Notes
Article history
The research reported in this issue of the journal was funded by the HTA programme as project number 14/158/02. The contractual start date was in December 2015. The draft report began editorial review in March 2019 and was accepted for publication in March 2020. The authors have been wholly responsible for all data collection, analysis and interpretation, and for writing up their work. The HTA editors and publisher have tried to ensure the accuracy of the authors’ report and would like to thank the reviewers for their constructive comments on the draft document. However, they do not accept liability for damages or losses arising from material published in this report.
Permissions
Copyright statement
© Queen’s Printer and Controller of HMSO 2020. This work was produced by Allotey et al. under the terms of a commissioning contract issued by the Secretary of State for Health and Social Care. This issue may be freely reproduced for the purposes of private research and study and extracts (or indeed, the full report) may be included in professional journals provided that suitable acknowledgement is made and the reproduction is not associated with any form of advertising. Applications for commercial reproduction should be addressed to: NIHR Journals Library, National Institute for Health Research, Evaluation, Trials and Studies Coordinating Centre, Alpha House, University of Southampton Science Park, Southampton SO16 7NS, UK.
2020 Queen’s Printer and Controller of HMSO
Chapter 1 Background
Pre-eclampsia is a pregnancy-specific condition associated with hypertension and multiorgan dysfunction such as proteinuria, renal or hepatic impairment and fetal growth restriction. 1–5 It is a heterogeneous disorder with a wide spectrum of multiorgan involvement, which reflects its various pathophysiological pathways. Pre-eclampsia affects between 2% and 8% of pregnancies worldwide6 and is a leading cause of both maternal and perinatal morbidity and mortality. 7–10 Each year, 18% of all maternal deaths can be attributed to pre-eclampsia and its complications, with most of these occurring in low- and middle-income countries. 11,12 In the long term, pre-eclampsia is associated with an increased maternal risk of ischaemic heart disease, chronic hypertension, stroke and end-stage renal disease. 13,14 Children from pre-eclamptic pregnancies also have higher risks of cardiovascular diseases,15,16 mental health disorders and cognitive impairment. 17,18
Two subgroups of pre-eclampsia are well recognised: early-onset, requiring delivery before 34 weeks’ gestation, and late-onset, with delivery occurring at or after 34 weeks’ gestation. 19–21 Early-onset pre-eclampsia is considered to be a pathophysiologically different disease from late-onset pre-eclampsia in the mechanism leading to placental dysfunction and clinical timing during pregnancy. 22 Early-onset pre-eclampsia is associated with a considerably higher increased risk of maternal complications, such as a 20-fold higher rate of mortality, than the late-onset type, and early delivery is the only treatment. 23–25 In addition to the prematurity-related complications, the risks of stillbirth and adverse perinatal outcomes are much higher in women with early-onset disease. 26
Although the proportion of women with early-onset pre-eclampsia is < 1% of all pregnancies, the complexity of treatment gives rise to high health-care costs. 27,28 Affected women are often admitted to a tertiary care facility, and 30% experience complications that may necessitate management in an intensive care unit. 29 Infants usually need prolonged care for the management of complications, including lifelong disabilities, arising as a result of premature delivery. The additional NHS costs incurred in caring for a baby born at or before 28 weeks and a baby born between 28 and 33 weeks are £94,190 and £61,509, respectively. 30 The cost to the NHS of caring for preterm babies, linked to neonatal care, such as incubation, and hospital readmissions, has been estimated at £939M annually. 30
Late-onset pre-eclampsia, including pre-eclampsia at term, also poses a significant health burden. It accounts for the majority of pre-eclampsia diagnoses in pregnancy. One-fifth of all women with late-onset disease have maternal complications such as HELLP (haemolysis, elevated liver enzymes and low platelet count) syndrome, and more than half of eclamptic seizures occur at term. 28,31,32
Pregnant women who are at high risk of pre-eclampsia require close monitoring and are usually started on prophylactic aspirin in early pregnancy to reduce the risk of development of pre-eclampsia and occurrence of adverse outcomes. Early commencement of this has the potential for maximum benefit,33 which may be limited to early-onset disease. 34 It is important to be able to quantify a woman’s risk of developing pre-eclampsia during the course of pregnancy to help guide clinical decisions and monitoring strategies. The National Institute for Health and Care Excellence prioritises screening for early-onset pre-eclampsia in its research recommendations on antenatal care of women. 35
Currently, the assessment of a woman’s risk of developing pre-eclampsia is based mainly on clinical history,36 but such risk-based predictions have been shown to have limited accuracy. 37 Risk factors based on clinical characteristics have also been shown to have quantitatively different associations with early- and late-onset pre-eclampsia,26 and, similarly, biochemical and ultrasound markers have variations in their performance in predicting the two types of pre-eclampsia. 37–39 Prediction models incorporating additional tests for biochemical and ultrasound markers may improve the predictive performance of models. 40–42 It is, however, unlikely that a single model will accurately predict both early- and late-onset pre-eclampsia. 26
There are more than 60 multivariable prediction models developed to predict pre-eclampsia, using various combinations of clinical, biochemical and ultrasound risk factors. 43 Such models and tests for predicting pre-eclampsia have been based on findings from aggregate meta-analysis and primary studies, and none is recommended for use in routine clinical practice. This is because there is an absence of information about the reproducibility of the models or their predictive performance in different settings.
Although interventions such as aspirin have been found to significantly reduce the risk of early-onset pre-eclampsia in women predicted to be at ‘high risk’ of pre-eclampsia using a model, lack of robust information on the accuracy of this model means that we could not rule out potential benefit in women considered to be ‘low risk’. Before they can be used in clinical practice, prediction models need to be appropriately validated in multiple data sets external to that used to develop the model. This often takes many years to accomplish in a primary study, and, as a result, very few models have been externally validated to date. 43–46 Individual studies also often have an insufficient sample size to externally validate the relatively rare but serious condition of early-onset pre-eclampsia. 26
Meta-analysis of individual participant data (IPD), whereby the raw participant-level information is obtained and synthesised across multiple data sets, overcomes the limitations above. 47–50 The availability of the raw data substantially increases the sample size beyond what is achievable in a single study, and offers a unique opportunity to evaluate the generalisability of predictive performance of existing models across a range of clinical settings. Using IPD meta-analysis allows the standardisation of predictors and outcome definitions, takes into account the performance of many candidate prognostic variables, directly handles missing predictors and outcomes data, accounts for heterogeneity in baseline risks, and, most importantly, develops, validates and tailors the use of the most accurate prediction models to the appropriate population.
The unmet need for prediction models for pre-eclampsia, particularly early-onset pre-eclampsia, is mainly a result of lack of information on the generalisability of the models and their performances in external cohorts. Hence, before more resources are spent on developing further models, what is needed is external validation of existing models. If existing models’ performances are suboptimal, then further development of new models is warranted with sufficient sample size.
Chapter 2 Objectives
Material in this chapter has been adapted from Allotey et al. 51 This is an Open Access article distributed in accordance with the terms of the Creative Commons Attribution (CC BY 4.0) license, which permits others to distribute, remix, adapt and build upon this work, for commercial use, provided the original work is properly cited. See: http://creativecommons.org/licenses/by/4.0/. The text below includes minor additions and formatting changes to the original text.
We planned to externally validate and update prediction models for (1) early-onset (delivery at < 34 weeks’ gestation), (2) late-onset (delivery at ≥ 34 weeks’ gestation) and (3) any-onset pre-eclampsia, and to further develop new prediction models for the above outcomes, if required, using IPD meta-analysis.
Primary objectives
-
To validate and improve or tailor the performance of existing models in relevant population groups for predicting early-onset, late-onset and any-onset pre-eclampsia in our IPD data set based on:
-
clinical characteristics only
-
clinical and biochemical markers
-
clinical and ultrasound markers
-
clinical, biochemical and ultrasound markers.
-
-
Using IPD meta-analysis, to develop and externally validate (using internal–external cross-validation) multivariable prediction models for early, late and any-onset pre-eclampsia in the following circumstances:
-
where existing predictive strategies cannot be adjusted for the target population
-
where no such models exist for the relevant pre-eclampsia outcomes.
-
-
To estimate the prognostic value of individual clinical, biochemical and ultrasound markers for predicting pre-eclampsia by IPD meta-analysis.
Secondary objectives
-
To assess the differential performance of the existing models in various predefined subgroups based on population characteristics (unselected; selected) and timing of model use (first trimester; second trimester).
-
To study the added accuracy when novel metabolic and microRNA-based biochemical markers are added to the developed model based on clinical, ultrasound and biochemical markers.
Chapter 3 Methods
Material in this chapter has been adapted from Allotey et al. 51 This is an Open Access article distributed in accordance with the terms of the Creative Commons Attribution (CC BY 4.0) license, which permits others to distribute, remix, adapt and build upon this work, for commercial use, provided the original work is properly cited. See: http://creativecommons.org/licenses/by/4.0/. The text below includes minor additions and formatting changes to the original text.
