO2 or PaCO2 or HCO3 or (blood adj3 pH)).tw,kf. |
| 172 | ((oxygen adj2 (concentration or saturation)) or sats).tw,kf. |
| 173 | (partial pressure and oxygen).hw. |
| 174 | (partial pressure adj3 (oxygen or O2)).tw,kf. |
| 175 | or/168-174 |
| 176 | 91 and 175 |
| 177 | ((blood or plasma or serum) adj2 (sodium or Na)).tw,kf. |
| 178 | electrolyte blood level/ or sodium blood level/ |
| 179 | (177 or 178) and 91 |
| 180 | (il-5 or interleukin 5 or b-cell-growth-factor-ii or bcgf-ii or eosinophil differentiation factor or t-cell replacing factor or il-6 or interleukin-6 or b-cell differentiation factor or b-cell stimulatory factor-2 or bsf-2 or (differentiation-inducing protein adj1 myeloid) or hybridoma growth factor or plasmacytoma growth factor or hepatocyte stimulating factor or interferon beta-2 or ifn-beta-2 or mgi-2 or il-10 or interleukin-10 or cytokine synthesis inhibitory factor or csif-10).tw,kf. |
| 181 | 180 and 91 |
| 182 | fibrinogen/ |
| 183 | fibrinogen.tw,kf. |
| 184 | fibrin degradation product/ |
| 185 | (fibrin* adj2 degradation).tw,kf. |
| 186 | d dimer/ |
| 187 | (d-dimer? or ddimer?).tw,kf. |
| 188 | or/182-187 |
| 189 | 91 and 188 |
| 190 | (((urin* or urea) adj2 (analys* or test* or marker?)) or UAT).tw,kf. |
| 191 | ((nitrogen or nitrate? or nitrite? or “N” or N2) adj3 (urea or urin*)).tw,kf. |
| 192 | urea nitrogen blood level/ |
| 193 | urea/ec |
| 194 | or/190-193 |
| 195 | 194 and 91 |
| 196 | Adrenomedullin/ |
| 197 | (adrenomedullin or adrenomedullin or proadrenomedullin or proadrenomedullin or ADM or proADM).tw,kf. |
| 198 | (196 or 197) and 91 |
| 199 | Enzyme Blood Level/ |
| 200 | Aspartate Aminotransferase Blood Level/ or Aspartate Aminotransferase Level/ |
| 201 | ((aspartat* adj3 (aminotrans* or amino-trans* or apoaminotrans* or apo-aminotrans* or apo-amino-trans* or apoamino-trans* or transaminas* or trans-aminas*)) or ((glutam* aspart* or glutam* oxaloacet*) adj3 (transaminas* or trans-aminas*)) or sgot).tw,kf. |
| 202 | *Aspartate Aminotransferase/ or Aspartate Aminotransferase/ec [Endogenous Compound] |
| 203 | Alanine Aminotransferase Level/ or Alanine Aminotransferase Blood Level/ |
| 204 | *Alanine Aminotransferase/ or Alanine Aminotransferase/ec [Endogenous Compound] |
| 205 | ((alanine adj3 (aminotrans* or amino-trans* or transamin* or trans-amin*)) or (glutam* adj3 pyruvic adj3 trans*) or sgpt).tw,kf. |
| 206 | or/199-205 |
| 207 | 91 and 206 |
| 208 | Lipopolysaccharide Binding Protein/ec [Endogenous Compound] |
| 209 | ((lipopolysac* or lipo-polysac* or lipo-poly-sac* or lipopoly-sac* or LPS) adj3 (bind* or bound*)).tw,kf. |
| 210 | (208 or 209) and 91 |
| 211 | Chitinase 3 Like Protein 1/ |
| 212 | (kitinase-3-like-1 or chitinase-3-like-1 or chitinase-3-like-protein-1 or CHI3L1).tw,kf. |
| 213 | (211 or 212) and 91 |
| 214 | (176 or 179 or 181 or 189 or 195 or 198 or 207 or 210 or 213) |
| 215 | limit 214 to conference abstract status |
| 216 | 214 not 215 |
| 217 | 120 or 137 or 157 or 167 or 216 |
Database: Cochrane Database of Systematic Reviews
https://www.cochranelibrary.com/cdsr/reviews
Issue 5 of 12, May 2023 (searched 18 May 2023)
Records screened in situ for potentially relevant reviews
| S1 | All-Text: * Limit CDSR to Review Type: <Diagnostic> |
| S2 | All-Text: * Limit CDSR to Protocol Type: <Diagnostic> |
Database: NIHR Journal Library
https://www.journalslibrary.nihr.ac.uk/advancedsearch/
Browsed online, using NHIR Library indexing categories to help identify relevant DTA reviews. A series of short iterative searches were also conducted. Records were screened in situ (30 May 2023).
Browsing
| S1 | NIHR Programme: <Systematic Reviews> Limited by: (i) HRCS Health Category: <Respiratory> or (ii) HRCS Health Category: <Infection> |
| S2 | NIHR Programme: <HTA> Limited by:(i) HRCS Health Category: <Respiratory> or (ii) HRCS Health Category: <Infection> |
| S3 | Research Type: <Evidence Synthesis> Limited by: (i) HRCS Health Category: <Respiratory> or (ii) HRCS Health Category: <Infection> |
| S4 | Research Type: NICE DAR (Diagnostic Assessment Report) |
Searching
| S1 | diagnos* AND review |
| S2 | diagnos* AND accuracy |
| S3 | diagnos* AND test* |
| S4 | rapid* AND test* |
| S5 | “point of care” |
Database: Epistemonikos
https://www.epistemonikos.org/en/advanced_search
| S1a | (respiratory OR “ear nose and throat” OR ENT OR otorhinolaryng* OR RTI OR LRTI OR URTI OR ARTI OR AURI OR ALRI OR airway* OR bronchopulmonar* OR broncho-pulmonar* OR tracheobronch* OR tracheo-bronch* OR “pulmonary tract” OR ((chest OR lung OR lungs OR lobar OR pleura*) AND (absces* OR infect* OR coinfect* OR inflamm*)) OR bronchit* OR bronchiolit* OR bronchopneumon* OR “common cold” OR coryza OR croup OR empyem* OR epipharyngit* OR epiglottit* OR epiglotit* OR flu OR influenza OR laryngit* OR laryngotracheobronchit* OR (laryngo AND tracheo AND bronchit*) OR (laryngo AND tracheobronchit*) OR laryngotracheit* OR nasopharyngit* OR “otitis media” OR parainfluenza OR pharyngit* OR pleurisy OR pneumoni* OR pleuropneumoni* OR rhinit* OR rhinopharyngit* OR rhinosinusit* OR sinusit* OR “sore throat” OR (throat AND infection*) OR supraglottit* OR supraglotit* OR tonsillit* OR tonsilit* OR tracheit* OR “whooping cough” OR pertussis OR pertussis OR asthma* OR “COPD” OR “COAD” OR “chronic obstructive pulmonary disease” OR “chronic obstructive airway disease” OR “chronic obstructive airways disease” OR “chronic obstructive lung disease” OR ((acute or subacute* or exacerbat* or prolonged) AND cough*)) Limit-1: Publication Type: <Systematic Review> AND Type of Study:<Diagnostic Accuracy> OR Limit-2: Publication Type: <Systematic Review> AND Type of Study: <Prediction (Diagnostic)> |
| S1b | SARS OR “severe acute respiratory syndrome” Limit-1: Publication Type: <Systematic Review> AND Type of Study: <Diagnostic Accuracy> [All SARS-CoV2, records not downloaded] Limit-2: Publication Type: <Systematic Review> AND Type of Study: <Prediction (Diagnostic)> |
| S1c | (rhinovir* OR (rhino* AND vir*) OR coryzavir* OR (coryza* AND vir*) OR influenzavir* OR (influenza* AND vir*) OR (H1N1 OR H3N2) OR parainfluenzavir* OR (parainfluenza* AND vir*) OR pneumovir* OR (pneumo* AND vir*) OR metapneumovir* OR meta-pneumovir* OR HMPV OR RSV OR (“respiratory syncytial” AND vir*) OR (strep* AND pneumon*) OR (diplococ* AND pneumon*) OR pneumococ* OR (staph* AND pneumon*) OR (chlamyd* AND pneumon*) OR (myco* AND pneumon*) OR (influenza AND bacil*) OR (bacteri* AND influenza*) OR (hemophil* AND influenza*) OR (haemophil* AND influenza*) OR (strep* AND (throat* OR pharyn* OR tonsil* OR airway* OR pulmonary OR brochopulmonar* OR brocho-pulmonar* OR respiratory*)) OR GABHS or (“group a” AND strep*) OR (strep* AND pyogen*)) Limit-1: Publication Type: <Systematic Review> AND Type of Study: <Diagnostic Accuracy> OR Limit-2: Publication Type: <Systematic Review> AND Type of Study: <Prediction (Diagnostic)> |
| S2a | ((“diagnostic accuracy” OR “diagnostic test accuracy” OR (diagnostic AND studies)) AND ((rapid* AND (detect* or method* or molecular or test*)) OR “near patient” OR “point of care” OR POCT* OR biomarker* OR panel OR panels) AND (“respiratory tract” or (respiratory AND infection*) OR “ear nose and throat” OR “ENT” OR otorhinolaryng* OR “RTI” OR “LRTI” OR “URTI” OR “ARTI” OR “AURI” OR “ALRI” OR airway* OR bronchopulmonar* OR broncho-pulmonar* OR tracheobronch* OR tracheo-bronch* OR “pulmonary tract” OR (pulmonary AND infection*) OR ((chest OR lung OR lungs OR lobar OR pleura*) AND (absces* OR infect* OR coinfect* OR inflamm*)) OR bronchit* OR bronchiolit* OR bronchopneumon* OR “common cold” OR coryza OR croup OR empyem* OR epipharyngit* OR epiglottit* OR epiglotit* OR flu OR influenza OR laryngit* OR laryngotracheobronchit* OR (laryngo AND tracheo AND bronchit*) OR (laryngo AND tracheobronchit*) OR laryngotracheit* OR nasopharyngit* OR “otitis media” OR parainfluenza OR pharyngit* OR pleurisy OR pneumoni* OR pleuropneumoni* OR rhinit* OR rhinopharyngit* OR rhinosinusit* OR sinusit* OR “sore throat” OR (throat AND infection*) OR supraglottit* OR supraglotit* OR tonsillit* OR tonsilit* OR tracheit* OR “whooping cough” OR pertussis OR pertussis OR asthma* OR “COPD” OR “COAD” OR “chronic obstructive pulmonary disease” OR “chronic obstructive airway disease” OR “chronic obstructive airways disease” OR “chronic obstructive lung disease” OR ((acute or subacute* or exacerbat* or prolonged) AND cough*))) Limit: Publication Type: <Systematic Review> |
| S2b | ((diagnos* OR detect*) AND (“clinical decision rule” OR “clinical decision rules” OR “prediction model” OR “prediction models” OR “predictive model” OR “predictive models” OR “prediction rule” OR “prediction rules” OR “predictive rule” OR “predictive rules”) AND (“respiratory tract” or (respiratory AND infection*) OR “ear nose and throat” OR “ENT” OR otorhinolaryng* OR “RTI” OR “LRTI” OR “URTI” OR “ARTI” OR “AURI” OR “ALRI” OR airway* OR bronchopulmonar* OR broncho-pulmonar* OR tracheobronch* OR tracheo-bronch* OR “pulmonary tract” OR (pulmonary AND infection*) OR ((chest OR lung OR lungs OR lobar OR pleura*) AND (absces* OR infect* OR coinfect* OR inflamm*)) OR bronchit* OR bronchiolit* OR bronchopneumon* OR “common cold” OR coryza OR croup OR empyem* OR epipharyngit* OR epiglottit* OR epiglotit* OR flu OR influenza OR laryngit* OR laryngotracheobronchit* OR (laryngo AND tracheo AND bronchit*) OR (laryngo AND tracheobronchit*) OR laryngotracheit* OR nasopharyngit* OR “otitis media” OR parainfluenza OR pharyngit* OR pleurisy OR pneumoni* OR pleuropneumoni* OR rhinit* OR rhinopharyngit* OR rhinosinusit* OR sinusit* OR “sore throat” OR (throat AND infection*) OR supraglottit* OR supraglotit* OR tonsillit* OR tonsilit* OR tracheit* OR “whooping cough” OR pertussis OR pertussis OR asthma* OR “COPD” OR “COAD” OR “chronic obstructive pulmonary disease” OR “chronic obstructive airway disease” OR “chronic obstructive airways disease” OR “chronic obstructive lung disease” OR ((acute or subacute* or exacerbat* or prolonged) AND cough*))) Limit: Publication Type: <Systematic Review> |
| S2c | ((“diagnostic accuracy” OR “diagnostic test accuracy” OR (diagnostic AND studies)) AND ((rapid* AND (detect* or method* or molecular or test*)) OR “near patient” OR “point of care” OR POCT* OR biomarker* OR panel OR panels) AND (rhinovir* OR (rhino* AND vir*) OR coryzavir* OR (coryza* AND vir*) OR influenzavir* OR (influenza* AND vir*) OR (H1N1 OR H3N2) OR parainfluenzavir* OR (parainfluenza* AND vir*) OR pneumovir* OR (pneumo* AND vir*) OR metapneumovir* OR meta-pneumovir* OR HMPV OR RSV OR (“respiratory syncytial” AND vir*) OR (strep* AND pneumon*) OR (diplococ* AND pneumon*) OR pneumococ* OR (staph* AND pneumon*) OR (chlamyd* AND pneumon*) OR (myco* AND pneumon*) OR (influenza AND bacil*) OR (bacteri* AND influenza*) OR (hemophil* AND influenza*) OR (haemophil* AND influenza*) OR (strep* AND (throat* OR pharyn* OR tonsil* OR airway* OR pulmonary OR brochopulmonar* OR brocho-pulmonar* OR respiratory*)) OR GABHS or (“group a” AND strep*) OR (strep* AND pyogen*))) Limit: Publication Type: <Systematic Review> |
| S2d | ((diagnos* OR detect*) AND (“clinical decision rule” OR “clinical decision rules” OR “prediction model” OR “prediction models” OR “predictive model” OR “predictive models” OR “prediction rule” OR “prediction rules” OR “predictive rule” OR “predictive rules”) AND (rhinovir* OR (rhino* AND vir*) OR coryzavir* OR (coryza* AND vir*) OR influenzavir* OR (influenza* AND vir*) OR (H1N1 OR H3N2) OR parainfluenzavir* OR (parainfluenza* AND vir*) OR pneumovir* OR (pneumo* AND vir*) OR metapneumovir* OR meta-pneumovir* OR HMPV OR RSV OR (“respiratory syncytial” AND vir*) OR (strep* AND pneumon*) OR (diplococ* AND pneumon*) OR pneumococ* OR (staph* AND pneumon*) OR (chlamyd* AND pneumon*) OR (myco* AND pneumon*) OR (influenza AND bacil*) OR (bacteri* AND influenza*) OR (hemophil* AND influenza*) OR (haemophil* AND influenza*) OR (strep* AND (throat* OR pharyn* OR tonsil* OR airway* OR pulmonary OR brochopulmonar* OR brocho-pulmonar* OR respiratory*)) OR GABHS or (“group a” AND strep*) OR (strep* AND pyogen*))) Limit: Publication Type: <Systematic Review> |
Searches for primary diagnostic accuracy studies
White cell differential count
A precision maximising search was conducted due to the limited timeframe and inherent noise retrieved when searching for white blood cells and inflammatory infections
Database: Ovid MEDLINE(R) ALL <1946 to June 6, 2023>
| 1 | Diagnosis/ |
| 2 | “Diagnostic Techniques and Procedures”/ |
| 3 | Diagnostic Test Approval/ |
| 4 | Diagnostic Tests, Routine/ |
| 5 | Molecular Diagnostic Techniques/ |
| 6 | exp Reagent Kits, Diagnostic/ |
| 7 | (diagnos* adj3 (analys* or assay* or immunoassay* or classif* or differenti* or method* or kit or kits or panel? or predict* or screen* or system* or technique* or test*)).ab. |
| 8 | diagnos*.ti,kf,hw. |
| 9 | (DTA or (diagnos* adj2 accura*)).tw,kf. |
| 10 | “sensitivity and specificity”/ or “predictive value of tests”/ or roc curve/ or signal-to-noise ratio/ or “limit of detection”/ |
| 11 | (sensitivity or specificity).tw,kf. |
| 12 | likelihood ratio*.tw,kf. |
| 13 | (predict* adj4 val*).tw,kf. or predict*.ti. |
| 14 | ((re-test or retest or test-retest) adj reliability).tw,kf. |
| 15 | ((accura* or reliab* or valid*) and (point-of-care or POC or (rapid adj2 (analys* or assay* or immunoassay* or classif* or detect* or diagnos* or differenti* or predict* or technique* or test*)))).tw,kf. |
| 16 | ((accura* or reliab* or valid*) and (bacteri* and (viral or virus*) and (analys* or assay* or immunoassay* or classif* or detect* or diagnos* or differenti* or predict* or technique* or test*))).tw,kf. |
| 17 | Validation Study/ |
| 18 | (validat* or validity).tw,kf. |
| 19 | area under curve/ |
| 20 | observer variation/ |
| 21 | (observer adj variation*).tw,kf. |
| 22 | ((degree? or rate* or rating) adj3 agreement?).tw,kf. |
| 23 | ((detect* or diagnos*) and agreement?).tw,kf. |
| 24 | Receiver Operating Characteristic/ |
| 25 | (receiver operating characteristic* or ROC).tw,kf. |
| 26 | likelihood functions/ |
| 27 | diagnostic error/ or false negative result/ or false positive result/ or missed diagnosis/ or false negative reactions/ or false positive reactions/ |
| 28 | (false adj (positiv* or negativ*)).tw,kf. |
| 29 | (QUADAS* or STARD).mp. |
| 30 | laboratory diagnosis/ |
| 31 | (reference standard? or gold standard?).tw,kf. |
| 32 | Diagnosis, Differential/ |
| 33 | (codetect* or co-detect* or codiagnos* or co-diagnos*).tw,kf. |
| 34 | ((discriminat* or differenti* or dual*) adj (detect* or diagnos*)).mp. |
| 35 | (bacteri* adj5 (viral or virus*) adj5 (analys* or assay* or immunoassay* or classif* or detect* or codetect* or determin* or diagnos* or codiagnos* or differenti* or discriminat* or distinguish* or identif* or method* or misdiagnos* or predict* or kit or kits or panel? or predict* or rapid or routine* or screen* or system* or technique* or test*)).tw,kf,hw. |
| 36 | or/1-35 |
| 37 | (((WBC or white blood cell? or white cell? or lymphocyte? or leukocyte? or monocyte? or CD4* or eosinophil? or neutrophil? or granulocyte?) adj3 (count* or distribution? or level? or number* or paramet* or ratio?)) or NLR).tw,kf. |