Part of this chapter have been reproduced from Snell et al. 52 Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
Our IPD meta-analysis followed existing recommendations on prognostic research model development and validation53–55 and adhered to reporting guidelines for prediction models and IPD meta-analysis. 56,57 We used a prospective protocol51 registered on the International Prospective Register of Systematic Reviews (PROSPERO) as CRD42015029349. 58
Eligibility criteria
Criteria for including relevant cohorts and studies in the individual participant data
We included primary studies (prospective and retrospective cohort studies, as well as cohorts nested within randomised trials), and large birth and population-based cohorts with information to assess the accuracy of clinical, biochemical and ultrasound predictors in women at low, high or any risk to predict early-, late- or any-onset pre-eclampsia and its associated complications. The process of identifying and selecting studies to invite to form the IPPIC network is described in detail in our published protocol, as well as in the following sections (see Literature search and study identification, The IPPIC pre-eclampsia network and Study selection, individual participant data collection and harmonisation). Studies or cohorts that addressed the structured question in Table 1 were included in the IPD. The predictors considered for evaluation were chosen a priori and were clearly defined and standardised. 39,60–72
| Question components | Definition |
|---|---|
| Population | Pregnant women |
| Predictors |
Maternal clinical characteristics at antenatal booking Maternal characteristics: age, BMI, height, weight, ethnicity, smoking, alcohol or substance misuse Medical history: pre-existing chronic kidney disease, heritable thrombophilias, autoimmune disease such as systemic lupus erythematosus and antiphospholipid syndrome, type 1 and 2 diabetes and hypertensive diseases Obstetric history: parity, previous pre-eclampsia, gestational diabetes, pregnancy interval of > 10 years, family history of pre-eclampsia, family history of cardiovascular disease, previous miscarriages, preterm birth, stillbirth or SGA fetus Current pregnancy: multiple pregnancy, mode of conception, early pregnancy bleeding, MAP, SBP and DBP, socioeconomic status, new partner, diet or exercise in pregnancy, urine dipstick, PCR, 24-hour protein Biochemical markers (first or second trimester): PAPP-A, sFlt-1, PlGF, AFP, human chorionic gonadotropin, sENG, CRP, hypertriglyceridaemia and PAI-1 Ultrasound markers (first or second trimester) CRL, abdominal circumference, expected fetal weight centile, uterine and umbilical artery Doppler (resistance index, pulsatility index, unilateral or bilateral notching) |
| Outcomes |
Primary outcomes Early-onset (delivery at < 34 weeks’ gestation), late-onset (delivery at ≥ 34 weeks’ gestation) and any-onset pre-eclampsia Secondary outcomes Maternal complications: eclampsia, HELLP syndrome, abruption, hepatic and renal failure, cortical blindness, pulmonary oedema, postpartum haemorrhage, disseminated intravascular coagulation, preterm delivery, admission to high-dependency unit/intensive care unit, maternal death, caesarean section, gestational diabetes mellitus Fetal and neonatal complications: birthweight in kg and centile (using the Gestation Network bulk centile calculator59), SGA fetus, stillbirth, neonatal death, hypoxic-ischaemic encephalopathy, respiratory distress syndrome, septicaemia, admission to neonatal unit |
| Study design | Observational studies and cohorts nested within randomised trials |
The primary outcomes were early-onset (delivery at < 34 weeks’ gestation), late-onset (delivery at ≥ 34 weeks’ gestation) and any-onset pre-eclampsia. The authors reported the definition of the primary outcome of pre-eclampsia along with gestational age at delivery, which was used to define early- and late-onset disease. Definitions of pre-eclampsia included proteinuric and non-proteinuric pre-eclampsia. 35,73
The secondary outcomes were composite adverse maternal or fetal and neonatal outcomes.
Literature search and study identification
We undertook a systematic review of reviews to identify relevant systematic reviews on clinical characteristics, biochemical and ultrasound markers for prediction of pre-eclampsia. 37 We adhered to the PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) guidelines on reporting, and the review was based on a prospective protocol. The systematic review methods have been published elsewhere, but, briefly, two reviewers independently screened abstracts, extracted data and carried out quality assessment. 37 We defined the inclusion and exclusion criteria for the systematic reviews, the outcome of interest (pre-eclampsia) and the predictors. We also updated our previous literature search of prediction models for pre-eclampsia43 (July 2012–December 2017) to identify additional models. We searched the following databases: MEDLINE, EMBASE, Bioscience Information Services (BIOSIS), Latin American and Caribbean Health Sciences Literature (LILACS), PASCAL, Science Citation Index, Cochrane Database of Systematic Reviews, Cochrane Central Register of Controlled Trials (CENTRAL), National Institute of Child and Human Development Data and Specimen Hub, Database of Abstracts of Reviews of Effects (DARE) and Health Technology Assessment database without any language restrictions. Research reported in grey literature was sought by searching a range of relevant databases including the Inside Conferences, Systems for Information in Grey Literature, MotherChild Link Registry (www.linkregistry.org/search.aspx), Dissertation Abstracts and ClinicalTrials.gov. Data extraction following the update of our literature search for prediction models for pre-eclampsia was carried out by two reviewers independently.
We used additional sources such as internet searches using general search engines (e.g. Google; www.google.co.uk/) and meta-search engines (e.g. Copernic; www.copernic.com/), and directly contacted researchers to identify relevant studies, birth cohorts or data sets that may have been missed. Collaborative groups such as the Global Pregnancy Collaboration (CoLab), Pre-eclampsia and Eclampsia Monitoring, Prevention and Treatment (PRE-EMPT) and the Global Obstetrics Network (GONet) were also approached to identify primary studies, unpublished research and birth cohorts. 74–76 We did not include studies, birth cohorts or data sets after October 2017, as we needed time to clean and format the data prior to any analysis. The details of the search strategy are provided in Appendix 2.
The IPPIC pre-eclampsia network
We established a collaborative network of investigators (IPPIC) from research groups that have undertaken studies on clinical characteristics, biochemical and ultrasound markers in the prediction of early- and any-onset pre-eclampsia. The network is a global effort bringing together 125 researchers, clinicians and epidemiologists from 25 countries and is supported by the World Health Organization (WHO). We invited authors of all primary studies identified from this review and also invited investigators of primary studies and large birth and population-based cohorts that were not included in existing reviews but were identified through our links with other collaborative groups74–76 if these provided relevant information to assess the accuracy of clinical, biochemical and ultrasound predictors of pre-eclampsia.
Study selection, individual participant data collection and harmonisation
The collaborative group agreed the minimum data to be requested for the IPD meta-analysis, and a custom-built database was set up based on these specifications. The minimum data requested were pre-eclampsia outcome with gestational age at delivery, as well as any of the clinical, biochemical and ultrasound predictors of pre-eclampsia listed in Table 1. Authors of the primary studies and data sets were contacted to ask if they would share their IPD in any format, along with data dictionaries or descriptions. We identified and invited authors and investigators from 180 data sets to join the project and share their IPD, with at least two further reminders to share data for the project. We continued to contact authors to request that they share their data until the October 2017 deadline for receiving new data sets was reached. When a data set received contained IPD from multiple studies, we checked the identity of each study to avoid duplication.
Original pseudonymised data sets were uploaded to a secure data storage environment (SafeHaven) at the Pragmatic Clinical Trials Unit, Queen Mary University of London, accessible only from a virtual desktop where manipulation of the data along with relevant data checks and documentation took place. The final merged data set, individual formatted files and documentation of all the transformations made were securely transferred to a web-based server at Centro Rosarino de Estudios Perinatales, Rosario, Argentina, a WHO Collaborative Centre in Child and Maternal Health. An independent Data Access Committee and data access process were established to facilitate access to and use of the data for future research.
Data extraction
We considered all recorded variables for inclusion, including those not reported in the published studies. At the study level, we extracted data on the providing collaborator, study design, data source, study period and study inclusion and exclusion criteria. At the participant level, we extracted information on individual participant characteristics and outcome data, as specified in Table 1.
Data harmonisation and recovery
Maternal age at baseline was recorded as a continuous variable in years in all data sets except three, in which age was calculated using the date of study or booking visit and the date of birth. Data on parity, defined as the number of pregnancies > 24 weeks’ gestation, were mostly recorded in the binary format (nulliparous/multiparous). Any continuous data for parity were therefore also transformed to the binary form. However, we retained the continuous data for any relevant analyses. Assumptions were made when harmonising the ethnicity variable, and this was recoded as white, black, Asian, Hispanic, mixed and other. Pre-gestational diabetes, type 1 diabetes and type 2 diabetes were harmonised as history of diabetes, and history of systemic lupus, multiple sclerosis, idiopathic thrombocytopenia, rheumatoid arthritis or antiphospholipid syndrome were harmonised as history of autoimmune disease. We also harmonised history of glomerulonephritis, nephrotic syndrome, nephritis or nephropathy as history of renal disease.
Maternal characteristic data, such as history of disease and previous pregnancy, were recovered by screening the participant inclusion and exclusion criteria of published articles when this information had not been provided in the original data set. We added data based on existing information contained in the data set and from published articles. For example, we inputted the data as ‘no previous history of pre-eclampsia’ if all participants in the data set were nulliparous.
Body mass index (BMI) (measured in kg/m2), systolic blood pressure (SBP), diastolic blood pressure (DBP) and mean arterial blood pressure (MAP) were recorded as continuous measures. Where MAP was not provided but SBP and DBP were, we derived MAP using the formula MAP = (SBP + 2 × DBP)/3. Where BMI was not provided in the data set it was derived using weight (kg)/height (m2). We derived the estimated fetal and birth weight centiles using the Perinatal Institute GROW centile calculator. 77 Mean values of uterine and umbilical artery pulsatility index were mostly reported in data sets. When these were not available, we derived mean uterine and umbilical artery pulsatility index by averaging the left and right pulsatility index measurements. Data on biochemical marker platform, assay and measurement range were obtained for each relevant recorded biochemical marker in the data sets. Conversion factors were applied where necessary to harmonise the units of measurement. Placental growth factor (PlGF) was mostly reported in the data sets as pg/ml and standardised as such, pregnancy-associated plasma protein A (PAPP-A) was standardised as mIU/l and soluble fms-like tyrosine kinase-1 (sFlt-1) was standardised as pg/ml. The authors’ reported definition of the primary outcome of pre-eclampsia along with gestational age at delivery was used to define early- and late-onset disease.