| 38 | (respiratory or (ear nose adj2 throat) or ENT or otorhinolaryng* or RTI or LRTI or URTI or ARTI or AURI or ALRI or airway* or bronchopulmonar* or broncho-pulmonar* or tracheobronch* or tracheo-bronch* or pulmonary tract or ((chest or lung or lungs or lobar or pleura*) and (absces* or infect* or coinfect* or inflamm*)) or bronchit* or bronchiolit* or bronchopneumon* or common cold or coryza or croup or empyem* or epipharyngit* or epiglottit* or epiglotit* or flu or influenza or laryngit* or laryngotracheobronchit* or (laryngo and tracheo and bronchit*) or (laryngo and tracheobronchit*) or laryngotracheit* or nasopharyngit* or otitis media or parainfluenza or pharyngit* or pleurisy or pneumoni* or pleuropneumoni* or rhinit* or rhinopharyngit* or rhinosinusit* or sinusit* or sore throat or (throat and infection*) or supraglottit* or supraglotit* or tonsillit* or tonsilit* or tracheit* or whooping cough or pertussis or pertussis or asthma* or COPD or COAD or chronic obstructive pulmonary disease or chronic obstructive airway disease or chronic obstructive airways disease or chronic obstructive lung disease or ((acute or subacute* or exacerbat* or prolonged) and cough*)).ti. |
| 39 | 36 and 37 and 38 |
| 40 | (differential diagnos* or codetect* or co-detect*).mp. |
| 41 | ((bacter* or bacilli* or bacili* or corynebac* or mycobac* or nonvir*) and (nonbacter* or viral* or virus* or adenovir*)).mp. |
| 42 | 40 or 41 |
| 43 | 39 and 42 |
| 44 | (((WBC or white blood cell? or white cell? or lymphocyte? or leukocyte? or monocyte? or CD4* or eosinophil? or neutrophil? or granulocyte?) and (count* or distribution? or level? or number* or paramet* or ratio?)) or NLR).ti. |
| 45 | 38 and 42 and 44 |
| 46 | 43 or 45 |
| 47 | (COVID19 or COVID-19 or COVID2019 or COVID-2019 or 2019 nCoV or 2019nCoV or 2019-novel CoV or “SARS-CoV-2” or “SARS-CoV2” or SARSCoV2 or “SARSCoV-2” or 2019 nCoV or 2019nCoV or 2019-novel CoV or “SARS coronavirus 2” or “Severe Acute Respiratory Syndrome Coronavirus-2”).ti. |
| 48 | 46 not 47 |
| 49 | ((neonat* or infant* or child* or p?ediatri*) not adult*).ti. |
| 50 | 48 not 49 |
Database: Ovid EMBASE <1980 to 2023 Week 22>
| 1 | Gold Standard/ |
| 2 | (reference standard? or gold standard?).tw,kf. |
| 3 | clinical diagnosis.mp. |
| 4 | Diagnostic Test Accuracy Study/ |
| 5 | Diagnostic Accuracy/ |
| 6 | (DTA or (diagnos* adj2 accura*)).tw,kf. |
| 7 | Validation Study/ |
| 8 | “Sensitivity and Specificity”/ |
| 9 | specificity.tw,kf. |
| 10 | Receiver Operating Characteristic/ |
| 11 | Reliability/ |
| 12 | Internal Validity/ |
| 13 | Internal Consistency/ |
| 14 | (validat* or validity).tw,kf. |
| 15 | likelihood ratio*.tw,kf. |
| 16 | predictive value/ |
| 17 | (predict* adj4 val*).tw,kf. or predict*.ti. |
| 18 | ((re-test or retest or test-retest) adj reliability).tw,kf. |
| 19 | diagnostic error/ or false negative result/ or false positive result/ or missed diagnosis/ |
| 20 | (false adj (positiv* or negativ*)).tw,kf. |
| 21 | receiver operating characteristic*.tw,kf. |
| 22 | ROC.tw,kf. |
| 23 | area under the curve/ |
| 24 | observer variation/ |
| 25 | (observer adj variation*).tw,kf. |
| 26 | ((degree? or rate* or rating) adj3 agreement?).tw,kf. |
| 27 | Diagnosis/ |
| 28 | diagnos*.ti,kf. |
| 29 | (diagnos* adj3 (analys* or assay* or immunoassay* or classif* or differenti* or method* or kit or kits or panel? or predict* or screen* or system* or technique* or test*)).ab. |
| 30 | diagnostic procedure/ or diagnostic test/ or diagnostic test approval/ or exp diagnostic kit/ or diagnosis time/ |
| 31 | laboratory diagnosis/ |
| 32 | molecular diagnosis/ |
| 33 | ((accura* or reliab* or valid*) and (point-of-care or POC or (rapid adj2 (analys* or assay* or immunoassay* or classif* or detect* or diagnos* or differenti* or predict* or technique* or test*)))).tw,kf. |
| 34 | ((accura* or reliab* or valid*) and (bacteri* and (viral or virus*) and (analys* or assay* or immunoassay* or classif* or detect* or diagnos* or differenti* or predict* or technique* or test*))).tw,kf. |
| 35 | “quality assessment of diagnostic accuracy studies”/ |
| 36 | QUADAS*.mp. |
| 37 | differential diagnosis/ |
| 38 | (codetect* or co-detect* or codiagnos* or co-diagnos*).tw,kf. |
| 39 | ((discriminat* or differenti* or dual*) adj (detect* or diagnos*)).mp. |
| 40 | (bacteri* adj5 (viral or virus*) adj5 (analys* or assay* or immunoassay* or classif* or detect* or codetect* or determin* or diagnos* or codiagnos* or differenti* or discriminat* or distinguish* or identif* or method* or misdiagnos* or predict* or kit or kits or panel? or predict* or rapid or routine* or screen* or system* or technique* or test*)).tw,kf,hw. |
| 41 | or/1-40 |
| 42 | (((WBC or white blood cell? or white cell? or lymphocyte? or leukocyte? or monocyte? or CD4* or eosinophil? or neutrophil? or granulocyte?) adj3 (count* or distribution? or level? or number* or paramet* or ratio?)) or NLR).tw,kf. |
| 43 | (respiratory or (ear nose adj2 throat) or ENT or otorhinolaryng* or RTI or LRTI or URTI or ARTI or AURI or ALRI or airway* or bronchopulmonar* or broncho-pulmonar* or tracheobronch* or tracheo-bronch* or pulmonary tract or ((chest or lung or lungs or lobar or pleura*) and (absces* or infect* or coinfect* or inflamm*)) or bronchit* or bronchiolit* or bronchopneumon* or common cold or coryza or croup or empyem* or epipharyngit* or epiglottit* or epiglotit* or flu or influenza or laryngit* or laryngotracheobronchit* or (laryngo and tracheo and bronchit*) or (laryngo and tracheobronchit*) or laryngotracheit* or nasopharyngit* or otitis media or parainfluenza or pharyngit* or pleurisy or pneumoni* or pleuropneumoni* or rhinit* or rhinopharyngit* or rhinosinusit* or sinusit* or sore throat or (throat and infection*) or supraglottit* or supraglotit* or tonsillit* or tonsilit* or tracheit* or whooping cough or pertussis or pertussis or asthma* or COPD or COAD or chronic obstructive pulmonary disease or chronic obstructive airway disease or chronic obstructive airways disease or chronic obstructive lung disease or ((acute or subacute* or exacerbat* or prolonged) and cough*)).ti. |
| 44 | 41 and 42 and 43 |
| 45 | (differential diagnos* or codetect* or co-detect*).mp. |
| 46 | ((bacter* or bacilli* or bacili* or corynebac* or mycobac* or nonvir*) and (nonbacter* or viral* or virus* or adenovir*)).mp. |
| 47 | 45 or 46 |
| 48 | 44 and 47 |
| 49 | (((WBC or white blood cell? or white cell? or lymphocyte? or leukocyte? or monocyte? or CD4* or eosinophil? or neutrophil? or granulocyte?) and (count* or distribution? or level? or number* or paramet* or ratio?)) or NLR).ti. |
| 50 | 43 and 47 and 49 |
| 51 | 48 or 50 |
| 52 | (COVID19 or COVID-19 or COVID2019 or COVID-2019 or 2019 nCoV or 2019nCoV or 2019-novel CoV or “SARS-CoV-2” or “SARS-CoV2” or SARSCoV2 or “SARSCoV-2” or 2019 nCoV or 2019nCoV or 2019-novel CoV or “SARS coronavirus 2” or “Severe Acute Respiratory Syndrome Coronavirus-2”).ti. |
| 53 | 51 not 52 |
| 54 | limit 53 to conference abstract status |
| 55 | 53 not 54 |
| 56 | ((neonat* or infant* or child* or p?ediatri*) not adult*).ti. |
| 57 | 55 not 56 |
Multiplex PCR
Database: Ovid MEDLINE(R) ALL <1946 to June 27, 2023> Final search strategy
| 1 | [Target Conditions: RTI] |
| 2 | exp Respiratory Tract Infections/ |
| 3 | exp Otorhinolaryngologic Diseases/ |
| 4 | ((airway* or bronchopulmonar* or broncho-pulmonar* or tracheobronch* or tracheo-bronch* or pulmonar* tract or pulmonary or respirat* tract or respiratory or (ear adj3 nose adj3 throat) or ENT or otorhinolaryng*) adj3 (infect* or coinfect* or inflamm*)).tw,kf. |
| 5 | ((chest or lung? or lobar or pleura?) adj3 (absces* or infect* or coinfect* or inflamm*)).tw,kf. |
| 6 | (bronchit* or bronchiolit* or allergic bronchopulmon* or bronchopneumon* or common cold* or coryza or croup or empyem* or epipharyngit* or epiglottit* or epiglotit* or flu or influenza or laryngit* or laryngotracheobronchit* or laryngo tracheo bronchit* or laryngo tracheobronchit* or laryngotracheit* or nasopharyngit* or otitis media or parainfluenza or pharyngit* or pleurisy or pneumoni* or pleuropneumoni* or rhinit* or rhinopharyngit* or rhinosinusit* or severe acute respiratory syndrome or SARS or sinusit* or sore throat* or throat infection* or supraglottit* or supraglotit* or tonsillit* or tonsilit* or tracheit* or whooping cough or pertussis or pertusis).mp. |
| 7 | ((acute* or exacerbat* or flare*) adj3 (asthma* or copd or coad or chronic obstructive pulmonary disease or chronic obstructive airway* disease or chronic obstructive lung disease)).mp. |
| 8 | ((acute* or subacute* or exacerbat* or prolonged) adj3 cough*).mp. |
| 9 | (RTI or LRTI or URTI or ARTI or AURI or ALRI).tw,kf. |
| 10 | or/2-9 |
| 11 | exp Respiratory System/ and (exp Viruses/ or exp Virus Diseases/) |
| 12 | exp pneumonia, viral/ or *orthomyxoviridae infections/ or influenza, human/ |
| 13 | ((airway* or respiratory or pulmonary or bronchopulmonar* or broncho-pulmonar* or tracheobronch* or tracheo-bronch* or (ear adj3 nose adj3 throat) or ENT or otorhinolaryng*) adj3 (nonbacter* or viral* or virus* or adenovir*)).tw,kf. |
| 14 | (rhinovir* or rhino* vir* or coryzavir* or coryza* vir* or influenzavir* or influenza* vir* or (H1N1 or H3N2) or parainfluenzavir* or parainfluenza* vir* or pneumovir* or pneumo* vir* or human metapneumovir* or human meta-pneumovir* or HMPV or respiratory syncytial vir*).mp. or RSV.tw,kf. |
| 15 | or/11-14 |
| 16 | exp Respiratory System/ and (exp Bacteria/ or exp Bacterial Infections/) |
| 17 | pneumonia, bacterial/ or chlamydial pneumonia/ or pneumonia, mycoplasma/ or pneumonia, pneumococcal/ or pneumonia, staphylococcal/ |
| 18 | ((airway* or respiratory or pulmonary or bronchopulmonar* or broncho-pulmonar* or tracheobronch* or tracheo-bronch* or (ear adj3 nose adj3 throat) or ENT or otorhinolaryng*) adj3 (bacter* or bacilli* or bacili* or corynebac* or mycobac* or nonvir* or pathogen*)).tw,kf. |
| 19 | (strep* pneumon* or diplococ* pneumon* or pneumococ* or staph* pneumon* or chlamyd* pneumon* or myco* pneumon* or influenza bacil* or bacteri* influenza* or h?emophil* influenza*).mp. |
| 20 | ((strep* adj3 (throat* or pharyn* or tonsil*)) or (strep* and (airway* or pulmonary or brochopulmonar* or brocho-pulmonar* or respiratory* or (ear adj3 nose adj3 throat) or ENT or Otorhinolaryng*))).mp. |
| 21 | (GABHS or (“group a” adj3 strep*)).tw,kf. |
| 22 | strep* pyogen*.mp. |
| 23 | or/16-22 |
| 24 | 10 or 15 or 23 |
| 25 | [Index Tests: Rapid Multiplex Tests] |
| 26 | (multiplex* and “sample to answer”).mp. |
| 27 | 24 and 26 |
| 28 | (maripoc* or mari-poc*).af. |
| 29 | (Rapid* and Diagnostic* and (MiniLab* or mini-lab*)).af. |
| 30 | (QIAstat* or QIA-stat* or (Qiagen* and (Resp* adj3 panel))).af. |
| 31 | (Biofire* Respiratory or Biofire* RP*).af. |
| 32 | (BioFire* adj (FilmArray* or Film-Array) adj (Respiratory Panel? or RP*)).af. |
| 33 | (Biofire* adj (FilmArray* or Film-Array*) adj Pneumo*).af. |
| 34 | (Biofire* adj (FilmArray* or Film-Array*)).ti. |
| 35 | (Biofire* and “sample to answer”).mp. |
| 36 | (Biofire* adj5 (rapid or real time or RT-PCR or rRT-PCR)).mp. |
| 37 | (34 or 35 or 36) and 24 |
| 38 | (Spotfire* or Spot-fire*).af. |
| 39 | 24 and 38 |
| 40 | (Cobas* adj5 ((lab* adj3 tube*) or liat*)).af. |
| 41 | 24 and 40 |
| 42 | (cobas* Influenza A* or cobas* Influenza B* or cobas* RSV or cobas* respiratory sync* virus).af. |
| 43 | ((Cepheid* adj3 GeneXpert* adj3 Xpress*) or (Cepheid* adj3 Gene-Xpert* adj3 Xpress*)).af. |
| 44 | (Xpert* adj3 Xpress* adj3 (influenza or flu or respiratory sync* virus or RSV)).af. |
| 45 | (Cepheid* adj3 Xpert* adj3 (influenza or flu or respiratory sync* virus or RSV)).af. |
| 46 | (ePlex* RP* or (ePlex* adj3 resp* adj3 panel?)).af. |
| 47 | ePlex*.af. |
| 48 | 24 and 47 |
| 49 | ((GenMark* or Gen-Mark*) and (RP* or (resp* adj3 panel?))).af. |
| 50 | (Simplexa* or Liaison* MDX*).af. |
| 51 | 24 and 50 |
| 52 | Aries*.mp. not (sheep or lamb or lambs or ram or rams or ewe or ewes or ovine or ovis aries).ti. |
| 53 | 24 and 52 |
| 54 | (Savanna* and (quidel* or molecular or multiplex* or rapid or real-time or RTPCR or RT-PCR or rRTPCR or rRT-PCR or test? or device? or panel? or PoCT or Point-of-Care or near-patient?)).mp. |
| 55 | 24 and 54 |
| 56 | ((RVP4* or RVP-4*) and (Savanna* or Quidel* or molecular or multiplex* or rapid or real-time or RTPCR or RT-PCR or rRTPCR or rRT-PCR or test? or device? or panel? or PoCT or Point-of-Care or near-patient?)).mp. |
| 57 | (Respiratory Vir* Panel4* or Respiratory Vir* Panel-4*).af. |
| 58 | Verigen*.af. |
| 59 | 24 and 58 |
| 60 | Panther* Fusion*.af. |
| 61 | 24 and 60 |
| 62 | “Flu A/B/RSV*”.af. |
| 63 | “AdV/hMPV/RV*”.af. |
| 64 | “SARS-CoV-2/Flu A/B*”.af. |
| 65 | “SARS-CoV-2/Flu A/B/RSV*”.af. |
| 66 | (paraflu or parafluTM or parafluR).af. |
| 67 | 27 or 28 or 29 or 30 or 31 or 32 or 33 or 37 or 39 or 41 or 42 or 43 or 44 or 45 or 46 or 48 or 49 or 51 or 53 or 55 or 56 or 57 or 59 or 61 or 62 or 63 or 64 or 65 or 66 |
| 68 | ((COVID19 or COVID-19 or COVID2019 or COVID-2019 or 2019 nCoV or 2019nCoV or 2019-novel CoV or “SARS-CoV-2” or “SARS-CoV2” or SARSCoV2 or “SARSCoV-2” or 2019 nCoV or 2019nCoV or 2019-novel CoV or “SARS coronavirus 2” or “Severe Acute Respiratory Syndrome Coronavirus-2”) not (rhinovir* or rhino* vir* or coryzavir* or coryza* vir* or influenzavir* or influenza* vir* or (H1N1 or H3N2) or parainfluenzavir* or parainfluenza* vir* or pneumovir* or pneumo* vir* or human metapneumovir* or human meta-pneumovir* or HMPV or respiratory sync* vir* or RSV)).ti. |
| 69 | 67 not 68 |
| 70 | ((“SARS-CoV-2” or “SARS-CoV2” or SARSCoV2 or “SARSCoV-2” or “SARS coronavirus 2” or “Severe Acute Respiratory Syndrome Coronavirus-2”) adj3 Flu* adj3 RSV).af. |
| 71 | 69 or 70 |
| 72 | ((neonat* or infant* or child* or p?ediatri*) not adult*).ti. |
| 73 | 71 not 72 |
Database: EMBASE <1974 to 2023 June 27> Final search strategy
| 1 | [Target Conditions: RTI] |
| 2 | respiratory tract infection/ or exp influenza/ or laryngotracheobronchitis/ or parainfluenza virus infection/ or respiratory syncytial virus infection/ or viral respiratory tract infection/ or lower respiratory tract infection/ or chest infection/ or pertussis/ or lung infection/ or exp infectious pneumonia/ or lung abscess/ or exp lung mycosis/ or exp viral bronchiolitis/ or upper respiratory tract infection/ or exp nose infection/ or oropharynx candidiasis/ or peritonsillar abscess/ or viral upper respiratory tract infection/ |
| 3 | ear nose throat disease/di or otorhinolaryngology/ or exp ear infection/ or exp otitis/ |
| 4 | ((airway* or bronchopulmonar* or broncho-pulmonar* or tracheobronch* or tracheo-bronch* or pulmonar* tract or pulmonary or respirat* tract or respiratory or (ear adj3 nose adj3 throat) or ENT or otorhinolaryng*) adj3 (infect* or coinfect* or inflamm*)).tw,kf. |
| 5 | ((chest or lung? or lobar or pleura?) adj3 (absces* or infect* or coinfect* or inflamm*)).tw,kf. |
| 6 | (bronchit* or bronchiolit* or allergic bronchopulmon* or bronchopneumon* or common cold* or coryza or croup or empyem* or epipharyngit* or epiglottit* or epiglotit* or flu or influenza or laryngit* or laryngotracheobronchit* or laryngo tracheo bronchit* or laryngo tracheobronchit* or laryngotracheit* or legionnair* disease or legionellos* or middle east respiratory syndrome or MERS or nasopharyngit* or otitis media or parainfluenza or pharyngit* or pleurisy or pneumoni* or pleuropneumoni* or rhinit* or rhinopharyngit* or rhinosinusit* or severe acute respiratory syndrome or SARS or sinusit* or sore throat* or throat infection* or supraglottit* or supraglotit* or tonsillit* or tonsilit* or tracheit* or whooping cough or pertussis or pertusis).mp. |
| 7 | ((acute* or exacerbat* or flare*) adj3 (asthma* or copd or coad or chronic obstructive pulmonary disease or chronic obstructive airway* disease or chronic obstructive lung disease)).mp. |
| 8 | ((acute* or subacute* or exacerbat* or prolonged) adj3 cough*).mp. |
| 9 | (RTI or LRTI or URTI or ARTI or AURI or ALRI).tw,kf. |
| 10 | or/2-9 |
| 11 | exp respiratory system/ and exp virus infection/ |
| 12 | ((airway* or respiratory or pulmonary or bronchopulmonar* or broncho-pulmonar* or tracheobronch* or tracheo-bronch* or (ear adj3 nose adj3 throat) or ENT or otorhinolaryng*) adj3 (nonbacter* or viral* or virus* or adenovir*)).tw,kf. |
| 13 | rhinovirus/ or exp human rhinovirus/ or exp rhinovirus infection/ |
| 14 | exp Influenza virus/ or orthomyxovirus infection/ |
| 15 | respirovirus/ or human parainfluenza virus 1/ or human parainfluenza virus 3/ or respirovirus infection/ |
| 16 | exp virus pneumonia/ |
| 17 | pneumovirus/ or pneumovirus infection/ or exp human respiratory syncytial virus/ or respiratory syncytial virus infection/ |
| 18 | metapneumovirus/ or metapneumovirus infection/ or human metapneumovirus/ or human metapneumovirus infection/ |
| 19 | (rhinovir* or rhino* vir* or coryzavir* or coryza* vir* or influenzavir* or influenza* vir* or (H1N1 or H3N2) or parainfluenzavir* or parainfluenza* vir* or pneumovir* or pneumo* vir* or human metapneumovir* or human meta-pneumovir* or HMPV or respiratory sync* vir*).mp. or RSV.tw,kf. |
| 20 | or/11-19 |
| 21 | exp respiratory system/ and (exp bacterium/ or exp bacterial Infection/) |
| 22 | ((airway* or respiratory or pulmonary or bronchopulmonar* or broncho-pulmonar* or tracheobronch* or tracheo-bronch* or (ear adj3 nose adj3 throat) or ENT or otorhinolaryng*) adj3 (bacter* or bacilli* or bacili* or corynebac* or mycobac* or nonvir* or pathogen*)).tw,kf. |
| 23 | bacterial pneumonia/ or chlamydial pneumonia/ or mycoplasma pneumonia/ or staphylococcal pneumonia/ or exp streptococcus pneumonia/ |
| 24 | (strep* pneumon* or diplococ* pneumon* or pneumococ* or staph* pneumon* or chlamyd* pneumon* or myco* pneumon* or influenza bacil* or bacteri* influenza* or h?emophil* influenza*).mp. |
| 25 | ((strep* adj3 (throat* or pharyn* or tonsil*)) or (strep* and (airway* or pulmonary or brochopulmonar* or brocho-pulmonar* or respiratory* or (ear adj3 nose adj3 throat) or ENT or Otorhinolaryng*))).mp. |
| 26 | streptococcus infection/ or streptococcus group a/ or exp group a streptococcal infection/ or streptococcal pharyngitis/ |
| 27 | (GABHS or (“group a” adj3 strep*)).tw,kf. |
| 28 | strep* pyogen*.mp. |
| 29 | or/21-28 |
| 30 | 10 or 20 or 29 |
| 31 | [DTA Filter] |
| 32 | Gold Standard/ |
| 33 | (reference standard? or gold standard?).tw,kf. |
| 34 | Diagnostic Test Accuracy Study/ |
| 35 | Diagnostic Accuracy/ |
| 36 | (DTA or (diagnos* adj2 accura*)).tw,kf. |
| 37 | Validation Study/ |
| 38 | “Sensitivity and Specificity”/ |
| 39 | (sensitivity or specificity).tw,kf. |
| 40 | Receiver Operating Characteristic/ |
| 41 | Reliability/ |
| 42 | Internal Validity/ |
| 43 | Internal Consistency/ |
| 44 | (validat* or validity).tw,kf. |
| 45 | likelihood ratio*.tw,kf. |
| 46 | predictive value/ |
| 47 | (predict* adj4 val*).tw,kf. or predict*.ti. |
| 48 | ((re-test or retest or test-retest) adj reliability).tw,kf. |
| 49 | diagnostic error/ or false negative result/ or false positive result/ or missed diagnosis/ |
| 50 | (false adj (positiv* or negativ*)).tw,kf. |
| 51 | receiver operating characteristic*.tw,kf. |
| 52 | ROC.tw,kf. |
| 53 | area under the curve/ |
| 54 | observer variation/ |
| 55 | (observer adj variation*).tw,kf. |
| 56 | ((degree? or rate* or rating) adj3 agreement?).tw,kf. |
| 57 | ((detect* or diagnos*) and agreement?).tw,kf. |
| 58 | diagnostic.ti,kf. |
| 59 | (diagnos* adj3 (classif* or differenti* or predict* or rapid* or RT-PCR or rRT-PCR)).ab. |
| 60 | diagnostic test approval/ or diagnosis time/ |
| 61 | laboratory diagnosis/ |
| 62 | molecular diagnosis/ |
| 63 | ((accura* or reliab* or valid*) and (point-of-care or POC or (rapid adj2 (analys* or assay* or immunoassay* or classif* or detect* or diagnos* or differenti* or predict* or technique* or test*)))).tw,kf. |
| 64 | ((accura* or reliab* or valid*) and (bacteri* and (viral or virus*) and (analys* or assay* or immunoassay* or classif* or detect* or diagnos* or differenti* or predict* or technique* or test*))).tw,kf. |
| 65 | “quality assessment of diagnostic accuracy studies”/ |
| 66 | (QUADAS* or STARD).mp. |
| 67 | differential diagnosis/ |
| 68 | (codetect* or co-detect* or codiagnos* or co-diagnos*).tw,kf. |
| 69 | ((discriminat* or differenti* or dual*) adj (detect* or diagnos*)).mp. |
| 70 | (bacteri* adj5 (viral or virus*) adj5 (analys* or assay* or immunoassay* or classif* or detect* or codetect* or determin* or diagnos* or codiagnos* or differenti* or discriminat* or distinguish* or identif* or method* or misdiagnos* or predict* or kit or kits or panel? or predict* or rapid or routine* or screen* or system* or technique* or test*)).tw,kf,hw. |
| 71 | “sample to answer”.mp. |
| 72 | or/32-71 |
| 73 | [Index Tests: Rapid Multiplex PCR] |
| 74 | rapid test/dc |
| 75 | (multiplex* and “sample to answer”).mp. |
| 76 | (74 or 75) and 30 |
| 77 | (maripoc* or mari-poc*).mp,ct,dv,dc,dm,mv,my,tn. |
| 78 | (Rapid* and Diagnostic* and (MiniLab* or mini-lab*)).mp,ct,dv,dc,dm,mv,my,tn. |
| 79 | (QIAstat* or QIA-stat* or (Qiagen* and (Resp* adj3 panel))).mp,ct,dv,dc,dm,mv,my,tn. |
| 80 | Biofire* Respiratory.mp,ct,dv,dc,dm,mv,my,tn. |
| 81 | BioFire* RP*.mp,ct,dv,dc,dm,mv,my,tn. |
| 82 | (Biofire* and “sample to answer”).mp,ct,dv,dc,dm,mv,my,tn. |
| 83 | (Biofire* adj5 (rapid or real time or RT-PCR or rRT-PCR)).mp,ct,dv,dc,dm,mv,my,tn. |
| 84 | or/77-83 |
| 85 | (BioFire* adj (FilmArray* or Film-Array) adj (Respiratory Panel? or RP*)).mp,ct,dv,dc,dm,mv,my,tn. |
| 86 | (Biofire* adj (FilmArray* or Film-Array*) adj Pneumonia).mp,ct,dv,dc,dm,mv,my,tn. |
| 87 | (85 or 86) and 72 |
| 88 | (Biofire* adj (FilmArray* or Film-Array*)).ti. |
| 89 | 88 and 30 and 72 |
| 90 | (Spotfire* or Spot-fire*).mp,ct,dv,dc,dm,mv,my,tn. |
| 91 | 90 and (30 or 72) |
| 92 | (Cobas* adj5 ((lab* adj3 tube*) or liat*)).mp,ct,dv,dc,dm,mv,my,tn. |
| 93 | (cobas* Influenza A* or cobas* Influenza B* or cobas* RSV or cobas* respiratory sync* virus).mp,ct,dv,dc,dm,mv,my,tn. |
| 94 | (92 and 30 and 72) or 93 |
| 95 | (Xpert* adj3 Xpress* adj3 (influenza or flu or respiratory sync* virus or RSV)).mp,ct,dv,dc,dm,mv,my,tn. |
| 96 | (Cepheid* adj3 Xpert* adj3 (influenza or flu or respiratory sync* virus or RSV)).mp,ct,dv,dc,dm,mv,my,tn. |
| 97 | ((Cepheid* adj3 GeneXpert* adj3 Xpress*) or (Cepheid* adj3 Gene-Xpert* adj3 Xpress*)).mp,ct,dv,dc,dm,mv,my,tn. |
| 98 | ((95 or 96) and 72) or 97 |
| 99 | (ePlex* RP* or (ePlex* adj3 resp* adj3 panel?)).mp,ct,dv,dc,dm,mv,my,tn. |
| 100 | ePlex*.mp,ct,dv,dc,dm,mv,my,tn. |
| 101 | (100 and 72) or 99 |
| 102 | ((GenMark* or Gen-Mark*) and (RP* or (resp* adj3 panel?))).mp,ct,dv,dc,dm,mv,my,tn. |
| 103 | 102 and 72 |
| 104 | 76 or 84 or 87 or 89 or 91 or 94 or 98 or 101 or 103 |
| 105 | (Simplexa* or Liaison* MDX*).mp,ct,dv,dc,dm,mv,my,tn. |
| 106 | 105 and 30 and 72 |
| 107 | Aries*.mp,ct,dv,dc,dm,mv,my,tn. |
| 108 | (sheep or lamb or lambs or ram or rams or ewe or ewes or ovine or ovis aries).ti. |
| 109 | 107 not 108 |
| 110 | 109 and 30 and 72 |
| 111 | (Savanna* and (quidel* or molecular or multiplex* or rapid or real-time or RTPCR or RT-PCR or rRTPCR or rRT-PCR or test? or device? or panel? or PoCT or Point-of-Care or near-patient?)).mp,ct,dv,dc,dm,mv,my,tn. |
| 112 | ((RVP4* or RVP-4*) and (Savanna* or Quidel* or molecular or multiplex* or rapid or real-time or RTPCR or RT-PCR or rRTPCR or rRT-PCR or test? or device? or panel? or PoCT or Point-of-Care or near-patient?)).mp,ct,dv,dc,dm,mv,my,tn. |
| 113 | (respiratory vir* Panel4* or respiratory vir* Panel-4*).mp,ct,dv,dc,dm,mv,my,tn. |
| 114 | (111 or 112 or 113) and 30 |
| 115 | Verigen*.mp,ct,dv,dc,dm,mv,my,tn. |
| 116 | 115 and 30 and 72 |
| 117 | Panther* Fusion*.mp,ct,dv,dc,dm,mv,my,tn. |
| 118 | 117 and 30 and 72 |
| 119 | Paraflu*.mp,ct,dv,dc,dm,mv,my,tn. |
| 120 | 119 and 72 |
| 121 | “Flu A/B/RSV*”.mp,ct,dv,dc,dm,mv,my,tn. |
| 122 | “AdV/hMPV/RV*”.mp,ct,dv,dc,dm,mv,my,tn. |
| 123 | 106 or 110 or 114 or 116 or 118 or 120 or 121 or 122 |
| 124 | 104 or 123 |
| 125 | ((COVID19 or COVID-19 or COVID2019 or COVID-2019 or 2019 nCoV or 2019nCoV or 2019-novel CoV or “SARS-CoV-2” or “SARS-CoV2” or SARSCoV2 or “SARSCoV-2” or 2019 nCoV or 2019nCoV or 2019-novel CoV or “SARS coronavirus 2” or “Severe Acute Respiratory Syndrome Coronavirus-2”) not (rhinovir* or rhino* vir* or coryzavir* or coryza* vir* or influenzavir* or influenza* vir* or (H1N1 or H3N2) or parainfluenzavir* or parainfluenza* vir* or pneumovir* or pneumo* vir* or human metapneumovir* or human meta-pneumovir* or HMPV or respiratory sync* vir* or RSV)).ti. |
| 126 | 124 not 125 |
| 127 | “SARS-CoV-2/Flu A/B*”.mp,ct,dv,dc,dm,mv,my,tn. |
| 128 | “SARS-CoV-2/Flu A/B*”.mp,ct,dv,dc,dm,mv,my,tn. |
| 129 | ((“SARS-CoV-2” or “SARS-CoV2” or SARSCoV2 or “SARSCoV-2” or “SARS coronavirus 2” or “Severe Acute Respiratory Syndrome Coronavirus-2”) adj3 Flu* adj3 RSV).mp,ct,dv,dc,dm,mv,my,tn. |
| 130 | or/126-129 |
| 131 | ((neonat* or infant* or child* or p?ediatri*) not adult*).ti. |
| 132 | 130 not 131 |
| 133 | limit 132 to conference abstract status |
| 134 | 132 not 133 |
Appendix 2
| Reference | Eligibility criteria | Databases searched (search date) | Tool used to assess the validity of primary studies | Total number of studies included in the review | Index test | Target condition | Number of studies included in most relevant analysis | Population | Clinical features | Setting | Funding/conflicts of interest | Notes |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bruning 201763 | ‘. . . all available rapid tests for the detection of respiratory viruses in patients of all ages with RTIs’ ‘Studies were considered for inclusion if they were written in English or Dutch and reported original data regarding the accuracy of a rapid test for ≥ 1 respiratory virus compared with PCR’ |
MEDLINE and EMBASE (January 2016) | QUADAS-2 | 179 | Any rapid test | RSV | 2 | Adults and children | Not stated | Not stated | Funded by EU’s Seventh Framework People Programme. The authors declare no conflicts of interest |
Both studies for RSV in mixed population. Excluded, as data superseded by more recent reviews (Gentilotti 202233 and Onwuchekwa 202335) |
| Any rapid test | Influenza A and/or B | 11 | Adults | Not stated | Not stated | |||||||
| Carlton 202132 | ‘Our review included diagnostic accuracy studies, reporting on point-of-care and rapid diagnostic tests consisting of more-than-one biomarker to identify bacterial or viral aetiology, in the general population presenting to primary or secondary care with acute RTI symptoms’ | MEDLINE, EMBASE, Web of Science (February 2021) | QUADAS-2 | 20 | Immuno-Xpert (TRAIL, IP-10 and CRP) | Bacterial or viral | 4 | Adults and children | Features of acute RTI | ‘… the general population presenting to primary or secondary care’ | Conducted as part of lead author’s undergraduate research project, without dedicated funding. Other authors’ time supported by NIHR ARC West. The authors declare no conflicts of interest |
Included |
| FebriDx (CRP and MxA) | Bacterial or viral | 4 | Adults and children | Features of acute RTI | ‘… the general population presenting to primary or secondary care’ | |||||||
| CRP and neopterin | Bacterial or viral | 1 | Adults | Features of acute RTI | ‘… the general population presenting to primary or secondary care’ | |||||||
| Chartrand 201264 | ‘Studies were included if they assessed the accuracy of an RIDT [rapid influenza diagnostic test] against 1 of the 2 accepted reference standards. [. . .] Acceptable reference standards included viral culture or RT-PCR’ | PubMed, EMBASE, BIOSIS and Web of Science (December 2011) | QUADAS | 159 | Any rapid test | Influenza A and/or B | 17 | Adults | Not stated | Not stated | Supported in part by the Canadian Institutes of Health Research Authors declare no conflicts of interest |
Superseded by more recent review (Gentilotti 202233) |
| Chartrand 201565 | ‘Studies were considered for inclusion if they assessed the diagnostic accuracy of a commercial rapid immunoassay for RSV in patients with suspected ARI’ | PubMed and EMBASE (April 2015) | QUADAS-2 | 71 | Any rapid test | RSV | 4 | Adults | People with suspected ARI | Any setting | No funding information or conflicts of interest reported | Not specific to primary/emergency care settings Superseded by more recent review (Onwuchekwa 202335) |
| Engel 201266 | ‘Studies using adult patients (> 16 years of age) consulting their GP with a probable LRTI were included if CRP was measured in (a part) of those patients’ | MEDLINE, EMBASE and the Cochrane Library (July 2010) | QUADAS and the ‘Cochrane Validity Score’ | 10 | CRP | Bacterial LRTI and pneumonia | Narrative synthesis of 5 relevant articles | Adults (> 16 years) | Suspected LRTI. People with URTI/confirmed pneumonia were excluded | Primary care | No funding received. The authors declare no conflicts of interest. |
No summary data are reported. Superseded by Gentilotti 202233 |
| Falk 200867 | ‘Population – participants in each study were to be recruited from a community, primary care setting or ambulatory setting, for example emergency departments, and have symptoms suggestive of acute respiratory infection suggestive of LRTI’ | PubMed, EMBASE, Google Scholar, the Cochrane database and the MEDION database (July 2008) | QUADAS | 8 | CRP | Pneumonia | 5–6 depending on threshold used | Adults (over 14 years) | ARI | Community and emergency care | Funded by Irish College of General Practitioners and Health Research Board Authors declare no conflicts of interest |
Superseded by Gentilotti 202233 |
| Gentilotti 202233 | ‘ … all the diagnostic test accuracy (DTA) studies [. . .] on patients of any age were eligible for inclusion.’ | PubMed, Web of Science, the Cochrane Library, EMBASE and Open Gray (May 2021) | QUADAS-2 | 421 | Symptoms and signs | Bacterial pneumonia | Between 4 and 26 studies, depending on symptoms/sign | Adults | Suspected LRTI | Community/emergency care settings | Funded by Innovative Medicines Initiative-2 Joint Undertaking | Included |
| CRP | Pneumonia or bacterial pneumonia | 4–6 (depending on threshold used) | Adults | Suspected LRTI | Community/emergency care settings | |||||||
| Procalcitonin | Pneumonia or bacterial pneumonia | 2–4 (depending on threshold used) | Adults | Suspected LRTI | Community/emergency care settings | The joint undertaking receives support from various pharmaceutical companies | ||||||
| Supplementary information: ‘A community-care setting was defined as the first point of contact with health services, including PC [primary care], | Immunochromatographic assay | Influenza A and/or B | 15 | Adults | Suspected LRTI | Community/emergency care settings | ||||||