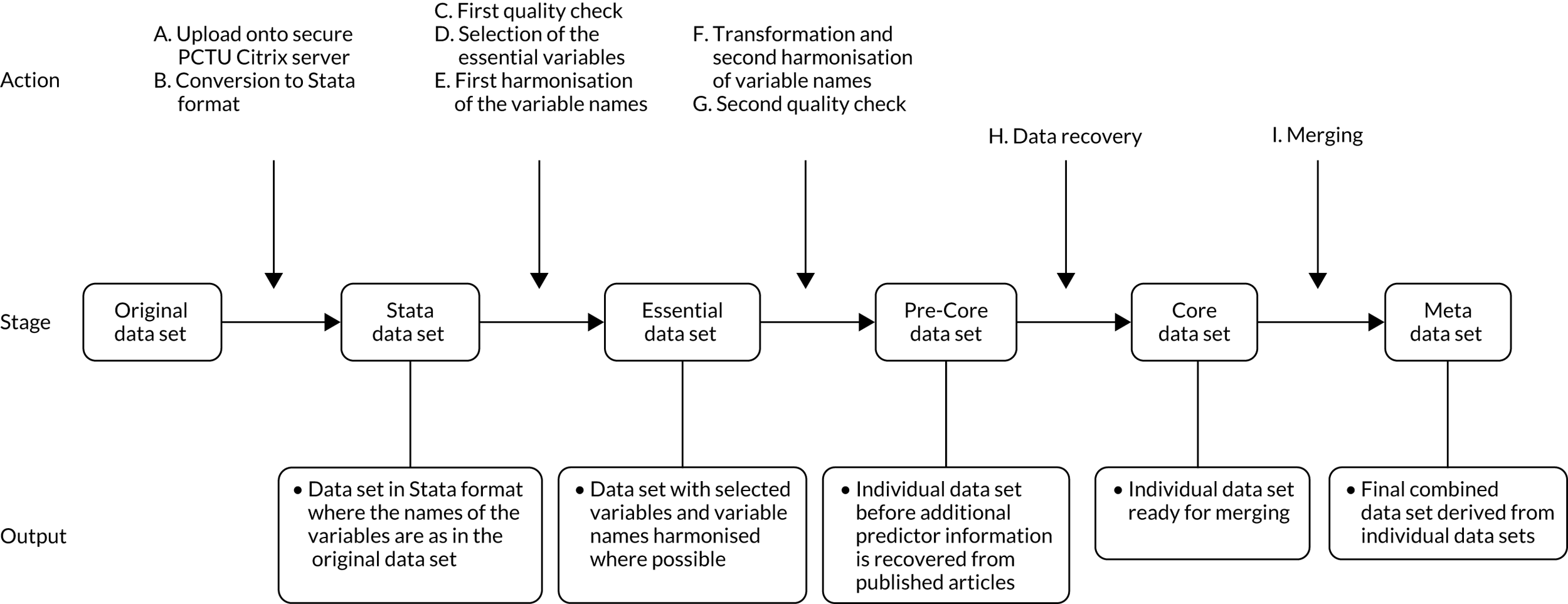
Clinical examinations, biochemical and ultrasound markers were further categorised into the trimester in which they were measured. We defined first-trimester values as ≤ 14 weeks, second-trimester values as > 14–28 weeks and third-trimester values as > 28 weeks. Where more than one variable measurement was available for a woman in a trimester, we chose the first or the earliest measurement. Harmonisation of the data sets followed the predefined process shown in Figure 1. A final list of the variables collected and harmonised for the project is provided in Appendix 3.
FIGURE 1.
Flow diagram of harmonisation of variables in the IPD data sets.
Data quality
Range and consistency checks were carried out on all data sets received, and summary tables were produced. Missing data > 10% for each variable, range checks for continuous variable measures, obvious errors, and inconsistencies between pre-identified variables or outlying values were queried and rectified with input from the original authors. Two reminders were sent to the original author to respond to queries, and if no response was received, a decision to exclude the variable in question was made by the project team.
Prioritisation of predictors
We carried out a prospective online prioritisation survey of IPPIC Network collaborators to identify the most clinically relevant predictors of pre-eclampsia for consideration in the development of the prediction models. Collaborators ranked the importance of the predictor variables identified on a scale from one (not important) to five (very important). The results were analysed as mode and interquartile range (IQR) to show variability and consensus in opinions. We a priori identified an IQR of ≤ 1 as indicating consensus between responders. Variables with mode of ≥ 4 and IQR of ≤ 1 were categorised as important, and those with mode of < 4 and IQR > 1 were categorised as unimportant. Variables with mode of 3 and IQR of ≤ 1 were reviewed and recategorised for importance at a consensus meeting.
Quality assessment
We assessed the risk of bias in individual studies and data sets using a modified version of the Prediction study Risk of Bias Assessment Tool (PROBAST). 78 The tool assessed the quality of data sets and individual studies across three domains: participant selection, predictors and outcomes. We classified the risk of bias to be low, high or unclear for each of the relevant domains. Each domain included signalling questions that were rated as ‘yes’, ‘probably yes’, ‘probably no’, ‘no’ or ‘no information’. Any signalling question rated as ‘probably no’ or ‘no’ indicated a potential for bias in the IPD received for that study, which was therefore classed as having a high risk of bias in that domain. The overall risk of bias of an IPD data set was considered low if the data set scored low in all domains, high if any one domain had a high risk of bias, and unclear for any other classifications.
Sample size considerations
No formal sample size requirements were necessary for the meta-analysis. However, to develop a sound prediction model, as a rule of thumb, 10 events are required for each candidate predictor variable. Early-onset pre-eclampsia is uncommon, occurring in only about 0.25–0.50% of all pregnancies. We conservatively estimated that our IPD data set of > 3 million pregnancies would allow us access to about 7500 women with pregnancies complicated by early-onset pre-eclampsia if they all recorded all of the predictors of interest. This would enable us to develop and robustly validate prediction models for the outcomes of any-onset, early-onset and late-onset pre-eclampsia.
Data synthesis
Analysis for prioritisation of pre-eclampsia predictors was carried out using SPSS version 24 (IBM SPSS Statistics for Windows, IBM Corporation, Armonk, NY, USA). IPD meta-analysis to estimate the prognostic value of individual predictors was carried out using Stata® version 12.1 (StataCorp LP, College Station, TX, USA) and R version 3.4.3 (The R Foundation for Statistical Computing, Vienna, Austria). External validation of existing models and model development were carried out using Stata MP version 15 (StataCorp LP, College Station, TX, USA) and R version 3.4.3. The combination of Stata and R was over a single software package to utilise the appropriate packages for each task. For example, multilevel imputation across data sets can be carried out using the ‘jomo’ package in R, which currently has no equivalent package in Stata.
External validation of existing pre-eclampsia prediction models
We validated each published pre-eclampsia prediction model that reported the full model equation with intercept and predictor effects in IPD from the UK. Analysis was restricted to the IPPIC-UK data sets to allow for the determination of the predictive performance of available models in the context of the UK health-care system and to reduce the heterogeneity in the definition of the outcome, which can vary across IPPIC data sets for different countries. 4,35 We included UK data sets or subsets of international data sets with UK participants if country of recruitment was recorded. We validated a prediction model only if at least one IPD data set contained values of all the predictors included in the model. We excluded data sets with no variation in the model predictions across individuals (i.e. every individual had the same predicted probability as a result of strict eligibility criteria). Smaller data sets with no outcome event or a single outcome event were also excluded, as were women with multifetal pregnancies, as the published models were intended to predict the risk of pre-eclampsia in women with singleton pregnancies only.
Missing data
Second-trimester measurements of BMI and MAP were used for model validation if the first-trimester values were missing from the data set.
Any predictors partially missing or outcome values missing for < 95% of individuals in a data set were multiply imputed under the missing at random assumption using multiple imputation by chained equations. 79,80 Imputation was carried out separately in each UK data set, which acknowledged the clustering of individuals within a data set. We generated 100 imputed data sets for each data set with any missing predictor or outcome values. Linear regression was used to impute for approximately normally distributed continuous variables, predictive mean matching for skewed continuous variables, logistic regression for binary variables, and multinomial logistic regression for categorical variables. Complete predictors were also included in the imputation models as auxiliary variables. To retain congeniality between the imputation and predictive models, the scale used to impute the continuous predictors was chosen to match the prediction models. 81 We undertook imputation checks by looking at histograms, summary statistics and tables of values across imputations, as well as by checking the trace plots for convergence issues.