| Direct immunofluorescence | Influenza A and/or B | 19 | Mixed adults and children | Suspected LRTI | Community/emergency care settings | The authors note that the commercial companies had no part in the design, analysis, writing or decision to publish the results. | ||||||
| Optical immunoassay | Influenza A and/or B | 9 | Mixed adults and children | Suspected LRTI | Community/emergency care settings | |||||||
| Chemiluminescent neuraminidase assay | Influenza A and/or B | 4 | Mixed adults and children | Suspected LRTI | Community/emergency care settings | |||||||
| LTCF [long term care facilities], OC [outpatient clinic], and ER [emergency room]. | PCR-based NAAT | Influenza A and/or B | 6 | Adults | Suspected LRTI | Community/emergency care settings | The authors declare no conflicts of interest | |||||
| Non-PCR- based NAAT | Influenza A and/or B | 2 | Mixed adults and children | Suspected LRTI | Community/emergency care settings | |||||||
| Rapid antigen detection test | RSV | 35 | Mixed adults and children | Suspected LRTI | Community/emergency care settings | |||||||
| PCR-based NAAT | RSV | 38 | Mixed adults and children | Suspected LRTI | Community/emergency care settings | |||||||
| PoCT was defined as a test to support clinical decision-making (signs and symptoms or imaging or host biomarkers or pathogen-based tests), which is performed on any part of the patient’s body or clinical samples, during or close to the time of consultation’ | Non-PCR- based NAAT | RSV | 5 | Mixed adults and children | Suspected LRTI | Community/emergency care settings | ||||||
| Hill 201968 | Adult outpatients with acute cough due to suspected pneumonia | PubMed, Scopus and the Cochrane Library (March 2017) | QUADAS and DART | Not stated | CRP | Pneumonia | Narrative synthesis of 6 articles | Adults | Suspected pneumonia | Not stated | No funding was received for the study. One author (RSI) reports they are Editor in Chief of the publishing journal. Remaining authors declare no conflicts of interest |
Superseded by Gentilotti 202233 |
| Procalcitonin | Pneumonia | Narrative synthesis of 6 articles | Adults | Suspected pneumonia | Not stated | |||||||
| Symptoms and signs | Pneumonia | Narrative synthesis of 2 articles | Adults | Suspected pneumonia | Not stated | |||||||
| Han 202069 | Diagnostic test accuracy studies of lateral flow assays for influenza with at least 40 participants | PubMed, EMBASE, Web of Science and the Cochrane Library (November 2019) | QUADAS-2 | 13 | Any lateral flow assay | Influenza A and/or B | 13 | Mixed adults and children | Not stated | Any | Funding information not reported. Authors declare no conflicts of interest |
Superseded by Gentilotti 202233 |
| Hoult 202270 | ‘Cross-sectional, cohort and randomised controlled studies that describe associations between serum or sputum molecular or cellular biomarkers and evidence of bacterial infection in people with acute exacerbation of COPD were eligible for inclusion’ | EMBASE and MEDLINE (March 2018) | QUADAS-2 | 39 | CRP | Bacterial exacerbation of COPD | Narrative synthesis of 8 articles | Adults with COPD | Not stated | Outpatient, hospitalised inpatients and ICU | No funding received for the study. Several authors report financial support from pharmaceutical companies, for work outside the study |
Excluded as setting not sufficiently similar in scope to this review, and unable to extract relevant data. Procalcitonin studies do not relate to people attending primary/emergency care |
| Procalcitonin | Bacterial exacerbation of COPD | Narrative synthesis of 5 articles | Adults with COPD | People with acute exacerbations of COPD | Hospitalised inpatients and ICU | |||||||
| Htun 201971 | ‘… published studies that assessed clinical predictors of community-acquired pneumonia [. . .]. Studies were included if participants aged ≥ 18 years without serious illness (e.g. mechanical ventilation) and pre-existing immune suppression (HIV, malnutrition, and immunosuppressant medication)’ | PubMed, EMBASE, Cochrane Library (March 2018) | QUADAS-2 | 13 | Symptoms and signs | Pneumonia | Between four and seven studies, depending on symptoms/sign | Adults | Acute respiratory symptoms | Outpatient, primary or emergency care settings | Supported by Centre of Infectious Disease Epidemiology and Research (funded by Singapore Ministry of Defence). The authors declare no conflicts of interest |
Superseded by Gentilotti 202233 |
| CRP | Pneumonia | 9 | Adults | Acute respiratory symptoms | Outpatient, primary or emergency care settings | |||||||
| Procalcitonin | Pneumonia | 4 | Adults | Acute respiratory symptoms | Outpatient, primary or emergency care settings | |||||||
| Huang 201872 | ‘Studies that evaluated the performance of FDA-approved mPCR systems for the detection of viral respiratory infection were included, as follow: (a) they assessed the accuracy of one or more the following systems: FilmArray, Nanosphere Verigene RV+ and Hologic Gen-Probe Prodesse assays [. . .] against reference standards’ | PubMed, EMBASE (July 2017) | QUADAS-2 | 20 | Multiplex PCR | Multiple single pathogens | 22 (influenza A) 13 (influenza B) 13 (RSV) 8 (adenovirus) 8 (hMPV) |
Adults and children | Mixture of symptomatic people and stored samples | Not stated | Supported in part by a National Taiwan University Hospital Research Grant The authors declare no conflicts of interest |
Scope too narrow for inclusion. Review limited to two rapid multiplex tests (and one laboratory- based multiplex test) |
| Lee 202173 | ‘… studies that evaluated the performance of the Quidel Sofia rapid influenza FIA, compared to a reference standard [. . .] studies that included patients with influenza-like illness.’ | MEDLINE, EMBASE and the Cochrane Central Register (July 2020) | QUADAS-2 | 17 | Quidel Sofia rapid influenza fluorescent immunoassay | Influenza A and B | 2 (influenza A) 1 (influenza B) |
Adults | People with influenza-like illness | Not stated | Supported by research grant from the Jeju National University Hospital The authors declare no conflicts of interest |
Scope too narrow. Superseded by Gentilotti 202233 |
| Merckx 201774 | ‘. . . studies [. . .] on the diagnostic accuracy of rapid influenza tests against an RT-PCR reference standard. Eligible participants were children and adults with clinically suspected influenza during periods of influenza activity’ | PubMed, EMBASE, BIOSIS Previews, Scopus, Web of Science and the Cochrane Central Register (May 2017) | QUADAS-2 | 162 | Traditional RIDT | Influenza A and B | 23 (influenza A) 5 (influenza B) |
Adults | Clinically suspected influenza | Mixed primary, emergency and hospital settings | Supported in part by the Quebec Health Research Fund and by an investigator-initiated study grant from BD Diagnostic Systems. Several authors report personal fees from funders and other pharmaceutical companies |
Superseded by Gentilotti 202233 |
| DIA | Influenza A and B | 8 (influenza A) 7 (influenza B) |
Adults | Clinically suspected influenza | Mixed primary, emergency and hospital settings | |||||||
| Rapid NAAT | Influenza A and B | 4 (influenza A) 4 (influenza B) |
Adults | Clinically suspected influenza | Mixed primary, emergency and hospital settings | |||||||
| Minnaard 201734 | ‘All studies on diagnostic accuracy of CRP for pneumonia (e.g. infiltrate on chest radiography as the reference standard) were eligible. Study participants had to be adults (≥ 18 years) suspected by their physician of having a lower respiratory tract infection presenting in a primary health care setting’ | MEDLINE, EMBASE, the Cochrane Library (Not stated. Most recent included study published in 2013) | QUADAS-2 | 8 | CRP and signs and symptoms | Pneumonia | 8 | Adults | Suspected LRTI | Primary and emergency care | Funding information not reported. Several authors report grants received from various sources, including pharmaceutical companies |
Included |
| Nicholson 201475 | ‘. . . publications on influenza PoCT diagnostic accuracy studies between 1991 and 2011 (inclusive) that met the following five criteria:1. Articles written in English.2. Commercially available test kits.3. Testing done in human seasonal and pandemic influenza... ’ | MEDLINE, BIOSIS and the Cochrane Library (May 2011) | QUADAS and STARD | 70 | Any PoCT for influenza | Influenza | 43 | Mixed adults and children | Not stated | Not stated | Funded by the NIHR Health Technology Assessment programme. Lead author previously consultant to GSK and Novartis. Various authors report paid work from pharmaceutical companies |
Superseded by Gentilotti 202233 |
| Onwuchekwa 202335 | ‘… primary studies were eligible if they reported on the diagnostic test performance or compared RSV detection rates using different specimens. We excluded [. . .] studies in children, and in vitro studies’ | EMBASE, MEDLINE, Web of Science (December 2021) | QUADAS-2 | 156 | DFA | RSV | 1 | Adults | Acute exacerbation of asthma | Any setting | Funded by Pfizer. Open access fees paid by Pfizer. Authors employees of Pfizer or P95 (the company contracted by Pfizer to conduct this work) |
Included data on DFA and RADT. Excluded data on multiplex tests, as new review of multiplex tests was conducted |
| RADT | RSV | 1 | Adults | LRTI and URTI | Any setting | |||||||
| Multiplex PCR | RSV | 1 | Adults | LRTI and URTI | Any setting | |||||||
| Pazmany 202136 | ‘(a) adult patients with bacterial and non-bacterial AECOPD; (b) results of microbiology tests (as the reference standard) with samples taken from sputum, tracheal aspirates or blood; and (c) at least one other on-admission diagnostic test performed from serum or sputum(index tests), were considered eligible’ | MEDLINE, EMBASE, CENTRAL, Scopus and Web of Science (October 2019) | QUADAS-2 | 21 | Symptoms and signs (sputum colour only) | Bacterial acute exacerbation of COPD | 3 | Adults | Acute exacerbation of COPD | Any setting | Funded by EU within the framework Programme Széchenyi 2020 and Human Resources Development Operational Programme. The authors declare no conflicts of interest |
Data on sputum included, as predominantly primary care setting. Other data relates to hospitalised participants. Not sufficiently close in scope to this review question (no data relating to outpatient/primary/emergency settings) |
| CRP | Bacterial acute exacerbation of COPD | 9 | Adults | Acute exacerbation of COPD | Any setting | |||||||
| Procalcitonin | Bacterial acute exacerbation of COPD | 8 | Adults | Acute exacerbation of COPD | Any setting | |||||||
| Neutrophil/lymphocyte ratio | Bacterial acute exacerbation of COPD | 1 | Adults | Acute exacerbation of COPD | Any setting | |||||||
| Eosinophil % | Bacterial acute exacerbation of COPD | 1 | Adults | Acute exacerbation of COPD | Any setting | |||||||
| Petrozzino 201076 | ‘Articles reporting RFT and clinical diagnostic performance, and effects on decision-making and diagnostic outcomes.’ Adults and children with influenza-like illness |
PubMed/MEDLINE; the Cochrane Library; British Medical Journal Clinical Evidence; Surveillance, Epidemiology and End Results; the World Health Organization website, the Agency for Healthcare Research and Quality website (2009) | US Preventive Services Task Force (USPSTF) evidence-based guidelines for internal validity of diagnostic accuracy studies | 16 | QuickVue RFT | Influenza A and B | 5 | Adults (≥ 15 years) | People presenting with influenza-like illness | Any setting | Supported by the Quidel Corporation, through the Aequitas Group. Several authors report being a consultant/employee of Aequitas during project |
Superseded by Gentilotti 202233 Data on symptoms and signs are outside the scope of the protocol: clinical symptoms and signs for a specific pathogen, rather than bacterial/viral infection |
| Symptoms and signs (clinical assessment) | Influenza A and B | 11 | Adults (≥ 15 years) | People presenting with influenza-like illness | Any setting | |||||||
| Schierenberg 201637 | ‘Models eligible for inclusion were logistic regression models including S&S [signs and symptoms] for predicting the probability of pneumonia in primary care patients with acute cough or suspected LRTI’ | PubMed, EMBASE and the Cochrane Library (August 2012) | QUADAS-2 | 8 | Any clinical prediction rule for pneumonia (signs and symptoms) | Pneumonia | 8 | Adults | Acute or worsened cough or LRTI symptoms | Primary or emergency care | No direct funding received for the study. The authors declare no conflicts of interest |
No summary estimates provided. Included |
| Van der Meer 200577 | ‘We aimed to include studies that compared C reactive protein with a chest radiograph [. . .] or microbiological work-up [. . .]. We excluded articles concerning immunocompromised patients, patients treated in intensive care units, or patients with hospital acquired pneumonia’ | MEDLINE and EMBASE (April 2004) | Lijmer criteria | 17 | CRP | Pneumonia | 5 | Adults | ARI | Primary/emergency care | No funding received. The authors declare no conflicts of interest |
Superseded by Gentilotti 202233 |
| Vos 201978 | Supplementary material: ‘We included peer-reviewed studies in English or Dutch providing original data on the diagnostic accuracy or clinical impact of a molecular rapid test for respiratory viruses, among which at least influenza virus and/or RSV, as compared to (non-rapid) molecular techniques. [. . .] The domain included patients of all ages with suspected (viral) RTI presenting in a hospital setting’ | MEDLINE, EMBASE, Cochrane Library (August 2017) | QUADAS-2 | 56 | Any molecular rapid test | Influenza A and/or B and/or RSV (pooled estimate) | 7 | Adults | Mixed (some studies with symptoms of ARI, some not reported) | Not stated | Funding information not reported The authors declare no conflicts of interest |
Superseded by Gentilotti 202233 |
| Wu, 201379 | ‘… articles [that provided an] evaluation of procalcitonin alone or compared with other laboratory markers, such as CRP, to diagnose bacterial pneumonia in patients with H1N1 influenza infection’ | MEDLINE, EMBASE and the Cochrane Library (November 2011) | QUADAS | 6 | Procalcitonin | Bacterial pneumonia | 6 | Adults | All diagnosed with H1N1 flu | Predominantly ICU or inpatient | Funding information not reported. The authors declare no conflicts of interest |
Two studies in emergency department or outpatient Superseded by Gentilotti 202233 |
Appendix 3
| Index test | Source of data | No. of included studies (participants) | Outcome | Result (95% CI) | Risk of bias | Indirectness | Inconsistency | Imprecision | Publication bias | Certainty of the body of evidence |
|---|---|---|---|---|---|---|---|---|---|---|
| Signs and symptoms | ||||||||||
| Cough | Gentilotti 202233 | 13 (8423) | Sensitivity | 89.1% (66.4 to 97.1) | Seriousa | Not seriousb | Unable to assessc | Very seriousd | Undetected | Very low |
| Specificity | 13.4% (2.5 to 48.4) | Seriousa | Not seriousb | Unable to assessc | Not serious | Undetected | Moderate | |||
| Sputum production | Gentilotti 202233 | 7 (6392) | Sensitivity | 63.9% (40.5 to 82.1) | Seriousa | Not seriousb | Unable to assessc | Seriouse | Undetected | Low |
| Specificity | 45.3% (25.9 to 66.3) | Seriousa | Not seriousb | Unable to assessc | Not serious | Undetected | Moderate | |||
| Discoloured sputum | Gentilotti 202233 | 9 (3014) | Sensitivity | 54.0% (39.8 to 67.7) | Seriousa | Not seriousb | Unable to assessc | Not serious | Undetected | Moderate |
| Specificity | 53.0% (39.0 to 66.5) | Seriousa | Not seriousb | Unable to assessc | Not serious | Undetected | Moderate | |||
| Purulent sputum (to detect bacterial exacerbations in people with COPD) | Pazmany 202136 | 3 (259) | Sensitivity | 71% (42 to 90) | Seriousf | No serious | Not serious | Very seriousd | Undetected | Very low |
| Specificity | 51% (30 to 73) | Seriousf | No serious | Not serious | Not serious | Undetected | Moderate | |||
| Chest pain | Gentilotti 202233 | 15 (8161) | Sensitivity | 33.9% (21.5 to 49.0) | Seriousa | Not seriousb | Unable to assessc | Not serious | Undetected | Moderate |
| Specificity | 73.0% (61.7 to 81.9) | Seriousa | Not seriousb | Unable to assessc | Seriouse | Undetected | Low | |||
| Dyspnoea | Gentilotti 202233 | 14 (6215) | Sensitivity | 62.6% (53.3 to 71.1) | Seriousa | Not seriousb | Unable to assessc | Not serious | Undetected | Moderate |
| Specificity | 45.5% (32.1 to 59.5) | Seriousa | Not seriousb | Unable to assessc | Not serious | Undetected | Moderate | |||
| Sore throat | Gentilotti 202233 | 5 (1096) | Sensitivity | 32.6% (20.2 to 48.0) | Seriousa | Not seriousb | Unable to assessc | Not serious | Undetected | Moderate |
| Specificity | 45.1% (33.1 to 57.6) | Seriousa | Not seriousb | Unable to assessc | Not serious | Undetected | Moderate | |||
| Runny nose | Gentilotti 202233 | 7 (4630) | Sensitivity | 45.3% (37.3 to 53.4) | Seriousa | Not seriousb | Unable to assessc | Not serious | Undetected | Moderate |
| Specificity | 41.8% (28.1 to 56.8) | Seriousa | Not seriousb | Unable to assessc | Not serious | Undetected | Moderate | |||
| Myalgia | Gentilotti 202233 | 6 (1430) | Sensitivity | 41.6% (19.0 to 68.5) | Seriousa | Not seriousb | Unable to assessc | Not serious | Undetected | Moderate |
| Specificity | 61.2% (40.7 to 78.4) | Seriousa | Not seriousb | Unable to assessc | Seriouse | Undetected | Low | |||
| Chill | Gentilotti 202233 | 8 (1933) | Sensitivity | 45.7% (31.5 to 60.8) | Seriousa | Not seriousb | Unable to assessc | Not serious | Undetected | Moderate |
| Specificity | 60.2% (48.5 to 70.8) | Seriousa | Not seriousb | Unable to assessc | Not serious | Undetected | Moderate | |||
| Diarrhoea | Gentilotti 202233 | 5 (4268) | Sensitivity | 10.8% (6.3 to 17.7) | Seriousa | Not seriousb | Unable to assessc | Not serious | Undetected | Moderate |
| Specificity | 89.5% (75.4 to 95.9) | Seriousa | Not seriousb | Unable to assessc | Seriouse | Undetected | Low | |||
| Impaired consciousness | Gentilotti 202233 | 4 (3208) | Sensitivity | 11.7% (9.3 to 14.5) | Seriousa | Not seriousb | Unable to assessc | Not serious | Undetected | Moderate |
| Specificity | 92.9% (90.5 to 94.7) | Seriousa | Not seriousb | Unable to assessc | Not serious | Undetected | Moderate | |||
| SpO2 | Gentilotti 202233 | 6 (2821) | Sensitivity | 22.8% (12.4 to 38.2) | Seriousa | Not seriousb | Unable to assessc | Not serious | Undetected | Moderate |
| Specificity | 86.6% (80.7 to 90.9) | Seriousa | Not seriousb | Unable to assessc | Seriouse | Undetected | Low | |||
| Fever > 37.8 °C | Gentilotti 202233 | 17 (11,219) | Sensitivity | 42.0% (26.7 to 58.9) | Seriousa | Not seriousb | Unable to assessc | Not serious | Undetected | Moderate |
| Specificity | 80.4% (59.8 to 91.9) | Seriousa | Not seriousb | Unable to assessc | Very seriousd | Undetected | Very low | |||
| Systolic BP | Gentilotti 202233 | 4 (3262) | Sensitivity | 9.6% (2.8 to 28.3) | Seriousa | Not seriousb | Unable to assessc | Not serious | Undetected | Moderate |
| Specificity | 95.0% (80.7 to 98.8) | Seriousa | Not seriousb | Unable to assessc | Seriouse | Undetected | Low | |||
| Tachycardia | Gentilotti 202233 | 11 (9474) | Sensitivity | 27.2% (15.1 to 43.9) | Seriousa | Not seriousb | Unable to assessc | Not serious | Undetected | Moderate |
| Specificity | 84.2% (71.5 to 91.9) | Seriousa | Not seriousb | Unable to assessc | Very seriousd | Undetected | Very low | |||
| Tachypnoea | Gentilotti 202233 | 12 (10,351) | Sensitivity | 27.9% (13.1 to 49.8) | Seriousa | Not seriousb | Unable to assessc | Not serious | Undetected | Moderate |
| Specificity | 80.2% (58.2 to 92.2) | Seriousa | Not seriousb | Unable to assessc | Very seriousd | Undetected | Very low | |||
| Reduced breath sounds | Gentilotti 202233 | 4 (459) | Sensitivity | 24.7% (8.3 to 54.4) | Seriousa | Not seriousb | Unable to assessc | Not serious | Undetected | Moderate |
| Specificity | 89.0% (75.0 to 95.6) | Seriousa | Not seriousb | Unable to assessc | Seriouse | Undetected | Low | |||
| Wheezing | Gentilotti 202233 | 6 (2403) | Sensitivity | 17.3% (9.6 to 29.2) | Seriousa | Not seriousb | Unable to assessc | Not serious | Undetected | Moderate |
| Specificity | 86.4% (70.5 to 94.4) | Seriousa | Not seriousb | Unable to assessc | Very seriousd | Undetected | Very low | |||
| Crackles | Gentilotti 202233 | 10 (6175) | Sensitivity | 40.3% (23.6 to 59.7) | Seriousa | Not seriousb | Unable to assessc | Not serious | Undetected | Moderate |
| Specificity | 83.1% (58.5 to 94.5) | Seriousa | Not seriousb | Unable to assessc | Very seriousd | Undetected | Very low | |||
| Combinations of signs and symptoms | ||||||||||
| Presence/absence of specific symptoms and signs | Schierenberg 201737 | 6 (not reported) | Area under the curve | Ranged from 53% to 79% depending on model used | Not serious | Not serious | Seriousg | Seriouse | Serioush | Very low |
| Symptoms, signs and CRP | ||||||||||
| Predicted risk threshold 2.5% | Minnaard 201734 | 8 (5308) | Sensitivity | 97% (95 to 98) | Not serious | Not serious | Unable to assessc | Not serious | Serioush | Moderate |
| Specificity | 36% (34 to 37) | Not serious | Not serious | Unable to assessc | Not serious | Serioush | Moderate | |||
| Predicted risk threshold 20% | Minnaard 201734 | 8 (5308) | Sensitivity | 70% (66 to 73) | Not serious | Not serious | Unable to assessc | Not serious | Serioush | Moderate |
| Specificity | 90% (89 to 91) | Not serious | Not serious | Unable to assessc | Seriouse | Serioush | Low | |||
| Index test | Source of data | No. of included studies (participants) | Outcome | Result (95% CI) | Risk of bias | Indirectness | Inconsistency | Imprecision | Publication bias | Certainty of the body of evidence |
|---|---|---|---|---|---|---|---|---|---|---|
| CRP | ||||||||||
| CRP > 10 mg/l | Gentilotti 202233 | 4 (944) | Sensitivity | 92% (56 to 99) | Seriousa | Not serious | Unable to assessb | Very Seriousc | Undetected | Very low |
| Specificity | 43% (22 to 66) | Seriousa | Not serious | Unable to assessb | Not serious | Undetected | Moderate | |||
| CRP > 20 mg/l | Gentilotti 202233 | 5 (3531) | Sensitivity | 83% (64 to 93) | Seriousa | Not serious | Unable to assessb | Very Seriousc | Undetected | Very low |
| Specificity | 55% (37 to 73) | Seriousa | Not serious | Unable to assessb | Not serious | Undetected | Moderate | |||
| CRP > 20 mg/l (primary care only, adults and children) | Gentilotti 202233 | 4 (3362) | Sensitivity | 78% (57 to 90) | Seriousa | Seriousd | Unable to assessb | Very Seriousc | Undetected | Very low |
| Specificity | 58% (36 to 78) | Seriousa | Seriousd | Unable to assessb | Seriouse | Undetected | Very low | |||
| CRP > 50 mg/l | Gentilotti 202233 | 5 (4219) | Sensitivity | 77% (51 to 91) | Seriousa | Not serious | Unable to assessb | Very Seriousc | Undetected | Very low |
| Specificity | 74% (51 to 88) | Seriousa | Not serious | Unable to assessb | Seriouse | Undetected | Low | |||
| CRP > 100 mg/l | Gentilotti 202233 | 6 (4418) | Sensitivity | 52% (31 to 72) | Seriousa | Not serious | Unable to assessb | Not serious | Undetected | Moderate |
| Specificity | 91% (79 to 97) | Seriousa | Not serious | Unable to assessb | Seriouse | Undetected | Low | |||
| Procalcitonin | ||||||||||
| Procalcitonin > 0.1 mcg/ml | Gentilotti 202233 | 4 (1092) | Sensitivity | 74% (38 to 93) | Seriousa | Not serious | Unable to assessb | Very seriousc | Undetected | Very low |
| Specificity | 74% (36 to 94) | Seriousa | Not serious | Unable to assessb | Very seriousc | Undetected | Very low | |||
| Procalcitonin > 0.25 mcg/ml | Gentilotti 202233 | 5 (4019) | Sensitivity | 44% (14 to 79) | Seriousa | Not serious | Unable to assessb | Seriouse | Undetected | Low |
| Specificity | 89% (50 to 98) | Seriousa | Not serious | Unable to assessb | Very seriousc | Undetected | Very low | |||
| Procalcitonin > 0.50 mcg/ml (adults and children) | Gentilotti 202233 | 4 (1195) | Sensitivity | 44% (19 to 73) | Seriousa | Seriousd | Unable to assessb | Not serious | Undetected | Low |
| Specificity | 93% (43 to 100) | Seriousa | Seriousd | Unable to assessb | Very seriousc | Undetected | Very low | |||
| TRAIL, IP-10 and CRP (ImmunoXpert) | ||||||||||
| TRAIL, IP-10 and CRP to diagnose bacterial infection (adults and children) | Carlton 202132 | 4 (1291) | Sensitivity | 85% (75 to 91) | Seriousf | Seriousg | Not serious | Seriouse | Undetected | Very low |
| Specificity | 86% (73 to 93) | Seriousf | Seriousg | Not serious | Very seriousc | Undetected | Very low | |||
| TRAIL, IP-10 and CRP to diagnose viral infection (adults and children) | Carlton 202132 | 3 (989) | Sensitivity | 90% (79 to 96) | Seriousf | Seriousg | Serioush | Seriouse | Undetected | Very low |
| Specificity | 92% (83 to 96) | Seriousf | Seriousg | Not serious | Seriouse | Undetected | Very low | |||
| CRP and MxA (FebriDx) | ||||||||||
| CRP and MxA to diagnose bacterial infection (adults and children) | Carlton 202132 | 4 (598) | Sensitivity | 84% (75 to 90) | No serious | Seriousg | No serious | Seriouse | Undetected | Low |
| Specificity | 93% (90 to 95) | No serious | Seriousg | No serious | Not serious | Undetected | Moderate | |||
| CRP and MxA to diagnose viral infection (adults and children) | Carlton 202132 | 4 (583) | Sensitivity | 87% (72 to 95) | No serious | Seriousg | No serious | Very seriousc | Undetected | Very low |
| Specificity | 82% (66 to 86) | No serious | Seriousg | No serious | Seriouse | Undetected | Low | |||
| Other host biomarkers | ||||||||||
| CRP and neopterin to diagnose bacterial infection | Carlton 202132 | 1 (198) | Sensitivity | 80% (71 to 86) | Seriousi | Seriousj | Not serious | Seriouse | Undetected | Very low |
| Specificity | 82% (71 to 89) | Seriousi | Seriousj | Not serious | Seriouse | Undetected | Very low | |||
| Index test | Source of data | No. of included studies (participants) | Outcome | Result (95% CI) | Risk of bias | Indirectness | Inconsistency | Imprecision | Publication bias | Certainty of the body of evidence |
|---|---|---|---|---|---|---|---|---|---|---|
| Single pathogen tests for influenza | ||||||||||
| Immunochromatography | Gentilotti 202233 | 15 (2897) | Sensitivity | 65% (47 to 79) | Seriousa | Not serious | Unable to assessb | Seriousc | Undetected | Low |
| Specificity | 96% (92 to 98) | Seriousa | Not serious | Unable to assessb | Not serious | Undetected | Moderate | |||
| Immunochromatography (adults and children, primary care only) | Gentilotti 202233 | 11 (3351) | Sensitivity | 56% (36 to 74) | Seriousa | Seriousd | Unable to assessb | Not serious | Undetected | Low |
| Specificity | 95% (89 to 98) | Seriousa | Seriousd | Unable to assessb | Seriousc | Undetected | Very low | |||
| Immunochromatography (adults and children, emergency department only) | Gentilotti 202233 | 25 (15,021) | Sensitivity | 71% (60 to 80) | Not serious | Seriousd | Unable to assessb | Seriousc | Undetected | Low |
| Specificity | 98% (96 to 99) | Not serious | Seriousd | Unable to assessb | Not serious | Undetected | Moderate | |||
| Immunochromatography (adults and children, outpatient department only) | Gentilotti 202233 | 17 (6110) | Sensitivity | 66% (55 to 76) | Not serious | Seriousd | Unable to assessb | Seriousc | Undetected | Low |
| Specificity | 97% (93 to 99) | Not serious | Seriousd | Unable to assessb | Not serious | Undetected | Moderate | |||
| Direct immunofluorescence (adults and children) | Gentilotti 202233 | 19 (7635) | Sensitivity | 78% (67 to 86) | Seriousa | Seriousd | Unable to assessb | Seriousc | Undetected | Very low |
| Specificity | 95% (90 to 98) | Seriousa | Seriousd | Unable to assessb | Not serious | Undetected | Low | |||
| Direct immunofluorescence (adults and children, emergency department only) | Gentilotti 202233 | 5 (1314) | Sensitivity | 82% (72 to 89) | Seriousa | Seriousd | Unable to assessb | Seriousc | Undetected | Very low |
| Specificity | 96% (93 to 97) | Seriousa | Seriousd | Unable to assessb | Not serious | Undetected | Low | |||
| Optical immunoassay (adults and children) | Gentilotti 202233 | 9 (3910) | Sensitivity | 68% (51 to 81) | Seriousa | Seriousd | Unable to assessb | Seriousc | Undetected | Very low |
| Specificity | 88% (81 to 93) | Seriousa | Seriousd | Unable to assessb | Seriousc | Undetected | Very low | |||
| MariPOC test (adults and children) | Gentilotti 202233 | 5 (1231) | Sensitivity | 78% (61 to 89) | Seriousa | Seriousd | Unable to assessb | Seriousc | Undetected | Very low |
| Specificity | 99% (97 to 99) | Seriousa | Seriousd | Unable to assessb | Not serious | Undetected | Low | |||
| Chemiluminescent neuraminidase assay (adults and children) | Gentilotti 202233 | 4 (787) | Sensitivity | 81% (51 to 94) | Seriousa | Seriousd | Unable to assessb | Very seriouse | Undetected | Very low |
| Specificity | 82% (65 to 91) | Seriousa | Seriousd | Unable to assessb | Very seriouse | Undetected | Very low | |||
| Nucleic acid amplification tests: standalone, single pathogen PCR (adults and children) | Gentilotti 202233 | 30 (25,027) | Sensitivity | 95.1% (89.3 to 97.8) | Seriousf | Seriousd | Unable to assessb | Seriousc | Undetected | Very low |
| Specificity | 97.5% (95.5 to 98.7) | Seriousf | Seriousd | Unable to assessb | Not serious | Undetected | Low | |||
| Nucleic acid amplification tests: non-PCR-based (adults and children) | Gentilotti 202233 | 23 (4863) | Sensitivity | 92% (88 to 94) | Seriousf | Seriousd | Unable to assessb | Seriousc | Undetected | Very low |
| Specificity | 98% (95 to 99) | Seriousf | Seriousd | Unable to assessb | Not serious | Undetected | Low | |||
| Nucleic acid amplification tests: non-PCR-based (adults and children, emergency department only) | Gentilotti 202233 | 14 (3138) | Sensitivity | 91% (87 to 94) | Seriousf | Seriousd | Unable to assessb | Seriousc | Undetected | Very low |
| Specificity | 98% (95 to 99) | Seriousf | Seriousd | Unable to assessb | Not serious | Undetected | Low | |||
| Single pathogen tests for RSV | ||||||||||
| Direct immunofluorescence | Onwuchekwa 202335 | 1 (49) | Sensitivity | 56% (31 to 78) | Not serious | Seriousg | Not serious | Very serioush | Undetected | Very low |
| Specificity | 100% (89 to 100) | Not serious | Seriousg | Not serious | Very serioush | Undetected | Very low | |||
| Rapid antigen test | Onwuchekwa 202335 | 1 (281) | Sensitivity | 18% (12 to 27) | Seriousi | Seriousj | Not serious | Not serious | Undetected | Low |
| Specificity | 98% (86 to 100) | Seriousi | Seriousj | Not serious | Seriousc | Undetected | Very low | |||
Appendix 4
FIGURE 4.
PRISMA flow diagram for white cell differential count.