We summarised the total number of participants, the number of events for each data set, and the overall numbers available for each model validation. We applied the model to each individual i in each (imputed) data set by calculating the linear predictor (LPi = α + β1 × x1 + β2 × x2 + . . .), and the predicted probability of pre-eclampsia,
for logistic regression equations; others detailed separately). For each prediction model, we summarised the overall distribution of the LP by data set using the median, IQR and full range, averaging statistics across imputations. 82
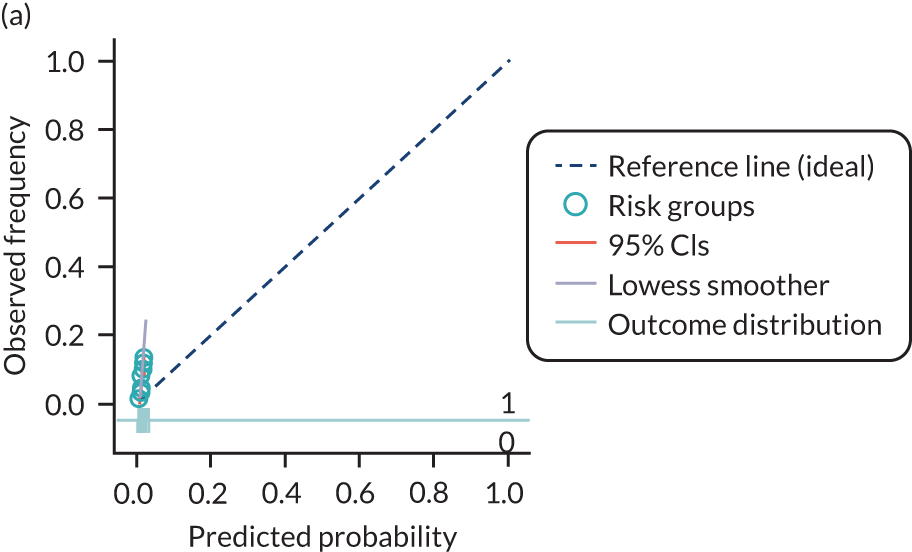
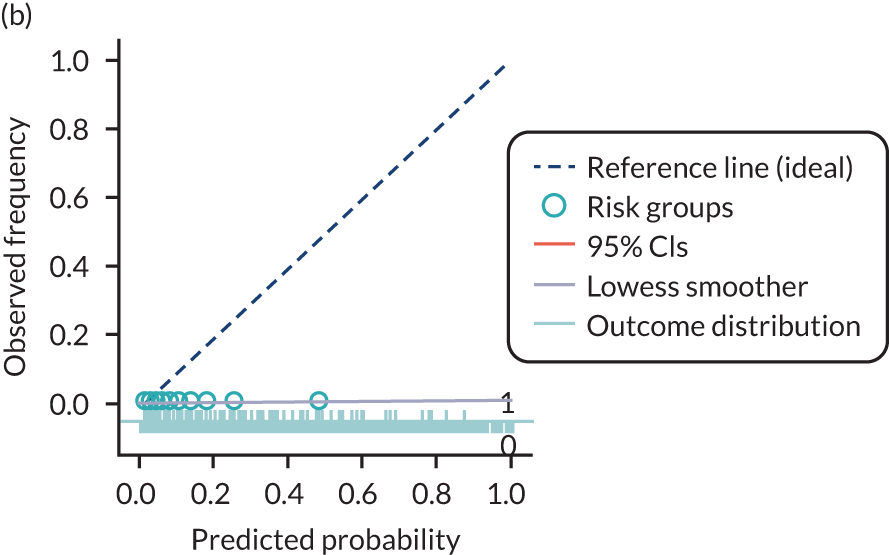
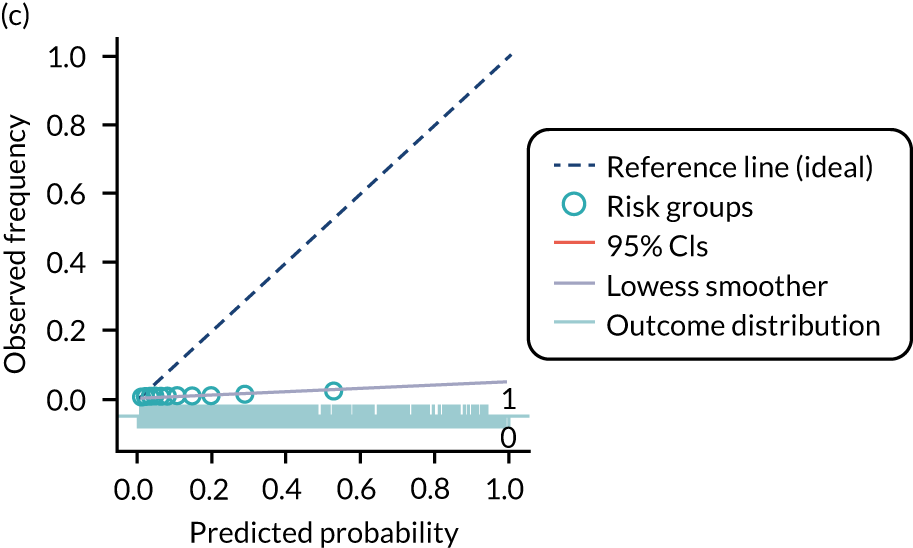
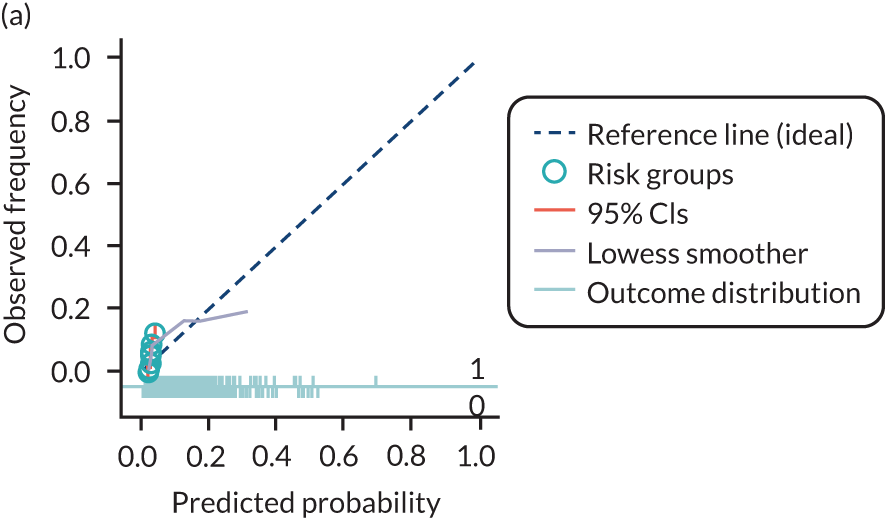
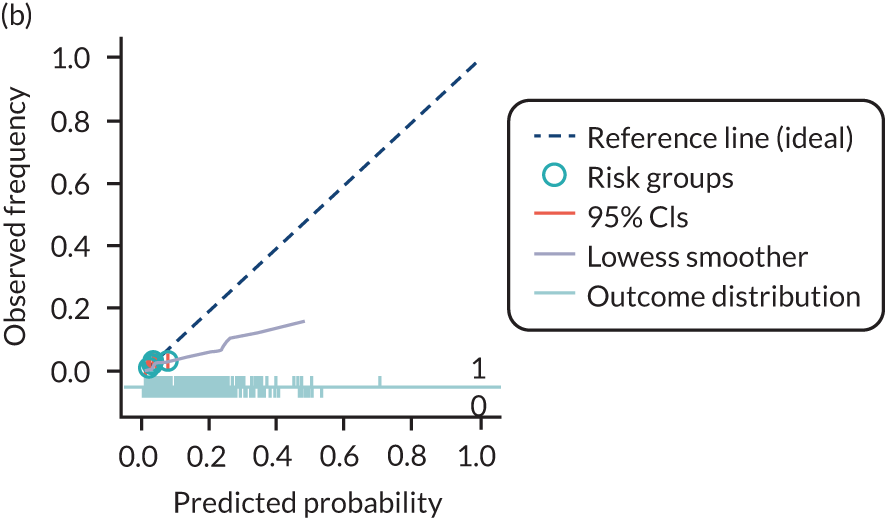
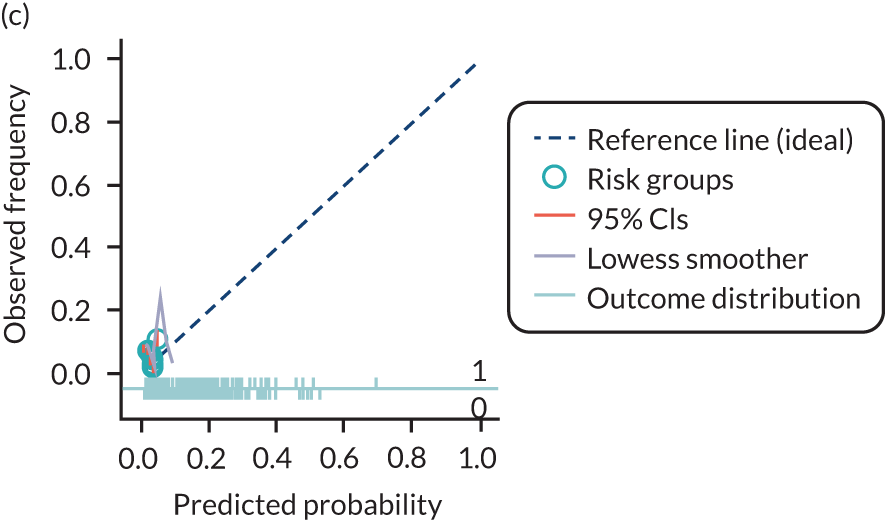
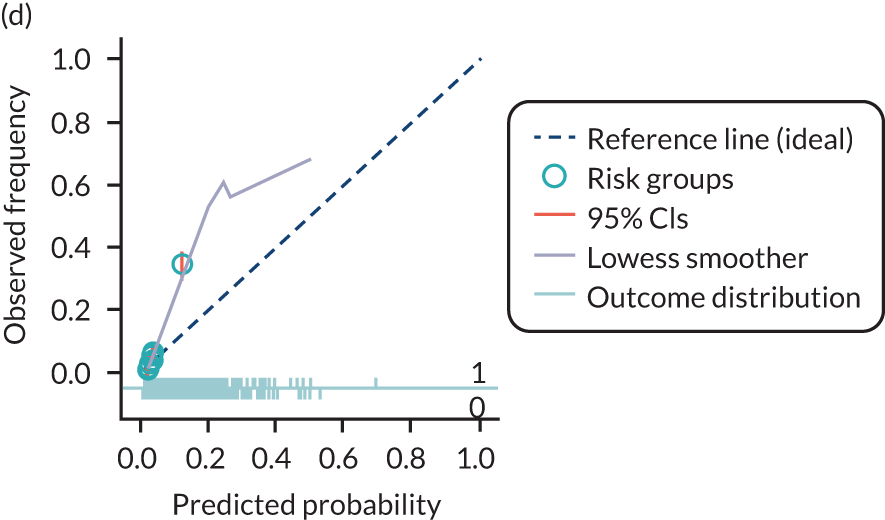
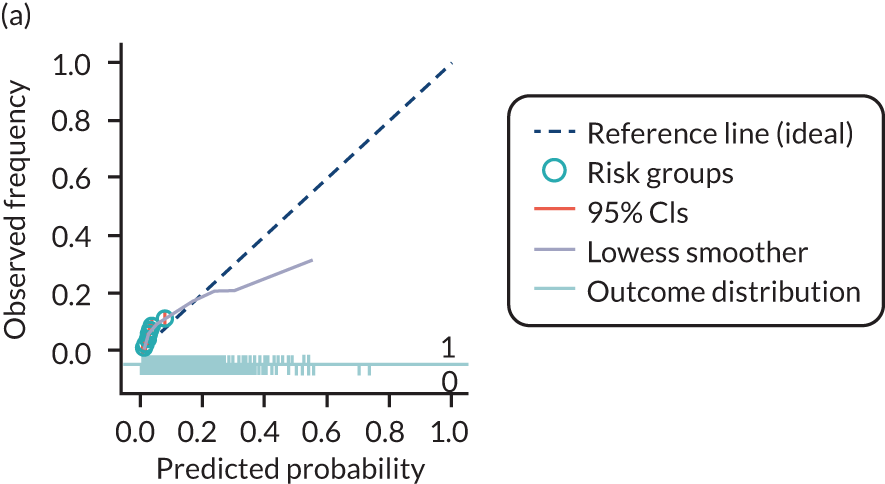
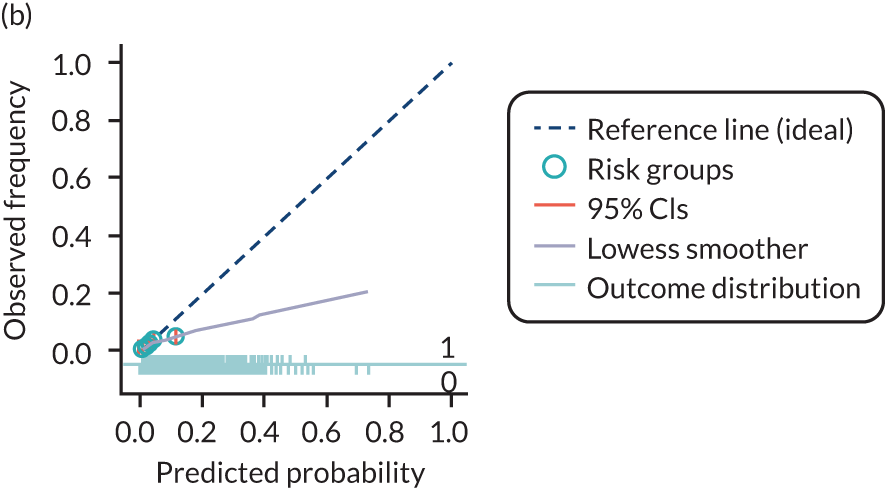
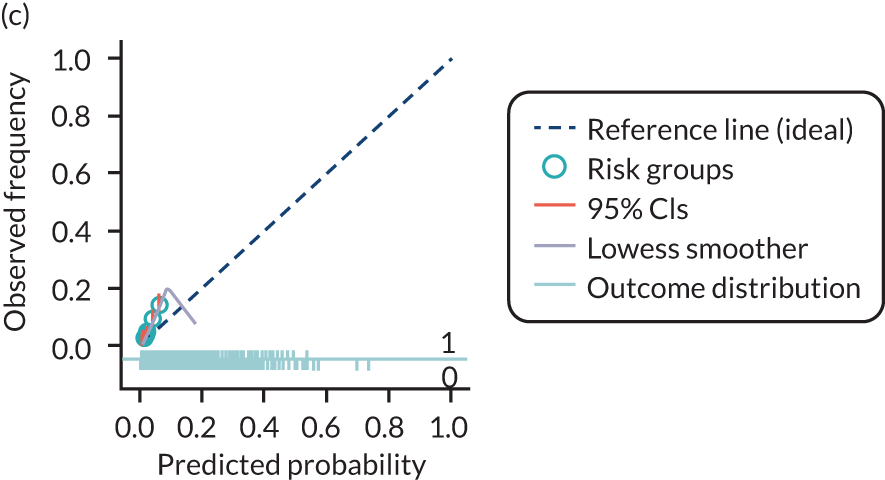
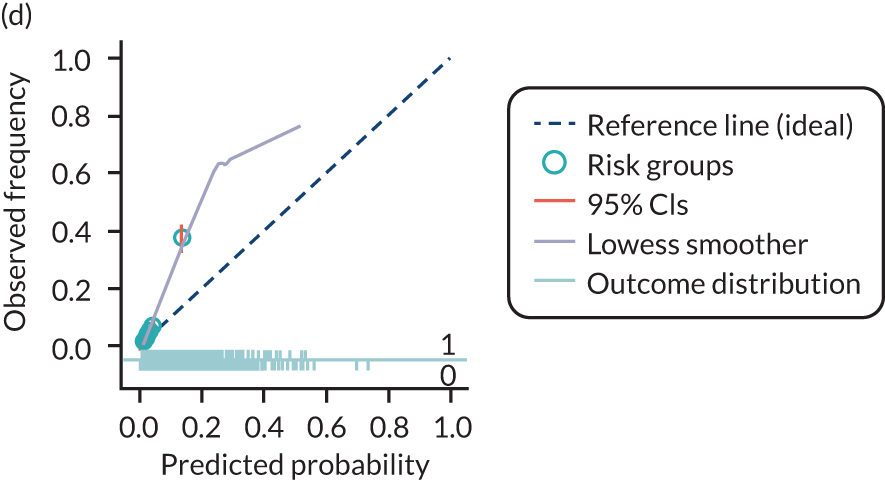
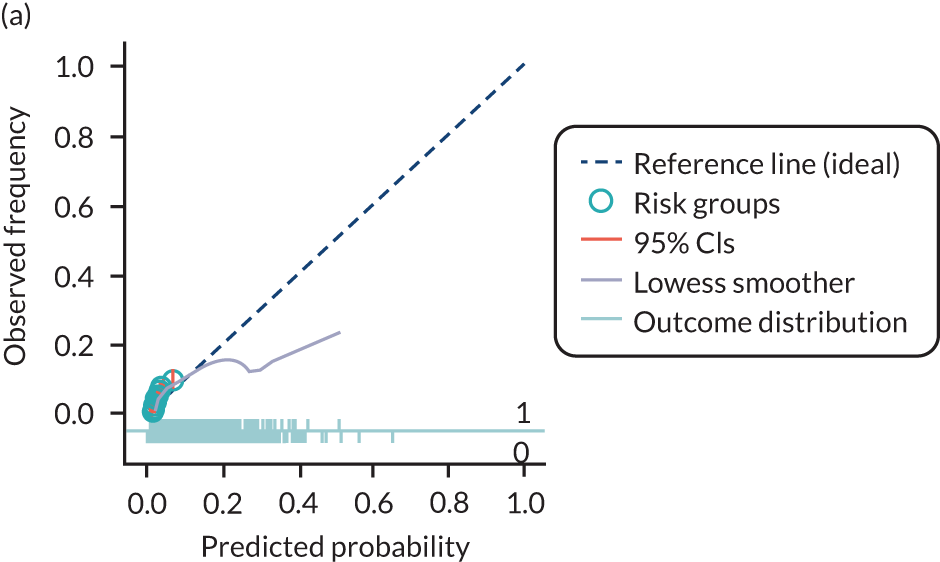
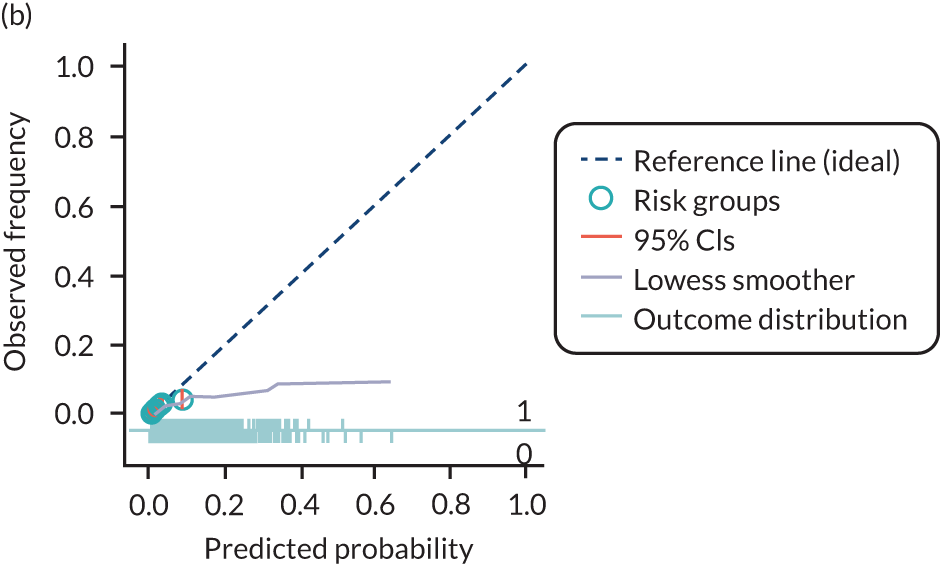
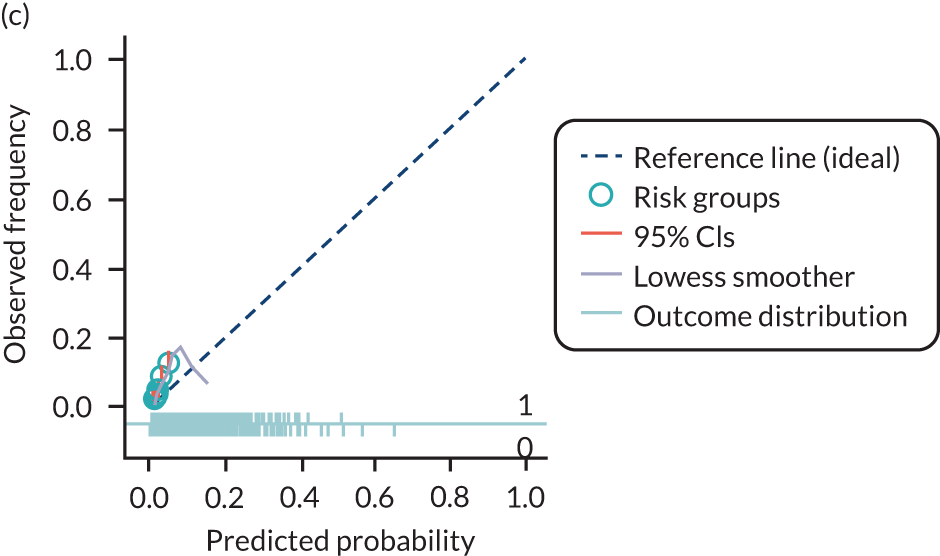
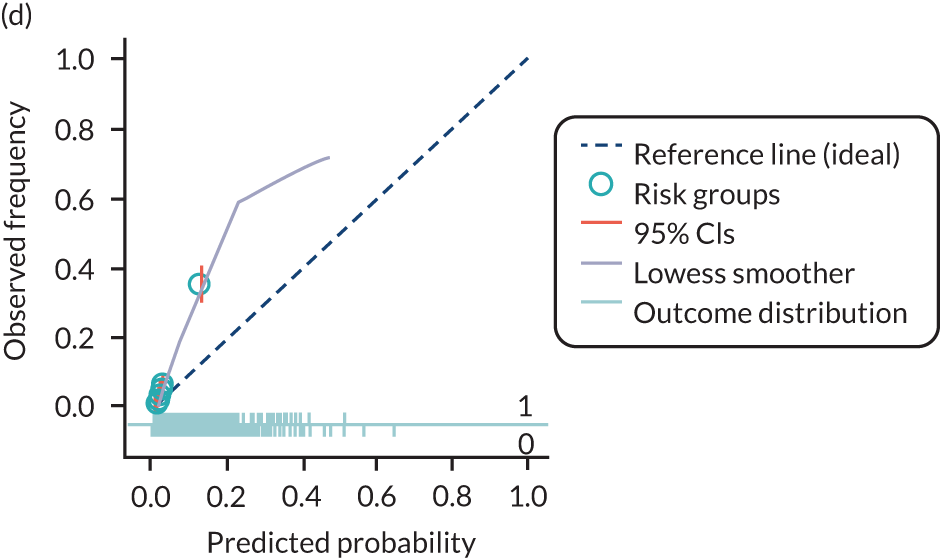
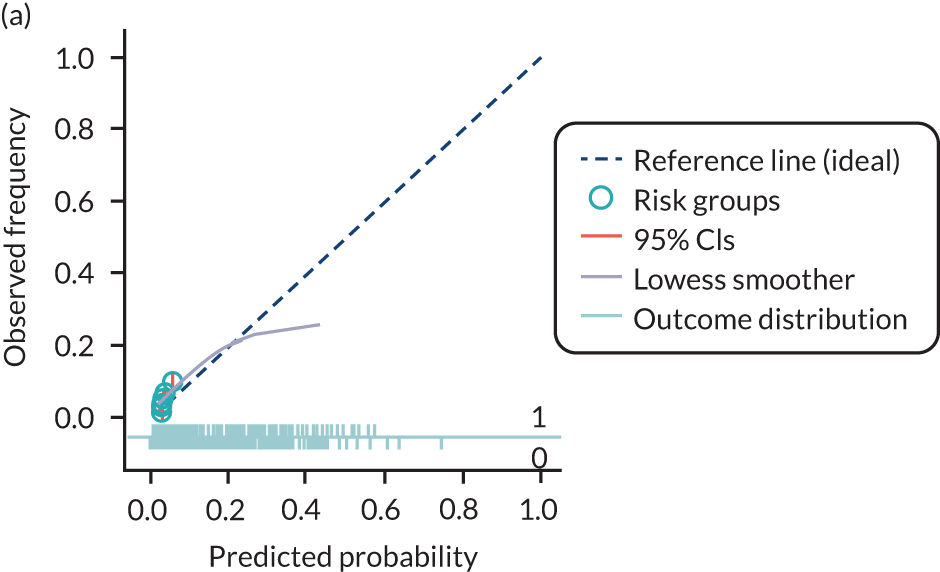
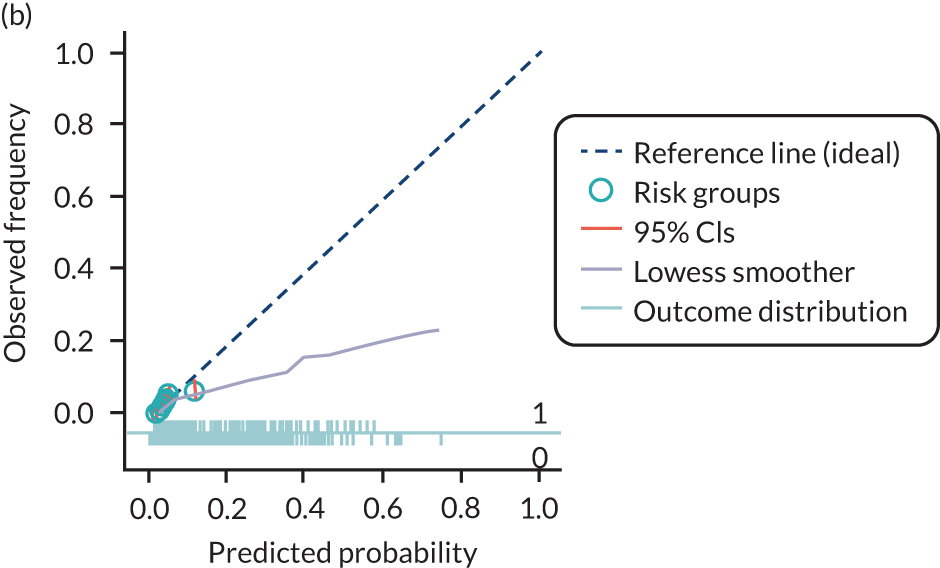
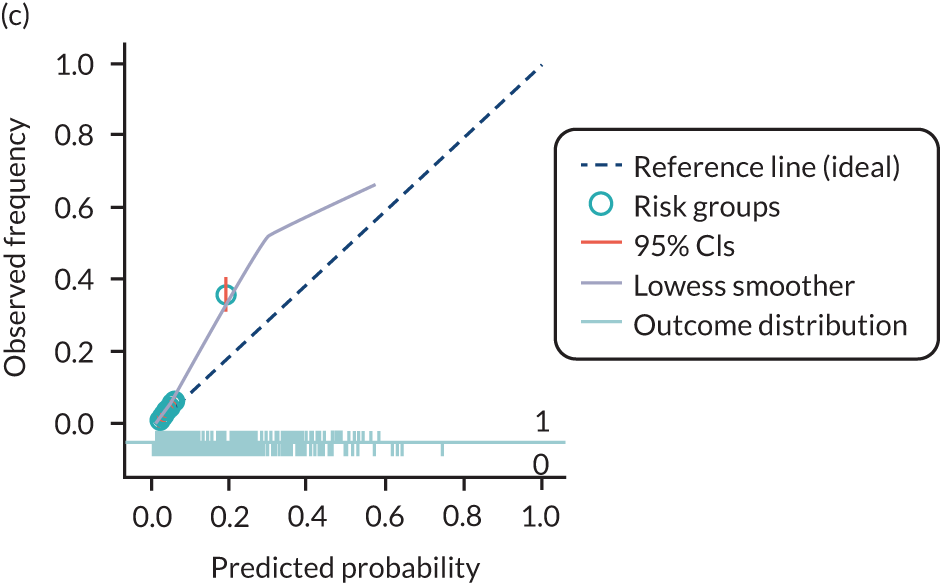
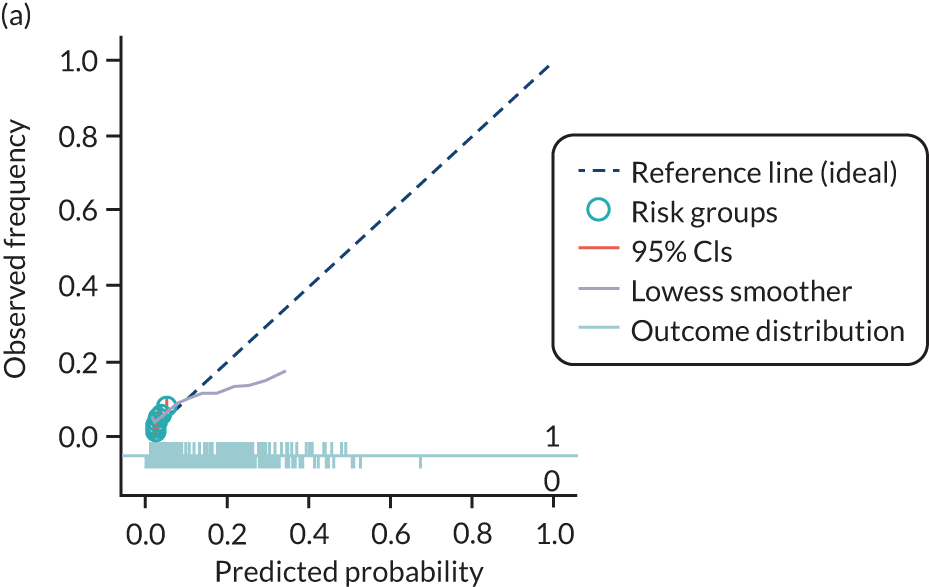
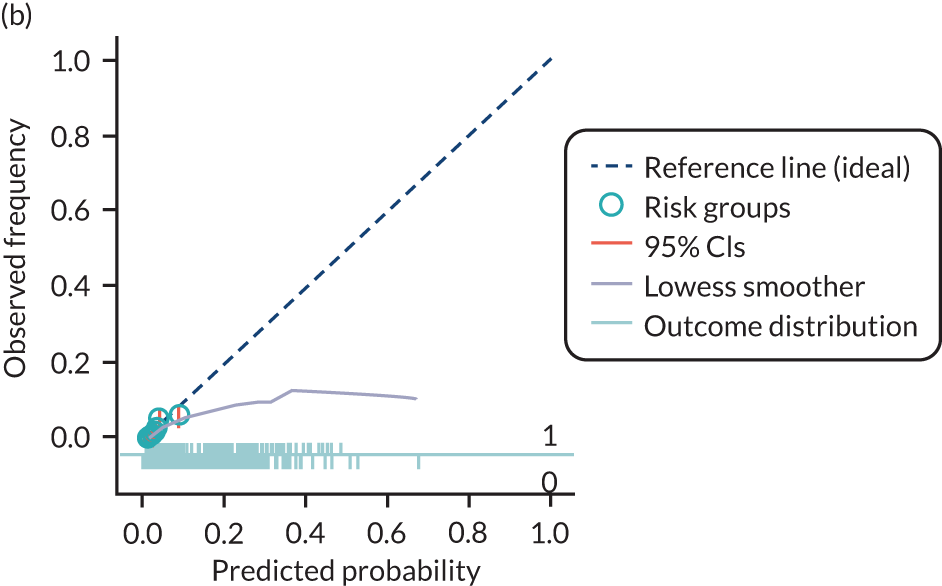
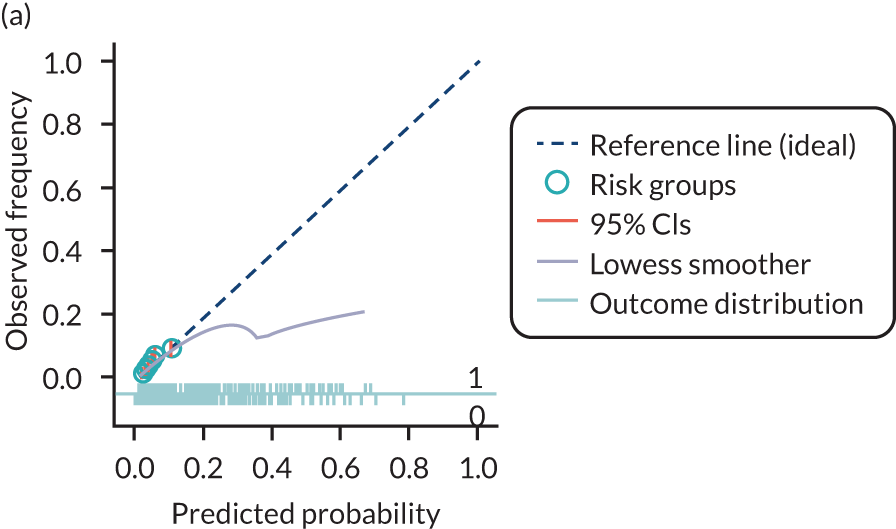
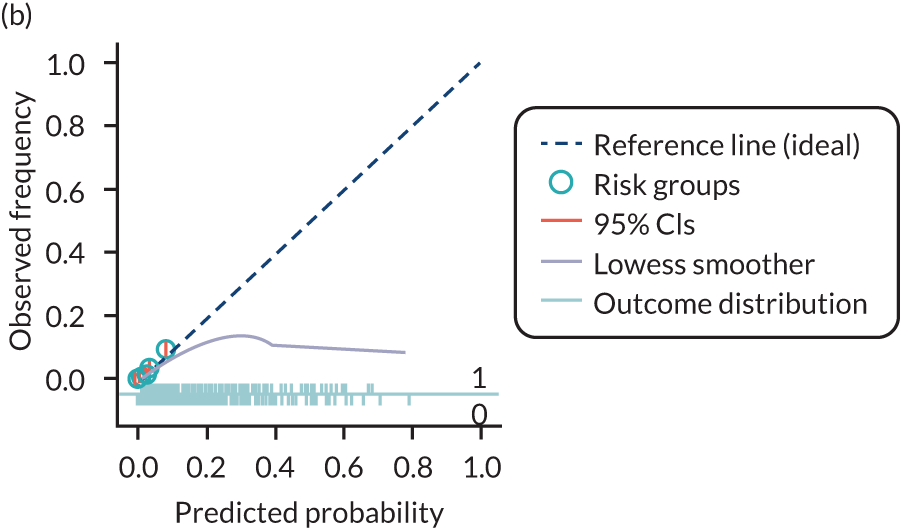
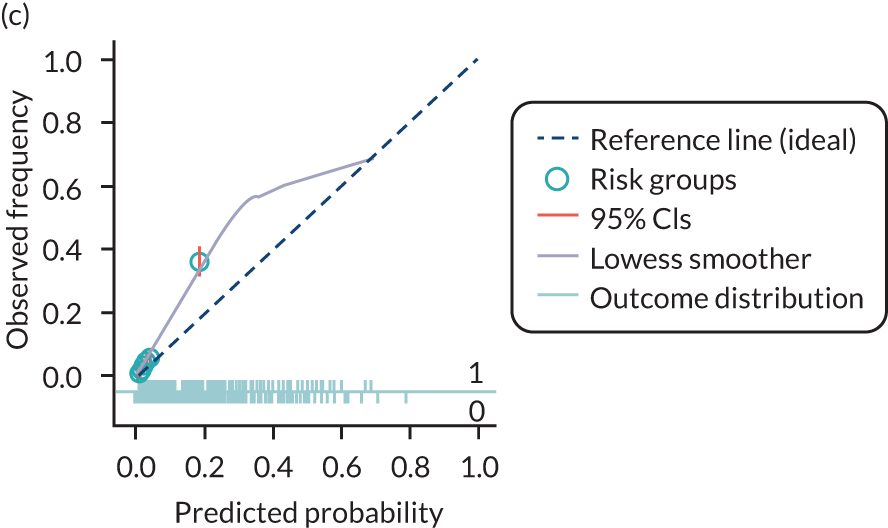
The predictive performance of each model was examined using measures of discrimination and calibration, first in the IPD for each available data set and then across data sets at the meta-analysis level. Discrimination is the ability of the model to separate between women who develop pre-eclampsia and those who do not, and was summarised using the C-statistic (equivalent to the area under the receiver operating characteristic curve for a logistic regression prediction model), with a value of 1 indicating perfect discrimination and a value of 0.5 indicating no discrimination beyond chance. We considered values > 0.7 to be most promising, given previously reported values in the literature, while noting the width of the confidence intervals (CIs). 