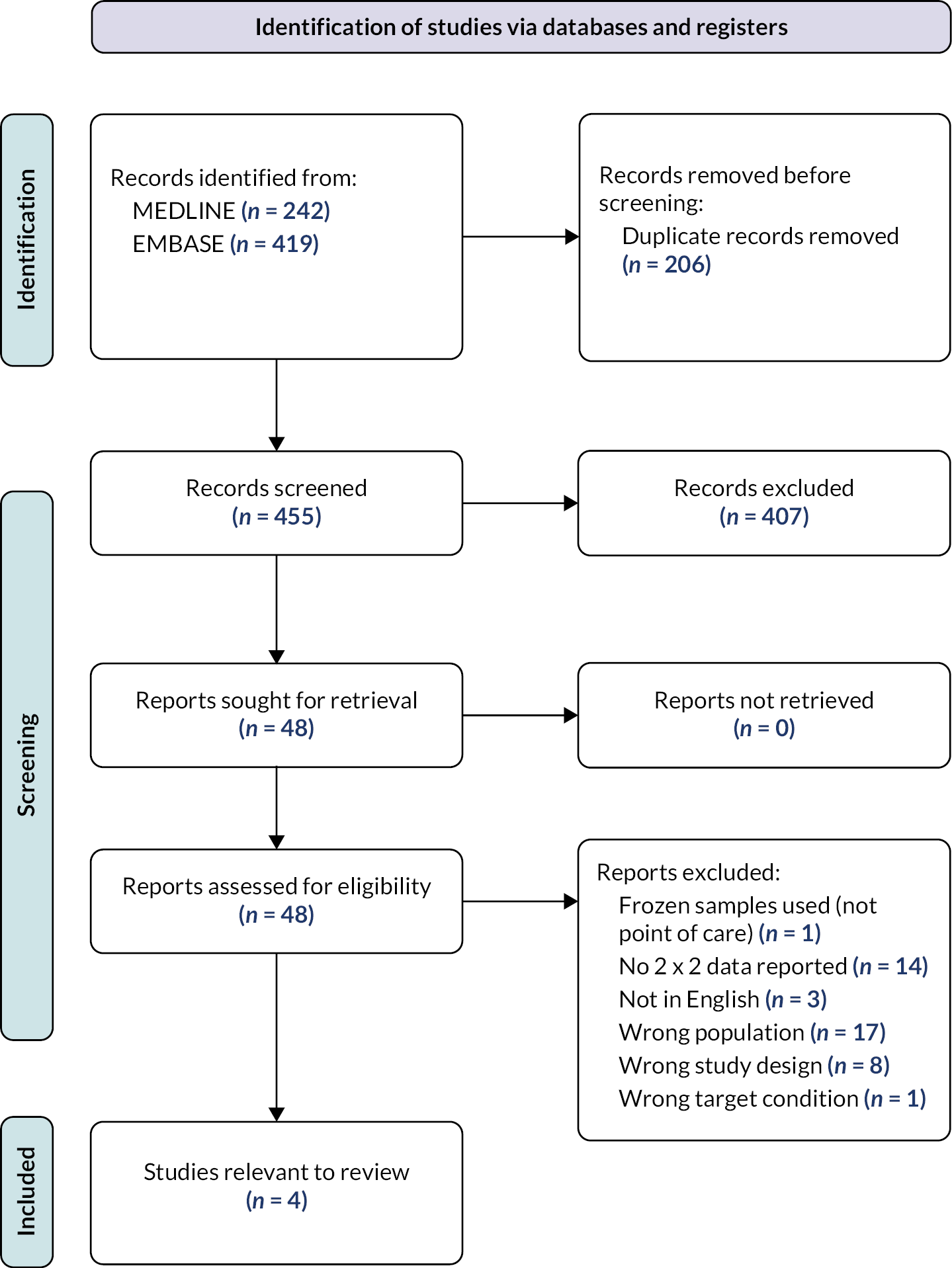
| Reference | Population | Clinical features | Setting | Target condition assessed | Index tests | Reference standard | Outcomes reported | Funding/conflicts of interest |
|---|---|---|---|---|---|---|---|---|
| Castro-Guardiola 200040 | Adults (n = 284) 62% male. Mean age 57.2 years [standard deviation (SD) 20] |
People who have been assessed by a clinician as having suspected pneumonia | Emergency department, Spain | Pneumonia | White blood cell count | Typical findings on a chest X-ray, plus at least two of the following features:
|
Area under the curve 0.65 | Not reported |
| Gulich 199943 | Adults (n = 179) 46.4% male. Mean age 34.3 years (SD 13.4) |
People presenting with a sore throat | Primary care, Germany | Bacterial pharyngitis | White blood cell count | Culture of group A or C beta-haemolytic streptococci, or Haemophilus influenzae | Area under the curve 0.68 | The study was supported by Bundesverband der Betriebskrankenkassen and by Nycomed GmbH, Munich |
| Holm 200741 | Adults (n = 364) 47% male. Median age 50 years |
People with symptoms of a lower respiratory tract infection | Primary care, Denmark | Pneumonia | White cell count ≥ 10 million/ml | Chest X-ray | Sensitivity 46% and specificity 80% (no confidence intervals reported) | Financial support received from the various contributors, including The Danish Lung Association, The Danish Medical Research Association and the Institute of Clinical Research. The authors declare no conflicts of interest |
| Liu 201342 | Adults (n = 500) 58% male. Mean age 42.7 years (range 18–94) |
People with a diagnosis of community-acquired pneumonia, based on findings from a chest X-ray and symptoms | Outpatient, China | Bacterial pneumonia | White cell count < 4 million/ml, 4–10 million/ml or > 10 million/ml | Microbiological culture and PCR | 2×2 data, sufficient to calculate sensitivity and specificity to diagnose bacterial infection at different thresholds of white cell count | Supported by grants from Beijing Science and Technology Key Projects Foundation. The authors declare no conflicts of interest |
| < 4 million/ml Sensitivity 10.07 (95% CI 5.74 to 16.06) Specificity 94.59 (95% CI 91.68 to 96.71) |
||||||||
| 4–10 million/ml Sensitivity 71.14 (95% CI 63.16 to 78.26) Specificity 31.34 (95% CI 26.52 to 36.48) |
||||||||
| > 10 million/ml Sensitivity 18.79 (95% CI 12.87 to 26) Specificity 74.07 (95% CI 69.16 to 78.58) |
FIGURE 5.
QUADAS-2 assessments: white cell differential count. aIndex test was incorporated as part of the reference standard, and was also not conducted in a point-of-care setting (central laboratory analysis of white cell count). bIndex test was not conducted in a point-of-care setting (central laboratory analysis of white cell count). cPeople with more severe illness (requiring hospital admission) were excluded from participation. Index test was not conducted in a point-of-care setting (central laboratory analysis of white cell count). Participants with possible malignancy were excluded from analysis. dUnclear whether sampling was consecutive/random. Unclear how white cell count was assessed, but not conducted in a point-of-care setting.
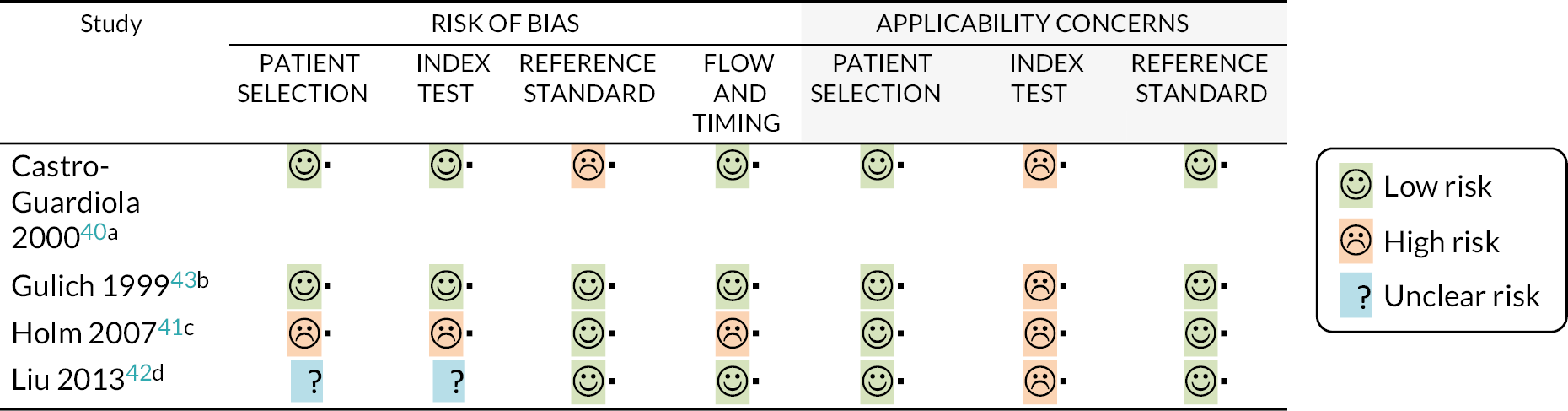
Appendix 5
| Index test | Source of data | No. of included studies (participants) | Outcome | Result (95% CI) | Risk of bias | Indirectness | Inconsistency | Imprecision | Publication bias | Certainty of the body of evidence |
|---|---|---|---|---|---|---|---|---|---|---|
| White cell differential count | ||||||||||
| White cell count to diagnose pneumonia | Castro-Guardiola 2000,40 Holm 2007,41 Liu 201342 | 3 (1148) | Two studies reported sensitivity estimates ranging from 10.1% to 71.1%, and specificity estimates ranging from 31.3% to 94.6%, depending on the threshold used (see Appendix 4, Table 6 for full details). One study reported an area under the curve of 0.65 | Seriousa | Seriousb | Seriousc | Very seriousd | Undetected | Very low | |
| White cell count to diagnose bacterial pharyngitis | Gulich 199943 | 1 (179) | Area under the curve | 0.68 (no confidence intervals) | Not serious | Seriousb | Not serious | Seriouse | Undetected | Low |
Appendix 6
FIGURE 6.
PRISMA flow diagram for multiplex PCR tests.
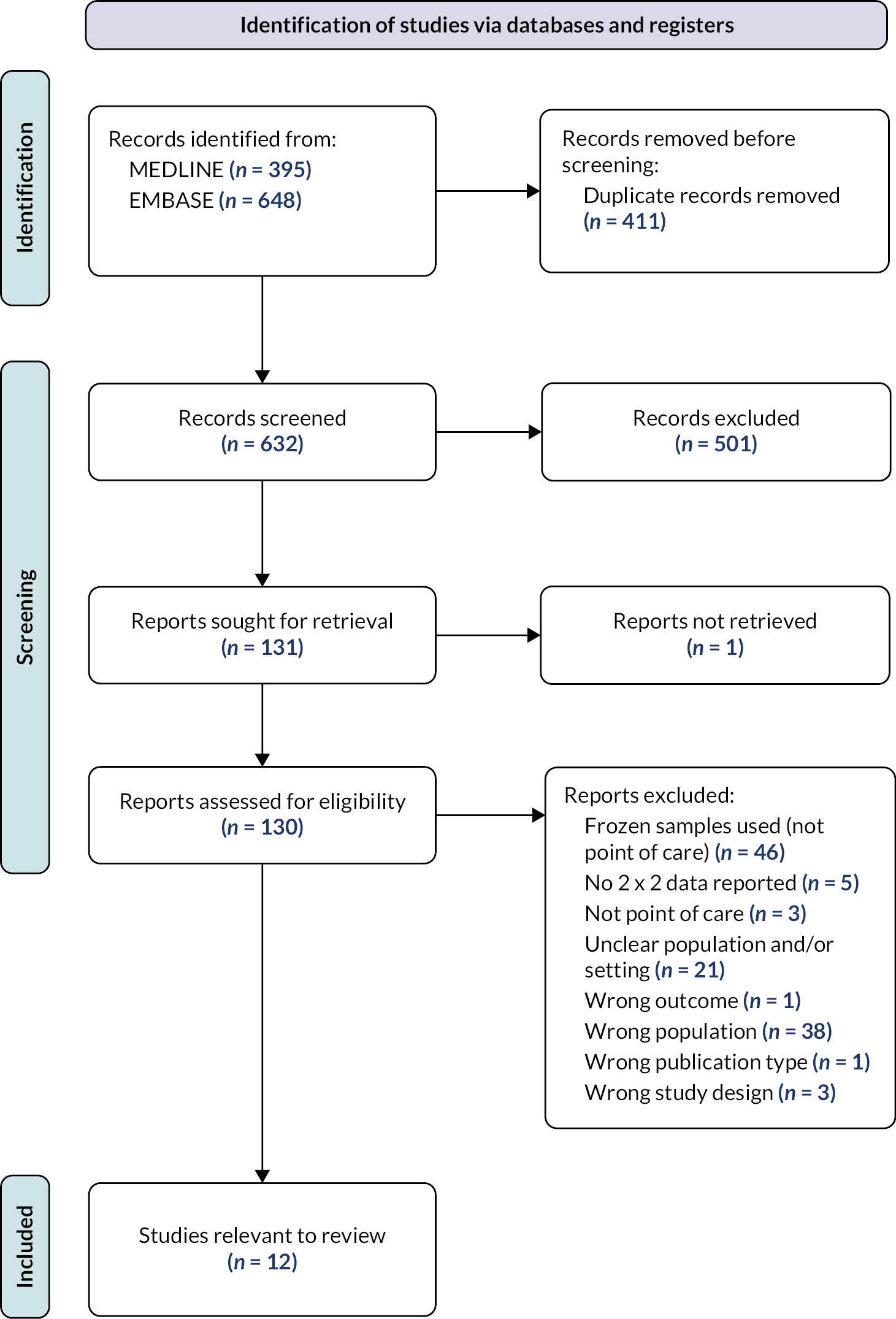
| Reference | Population | Clinical features | Setting | Target condition assessed | Index tests | Reference standard | Notes | Funding/conflicts of interest |
|---|---|---|---|---|---|---|---|---|
| Boku 201351 | Adults. Mean age 34.4 years (range 20–63) 53.1% male |
Symptoms of acute respiratory infection, or presence of fever and known contact with influenza | Hospital outpatient setting, Japan | Flu A/B | Verigene system RV + on nasopharyngeal swabs | Viral culture plus laboratory PCR | Not reported | |
| Escarate 202244 | Adults. Aged ≥ 65 years. Sex not reported |
Tested due to an outbreak of a respiratory illness. Symptoms of acute respiratory infection | Outpatient/primary care (long-term care facilities), Australia | Flu A, Flu B and RSV | Xpert Xpress Flu/RSV on nasopharyngeal swabs or combined nose and throat swabs | Primary reference standard: PCR from central laboratory Secondary reference standard: included expert opinion assessment of discordant specimens |
Note that data are not included in the meta-analysis, as the authors only report specificity (not sensitivity) and the bivariate model requires both parameters | The authors declare no conflicts of interest |
| Farfour 202245 | Adults. Age not reported. Sex not reported |
Suspected viral respiratory infection | Emergency department, France | Flu A, RSV | Idylla SARS CoV/Flu/RSV on nasopharyngeal swabs | Laboratory-based multiplex PCR | No external funding received | |
| Hansen 201852 | Adults and children (children comprised 20% of total population) Age not reported. Sex not reported |
Presenting with at least one sign of influenza | Emergency department, USA | Flu A/B | Cobas Liat Influenza A/B assay on nasopharyngeal swabs | Primary reference standard: PCR from central laboratory. Secondary reference standard: included analysis of discordant specimens with a second multiplex rapid test |
Partial funding for this study was provided by an unrestricted educational grant from Roche molecular to GTH and from the Minneapolis Medical Research | |
| Maignan 201646 | Adults. Median 70 years (IQR 44–84). 51% male |
Presenting with fever and at least one sign of a respiratory tract infection | Emergency department, France | Flu A, Flu B, Flu A/B | Cobas Liat Influenza A/B assay on nasopharyngeal swabs | Primary reference standard: PCR from central laboratory, with analysis of discordant results with Xpert Xpress Flu/RSV assay and results from the national influenza virus reference centre | Partially funded by Roche Diagnostics. Roche Diagnostics had no access to the data and were not involved in the interpretation of the data or the writing of the manuscript | |
| Morris 202147 | Adults and children included in the study. Data were extracted which relate to adults only. Median 55 years (IQR 29–73). 44.7% male |
Symptoms of acute respiratory infection | Emergency department, respiratory admissions unit and bone marrow transplant unit were included in the study, UK. Extracted data relate to adults in an emergency department setting only | Flu A, RSV | Xpert Xpress Flu/RSV. Sample type unclear | Primary reference standard: laboratory-based PCR | No funding required. The authors declare no conflicts of interest | |
| Peretz 202053 | Adults. Aged 18 to 97. 57% male |
People with suspected influenza | Emergency department, Israel | Flu A/B | Xpert Xpress Flu A/B and Simplexa Flu A/B and RSV on nasopharyngeal swabs | Comparator: rapid antigen test | Note that this study provides data on concordance between multiplex PCR and a rapid antigen test. However, as the rapid antigen test is not regarded as a reference standard by the authors, these data were not included in the analysis | No funding required. The authors declare no conflicts of interest |
| Comparison of Xpert Xpress Flu with Influ A+B K-SeT rapid antigen test: Percentage positive agreement: 96.3% (87.3 to 99.6) Percentage negative agreement: 95.7% (90.2 to 98.6) |
||||||||
| Comparison of Simplexa Flu A/B and RSV with Influ A+B K-SeT rapid antigen test: Percentage positive agreement: 96.3% (87.3 to 99.6) Percentage negative agreement: 97.4% (92.5 to 99.5) |
||||||||
| Tanei 201454 | Adults. Median 30.5 years, range 20–63. 42.7% male |
Symptoms of acute respiratory infection plus a fever of ≥ 37 °C | Outpatients in a hospital general medical department, Japan | Flu A/B | Verigene RV+ | Primary reference standard: rapid antigen test | Note that this study provides data on concordance between multiplex PCR and a rapid antigen test. However, as the rapid antigen test is not regarded as a reference standard by the authors, these data were not included in the analysis | This study was supported in part by a Grant-in-Aid from the MEXT Strategic Research Foundation Project for Private Universities. The authors declare no conflicts of interest |
| Comparison of Verigene RV+ with RapidTesta FLU II rapid antigen test: Percentage positive agreement: 95.6% (84.9 to 99.5) Percentage negative agreement:56.8% (39.5 to 72.9) |
||||||||
| Valentin 201948 | Adults. Age not reported. Sex not reported |
Adult patients suffering from acute febrile respiratory tract infection with at least one risk factor for complications of seasonal influenza | Emergency department, Austria | Flu A, Flu B, Flu A/B | Xpert Xpress Flu/RSV and Cobas Liat Influenza A/B assay on nasopharyngeal swabs | Primary reference standard: laboratory-based PCR | Reagents used for the tests were partly supplied by Roche and Cepheid. No other funding was received. The authors declare no conflicts of interest | |
| Yin 202249 | Adults and children (23% of participants were children). Age not reported. 58% male |
Symptoms of acute respiratory infection | Emergency department, Belgium | Flu A, Flu B, RSV | Cobas Liat Influenza A/B assay on nasopharyngeal swabs | Primary reference standard: composite of rapid antigen tests plus culture. Samples were considered positive if they were positive on at least two of the three tests used (including the index test) | Roche Diagnostics supplied instruments and reagents needed for this study. No personal grants or funding was received by the authors for this study. The authors declare no conflicts of interest | |
| Youngs 201950 | Adults. Age not reported. Sex not reported |
Suspected influenza | Emergency department, UK | Flu A, Flu B, Flu A/B | Cobas Liat Influenza A/B assay on throat swabs | Primary reference standard: composite of laboratory-based PCR method and an alternative multiplex test (Xpert Xpress Flu/RSV). Secondary reference standard: as above, but including expert opinion |
The authors declare no conflicts of interest | |
| Zuurbier 202255 | Adults. 45.9% male. Median age 75 years (IQR 67–80) |
Symptoms of acute respiratory tract infection | Home setting/primary care, Belgium, Netherlands, and UK | RSV | Xpert Xpress Flu/RSV on nasopharyngeal swabs | Primary reference standard: laboratory-based PCR | RESCEU has received funding from the Innovative Medicines Initiative 2 Joint Undertaking. Several authors declare they received personal fees from Roche, GSK and other pharmaceutical companies, outside the submitted work. Additionally, University Medical Centre Utrecht received funding from various pharmaceutical companies |
FIGURE 7.
QUADAS-2 assessments: multiplex PCR tests. aUnclear if a consecutive/random sample of participants was enrolled. Unclear if reference standard results were interpreted without knowledge of the index test. bUnclear if a consecutive/random sample of participants was enrolled. Unclear if all participants would have attended for assessment by a healthcare provider if they were not enrolled in the study (care home residents). Some participants did not receive a reference standard and were excluded from the analysis. cUnclear whether the index test was carried out at point of care. Some participants were excluded from analysis. dUnclear if a consecutive/random sample of participants was enrolled. Participants with multiple conditions were excluded from this study. eUnclear if a consecutive/random sample of participants was enrolled. Staff were asked to collect samples ‘as per usual practice’. No guidance on specific inclusion/exclusion criteria for sampling. fUnclear if a consecutive/random sample of participants was enrolled. Rapid antigen test was used as the reference standard. Unclear if the index test was interpreted without knowledge of the reference standard. Index test not actually conducted in a point-of-care setting. gUnclear if the index test was interpreted without knowledge of the reference standard. Rapid antigen test was used as the reference standard. Index test not actually conducted in a point-of-care setting. hNot all participants were included in the analysis. Index test not analysed at point of care. iUnclear if a consecutive/random sample of participants was enrolled. Index tests were a component of the reference standard. Not all samples analysed in point-of-care setting. jReference standard results were interpreted with knowledge of the index test results. High number of exclusions. kParticipants were undergoing home assessment for respiratory illness as part of a larger study. Many would not have sought medical care had it not been that they were participating in this study. High number of exclusions from analysis.
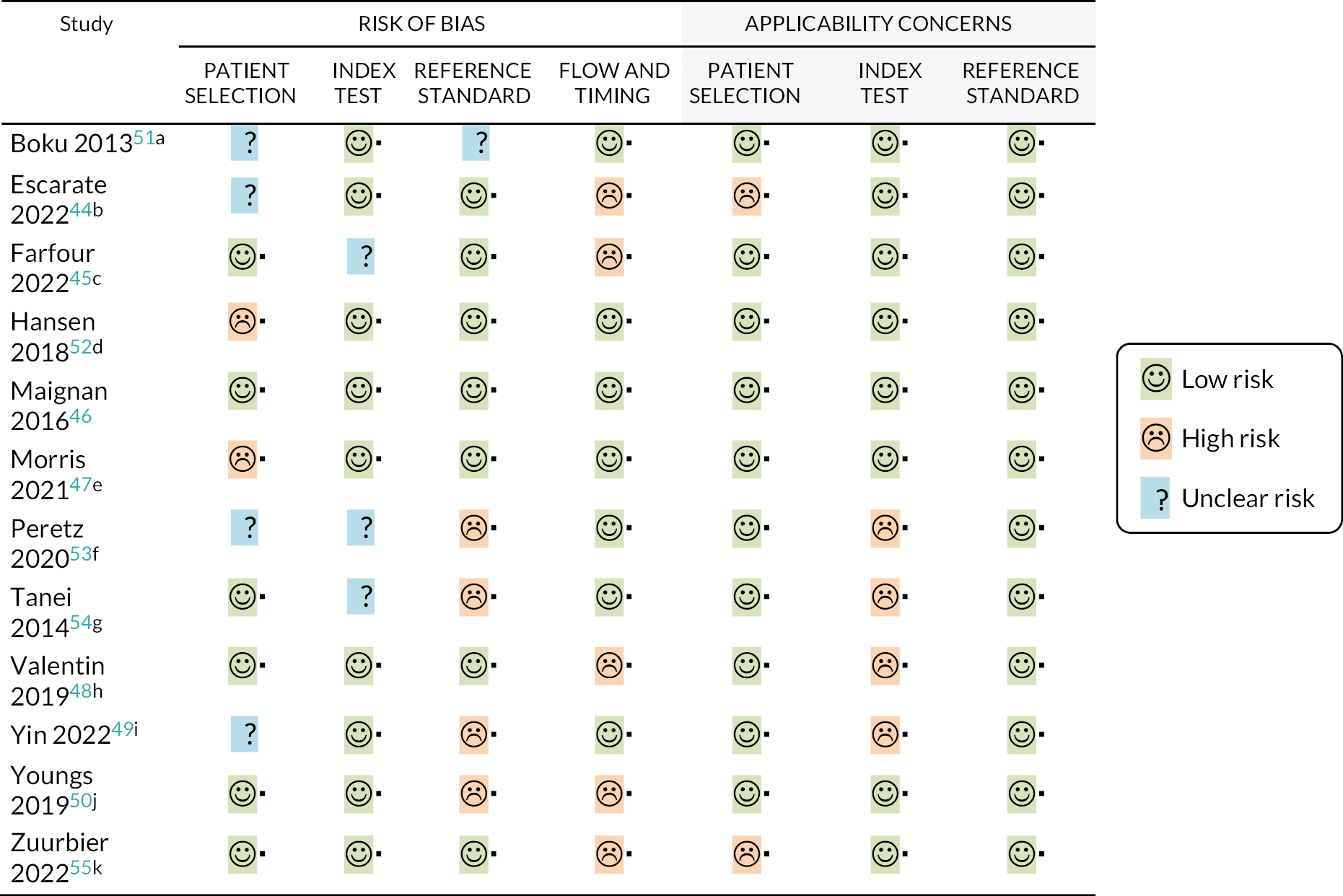
Appendix 7
| Index test | Source of data | No. of included studies (participants) | Outcome | Result (95% CI) | Risk of bias | Indirectness | Inconsistency | Imprecision | Publication bias | Certainty of the body of evidence |
|---|---|---|---|---|---|---|---|---|---|---|
| Multiplex tests | ||||||||||
| All multiplex tests for influenza A | Escarate 2022,44 Farfour 2022,45 Morris 2021,47 Maignan 2016,46 Valentin 201948 (two tests included), Yin 2022,49 Youngs 201950 | Eight studies (2212) | Sensitivity | 98.2% (90.7 to 99.7) | Seriousa | Not serious | Seriousb | Not serious | Undetected | Low |
| Specificity | 98.6% (96.6 to 99.4) | Seriousa | Not serious | Seriousb | Not serious | Undetected | Low | |||
| Cobas Liat tests for influenza A | Maignan 2016,46 Valentin 2019,48 Yin 2022,49 Youngs 201950 | Four studies (1259) | Sensitivity | 99.8% (18.8 to 100) | Not serious | Not serious | Seriousb | Very seriousc | Undetected | Very low |
| Specificity | 97.9 (94.0 to 99.3) | Not serious | Not serious | Seriousb | Not serious | Undetected | Moderate | |||
| Xpert Xpress tests for influenza A | Escarate 2022,44 Morris 2021,47 Valentin 201948 | Three studies (754) | Sensitivity | 97.0% (92.9 to 98.7) | Seriousa | Not serious | Not serious | Not serious | Undetected | Moderate |
| Specificity | 98.5% (96.2 to 99.4) | Seriousa | Not serious | Not serious | Not serious | Undetected | Moderate | |||
FIGURE 8.
Meta-analysis for influenza A.
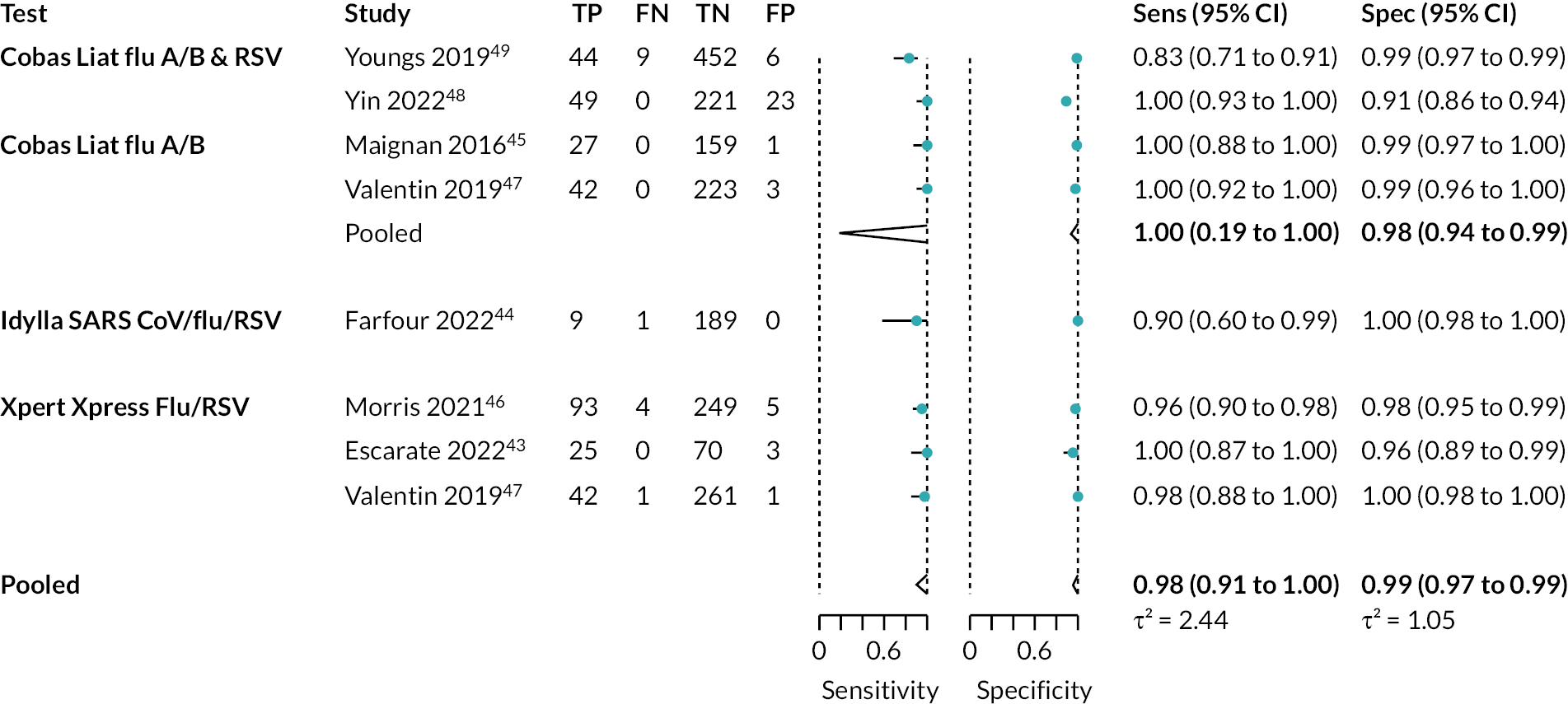
FIGURE 9.
Influenza A data and meta-analysis results in ROC space.
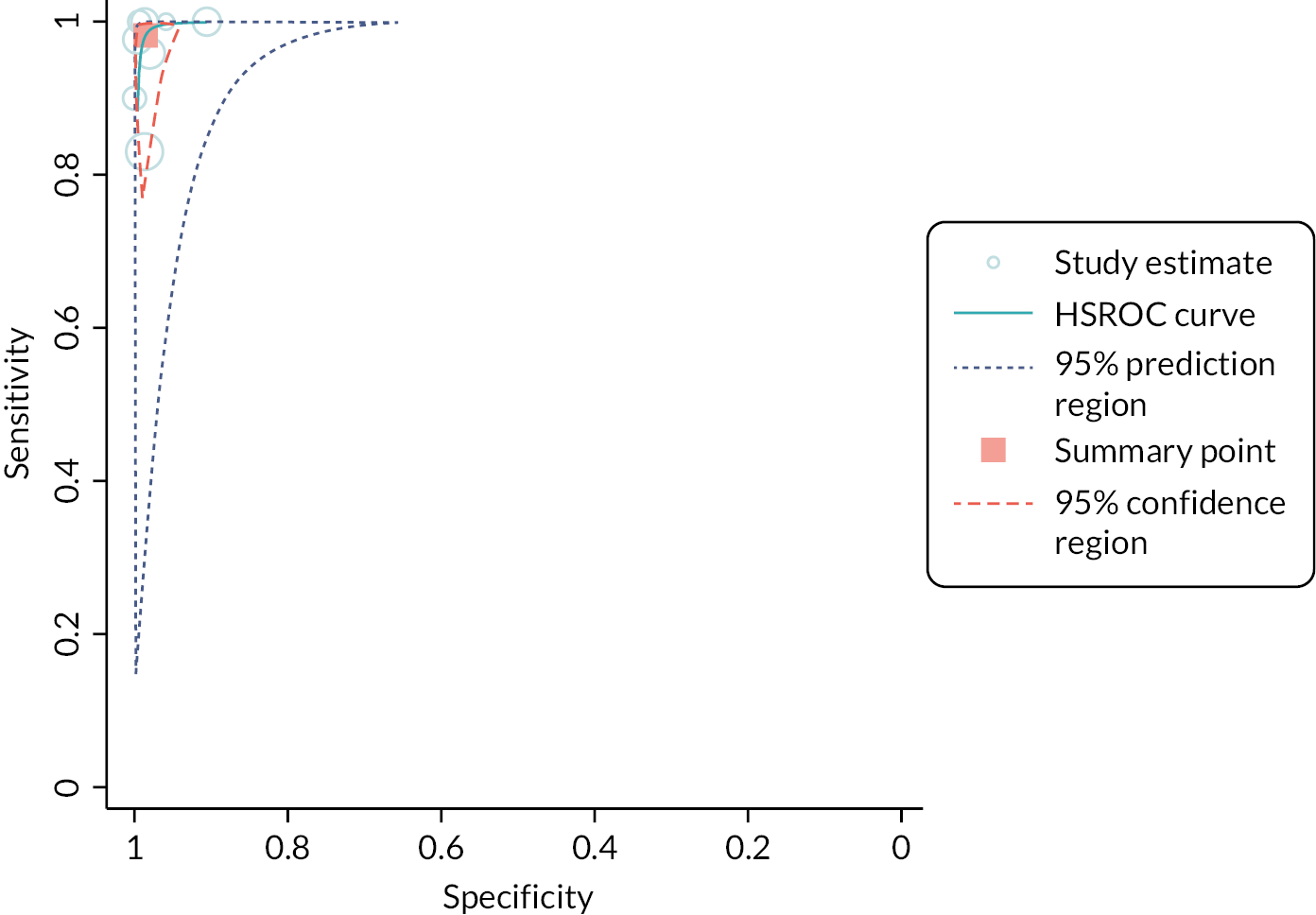
Appendix 8
| Index test | Source of data | No. of included studies (participants) | Outcome | Result (95% CI) | Risk of bias | Indirectness | Inconsistency | Imprecision | Publication bias | Certainty of the body of evidence |
|---|---|---|---|---|---|---|---|---|---|---|
| Multiplex tests | ||||||||||
| All multiple × tests for influenza B | Escarate 2022,44 Maignan 2016,46 Valentin 201948 (two tests included), Yin 2022,49 Youngs 201950 | Six studies (1823) | Sensitivity | 94.5% (88.6 to 97.5) | Seriousa | Not serious | Seriousb | Seriousc | Undetected | Very low |
| Specificity | 99.1 (98.1 to 99.6) | Seriousa | Not serious | Seriousb | Not serious | Undetected | Low | |||
| Cobas Liat tests for influenza B | Maignan 2016,46 Valentin 2019,48 Yin 2022,49 Youngs 201950 | Four studies (1420) | Sensitivity | 92.9% (84.3 to 96.9) | Not serious | Not serious | Seriousb | Seriousc | Undetected | Low |
| Specificity | 99.0% (97.6 to 99.6) | Not serious | Not serious | Seriousb | Not serious | Undetected | Moderate | |||
| Xpert Xpress tests for influenza B | Escarate 2022,44 Valentin 201948 | Two studies (403) | Sensitivity | 96.4% (90.7 to 99.0) | Seriousa | Not serious | Not serious | Not serious | Undetected | Moderate |
| Specificity | 99.4% (97.4 to 99.8) | Seriousa | Not serious | Not serious | Not serious | Undetected | Moderate | |||
FIGURE 10.
Meta-analysis for influenza B.
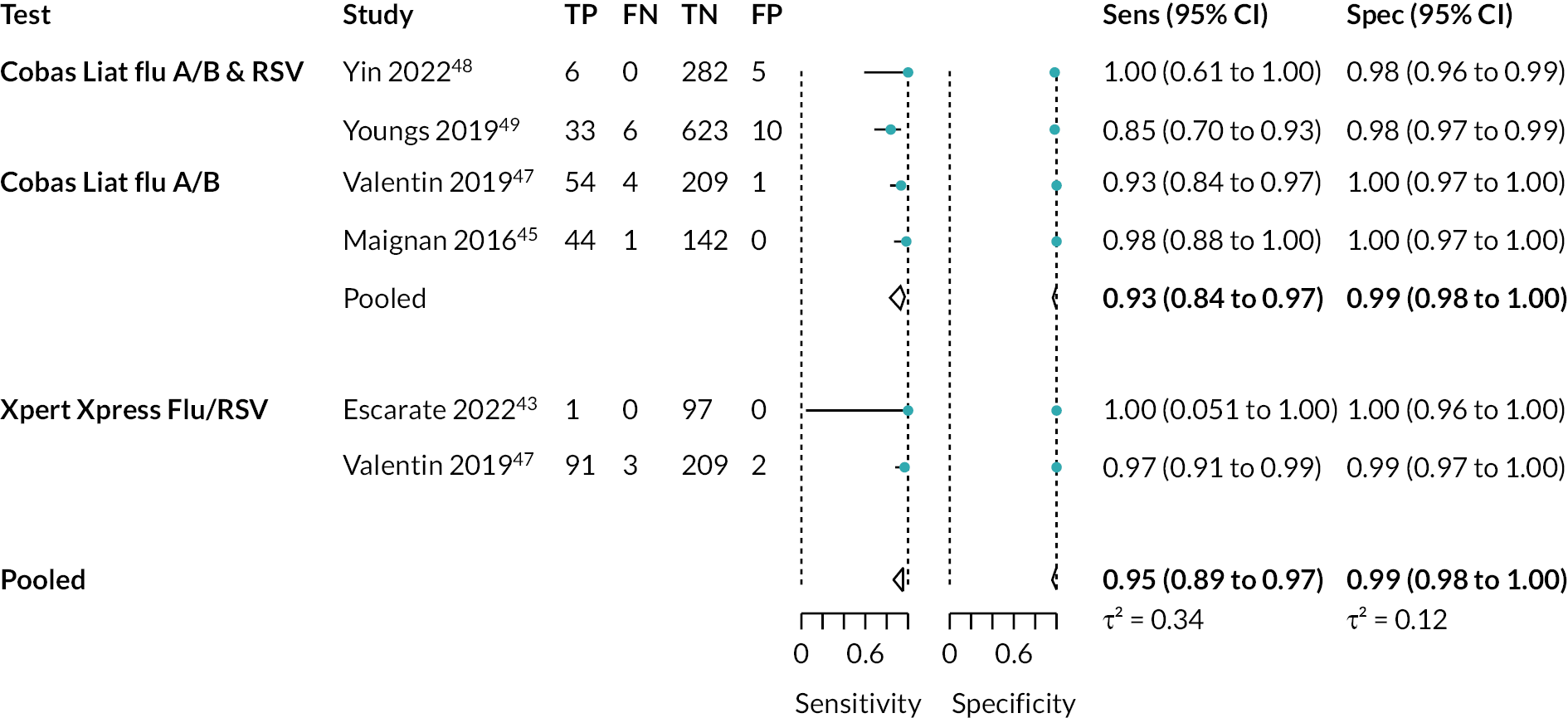
FIGURE 11.