83 Calibration refers to how well the risk predictions from the model agree with the observed outcome risks for individuals in a data set. Calibration was assessed using two measures: calibration slope, which is the slope of the regression line fitted to the relationship between predicted and observed risk probabilities on the logit scale (ideal value of 1); and calibration-in-the-large, which indicates whether risk predictions are systematically too high or too low (ideal value of 0). We produced calibration plots in each data set to visually compare observed and predicted probabilities when there were enough events to categorise participants into risk groups. The predicted probability of pre-eclampsia for each individual was obtained by pooling the imputation-specific estimates of the model’s LP and then applying the logit transformation. 82
Where data had been imputed in a particular IPD data set, the predictive performance measures were calculated in each of the imputed data sets, and Rubin’s rules were then applied to combine statistics (and corresponding standard errors) across imputations. 84 As the C-statistic is a proportion, it is unlikely to be normally distributed. Hence, we combined C-statistics across imputations on the logit scale,85 and standard errors for logit C-statistics were calculated using the delta method, as recommended. 86
When it was possible to validate a model in multiple data sets, we summarised the performance measures across data sets using a random-effects meta-analysis estimated using restricted maximum likelihood (for each performance measure separately). 86,87 Summary (average) performance statistics were reported with 95% CI (derived using the Hartung–Knapp–Sidik–Jonkman variance correction). 88 We also reported the estimate of between-study heterogeneity (τ2) and the proportion of variability due to between-study heterogeneity (I2). We generated plots to show and compare the average performance (across data sets) of multiple models, along with CIs.