Influenza B data and meta-analysis results in ROC space.
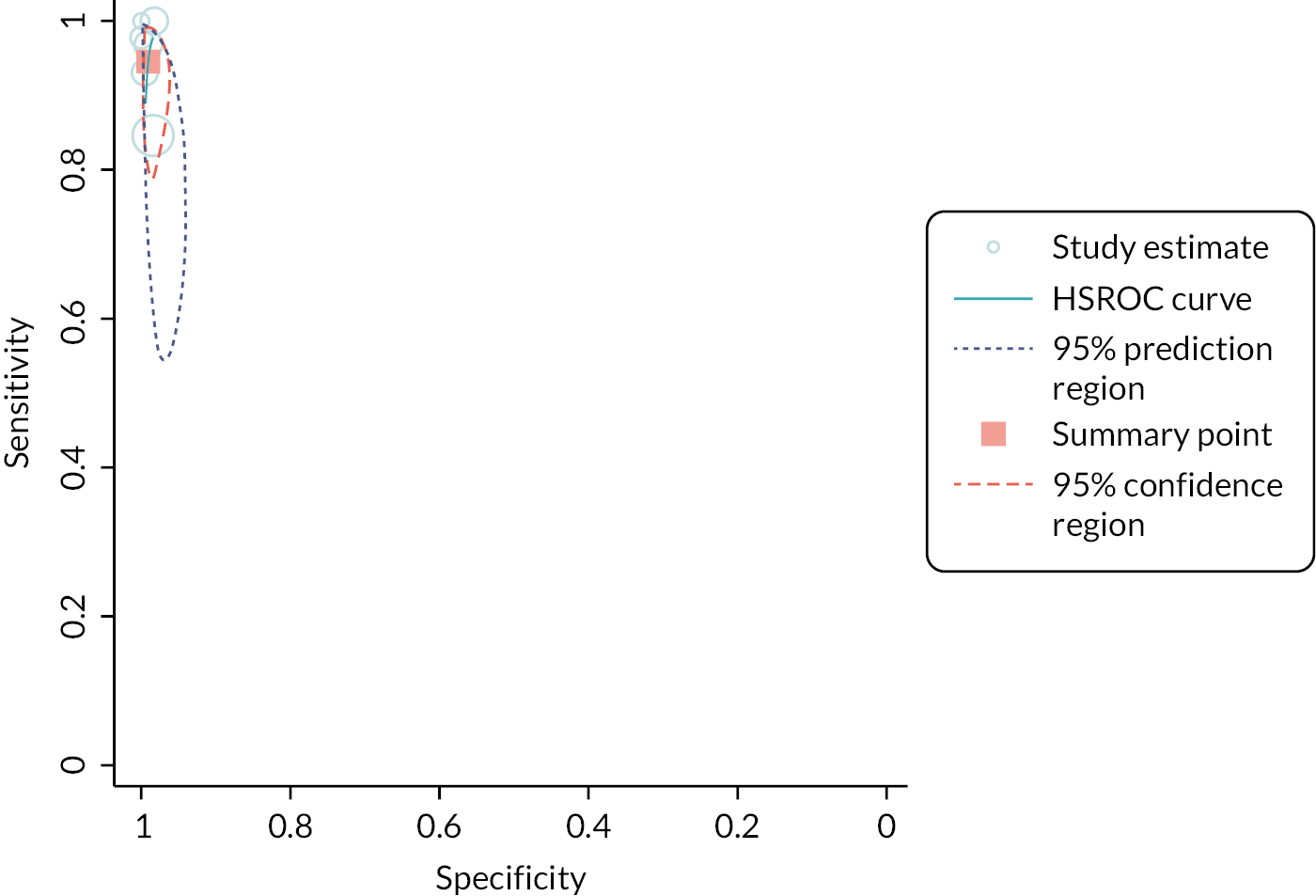
Appendix 9
| Index test | Source of data | No. of included studies (participants) | Outcome | Result (95% CI) | Risk of bias | Indirectness | Inconsistency | Imprecision | Publication bias | Certainty of the body of evidence |
|---|---|---|---|---|---|---|---|---|---|---|
| Multiplex tests | ||||||||||
| All multiple × tests for influenza A/B | Boku 2013,51 Escarate 2022,44 Hansen 2018,52 Maignan 2016,46 Valentin 201948 (two tests included), Yin 2022,49 Youngs 201950 | Eight studies (2162) | Sensitivity | 97.4% (92.9 to 99.0) | Seriousa | Not serious | Seriousb | Not serious | Undetected | Low |
| Specificity | 97.0% (94.5 to 98.4) | Seriousa | Not serious | Seriousb | Not serious | Undetected | Low | |||
| Cobas Liat tests for influenza A/B | Hansen 2018,52 Maignan 2016,46 Valentin 2019,48 Yin 2022,49 Youngs 201950 | Five studies (1712) | Sensitivity | 97.1% (88.6 to 99.3) | Not serious | Not serious | Seriousb | Seriousc | Undetected | Low |
| Specificity | 96.8% (93.2 to 98.5) | Not serious | Not serious | Seriousb | Not serious | Undetected | Moderate | |||
| Xpert Xpress tests for influenza A/B | Escarate 2022,44 Valentin 201948 | Two studies (403) | Sensitivity | 97.5% (93.6 to 99.1) | Seriousa | Not serious | Not serious | Not serious | Undetected | Moderate |
| Specificity | 97.5% (94.5 to 98.9) | Seriousa | Not serious | Not serious | Not serious | Undetected | Moderate | |||
FIGURE 12.
Meta-analysis for influenza A or B.
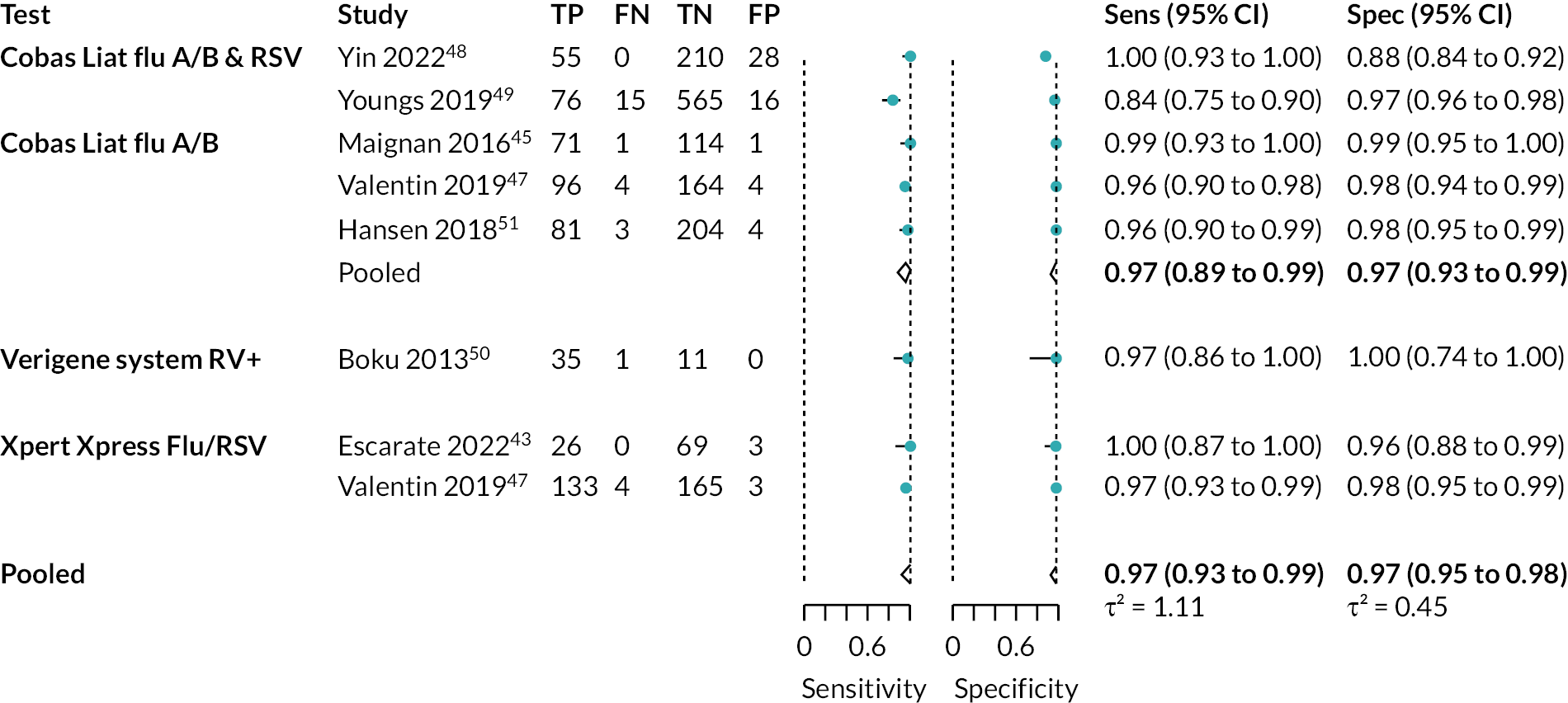
FIGURE 13.
Influenza A/B data and meta-analysis results in ROC space.
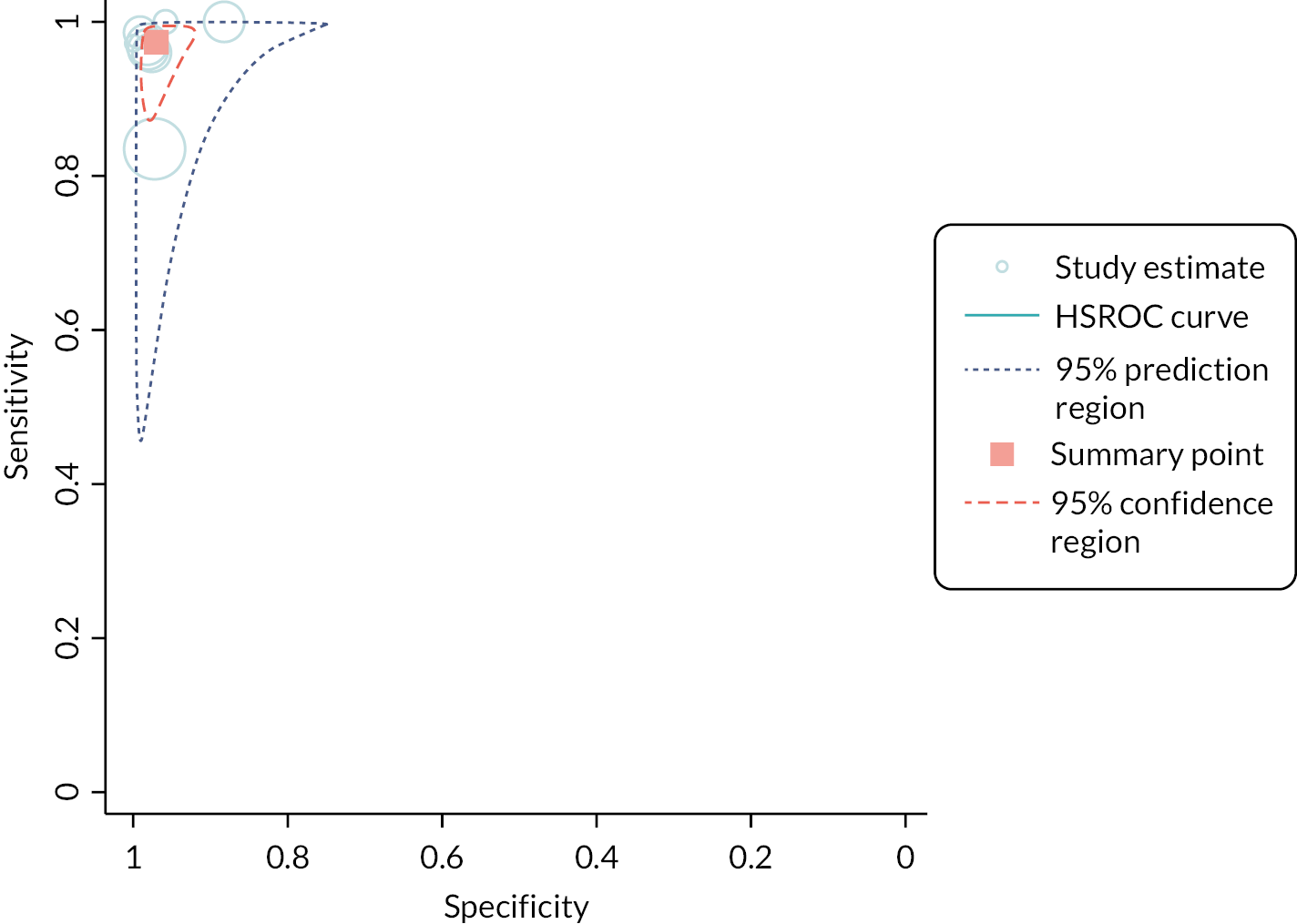
Appendix 10
| Index test | Source of data | No. of included studies (participants) | Outcome | Result (95% CI) | Risk of bias | Indirectness | Inconsistency | Imprecision | Publication bias | Certainty of the body of evidence |
|---|---|---|---|---|---|---|---|---|---|---|
| Multiplex tests | ||||||||||
| All multiplex tests for RSV | Farfour 2022,45 Morris 2021,47 Yin 2022,49 Youngs 2019,50 Zuurbier 202255 | Five studies (2273) | Sensitivity | 84.9% (73.5 to 91.9) | Seriousa | Not serious | Not serious | Very seriousb | Undetected | Very low |
| Specificity | 99.5% (99.1 to 99.7) | Seriousa | Not serious | Not serious | Not serious | Undetected | Moderate | |||
| Cobas Liat tests for RSV | Yin 2022,49 Youngs 201950 | Two studies (965) | Sensitivity | 86.7% (59.5 to 96.6) | Seriousa | Not serious | Not serious | Very seriousb | Undetected | Very low |
| Specificity | 99.3% (98.5 to 99.6) | Seriousa | Not serious | Not serious | Not serious | Undetected | Moderate | |||
| Xpert Xpress tests for RSV | Morris 2021,47 Zuurbier 202255 | Two studies (1109) | Sensitivity | 84.5% (69.4 to 92.9) | Seriousa | Not serious | Not serious | Very seriousb | Undetected | Very low |
| Specificity | 99.6% (99.0 to 99.9) | Seriousa | Not serious | Not serious | Not serious | Undetected | Moderate | |||
FIGURE 14.
Meta-analysis for RSV.
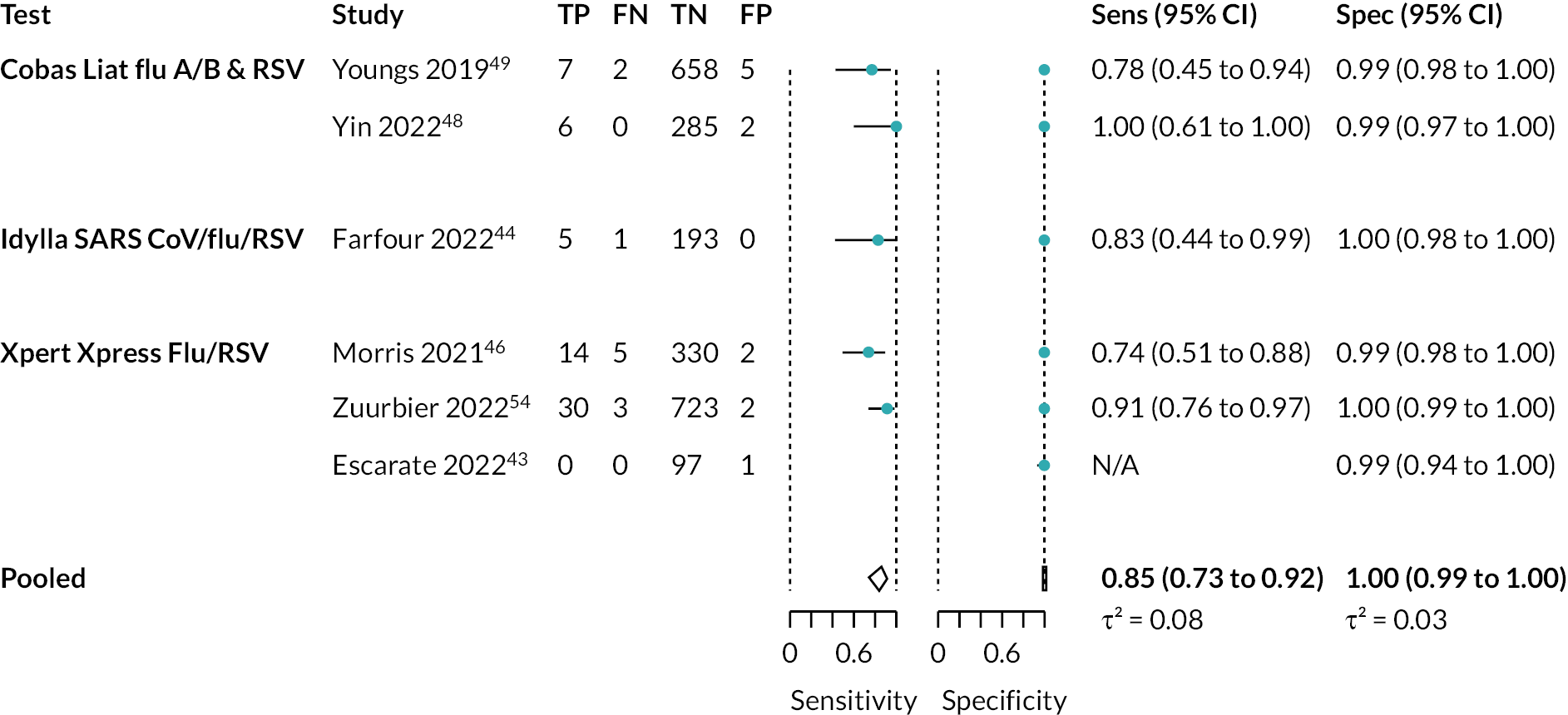
FIGURE 15.
RSV data and meta-analysis results in ROC space.