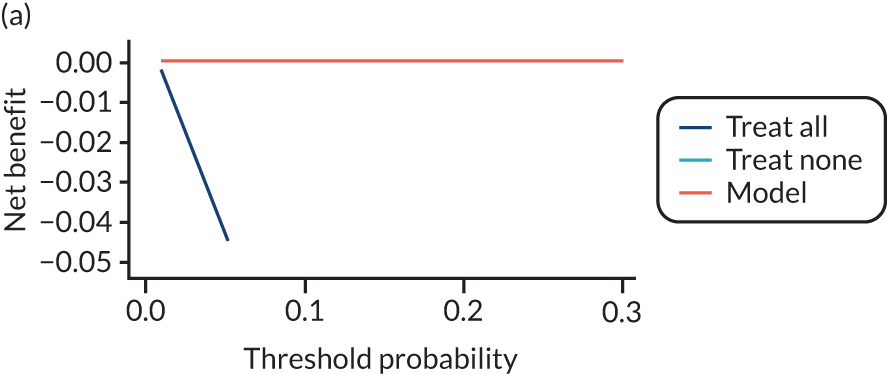
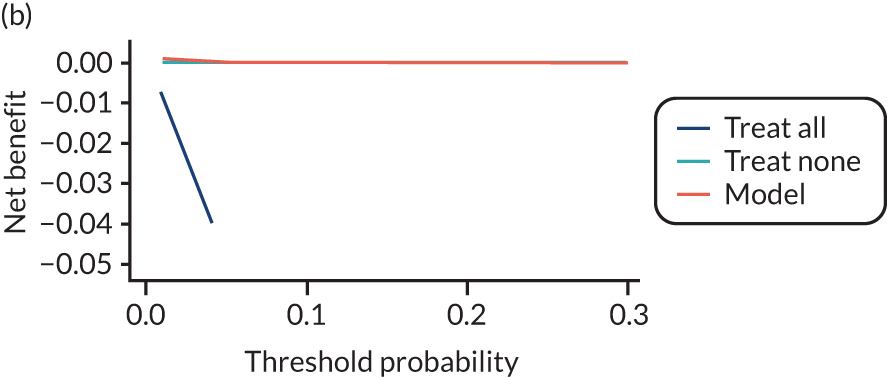
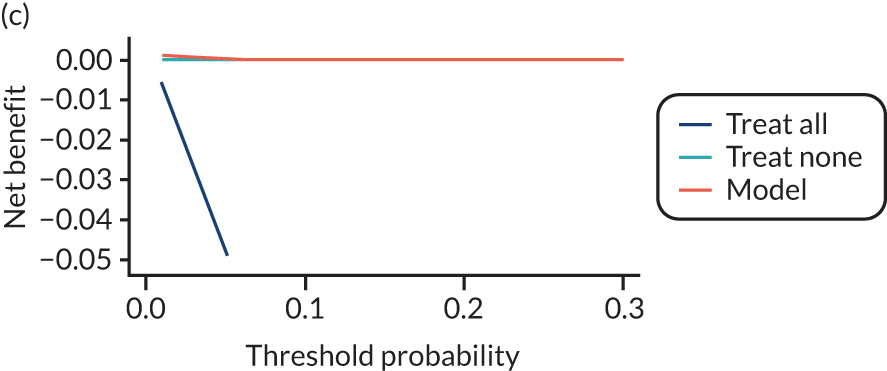
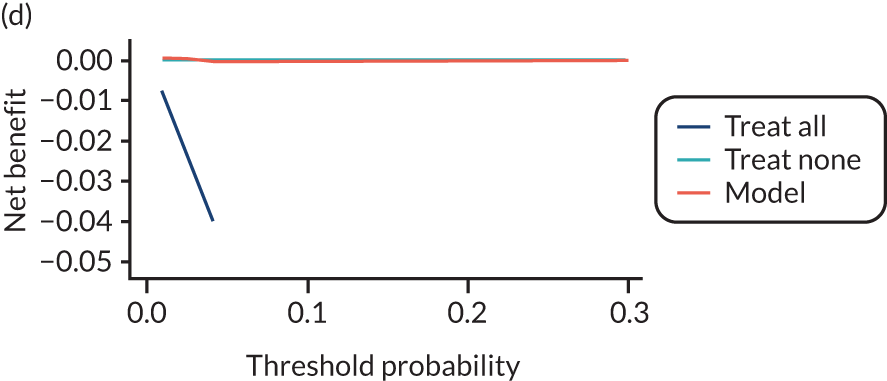
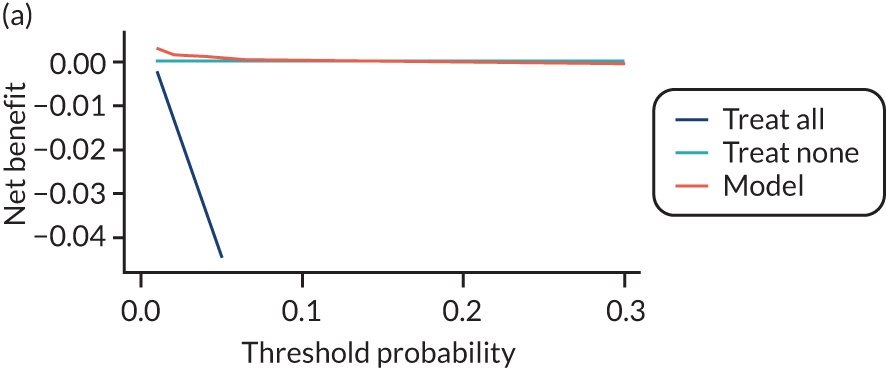
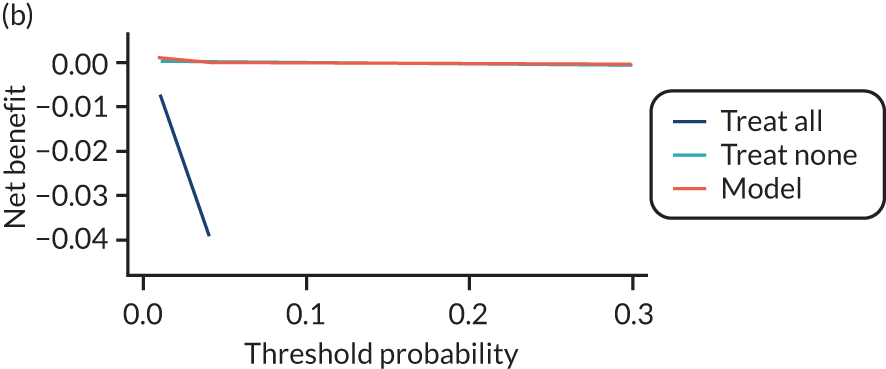
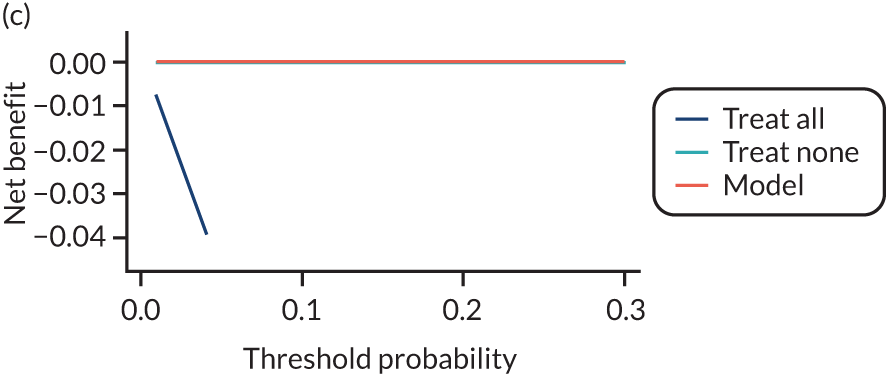
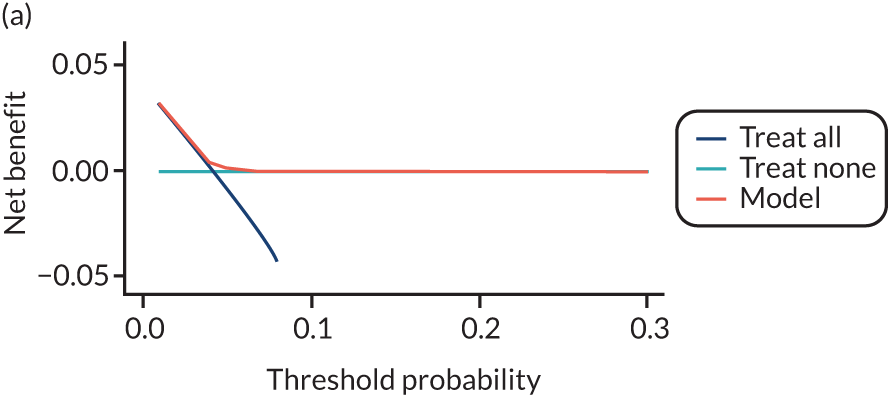
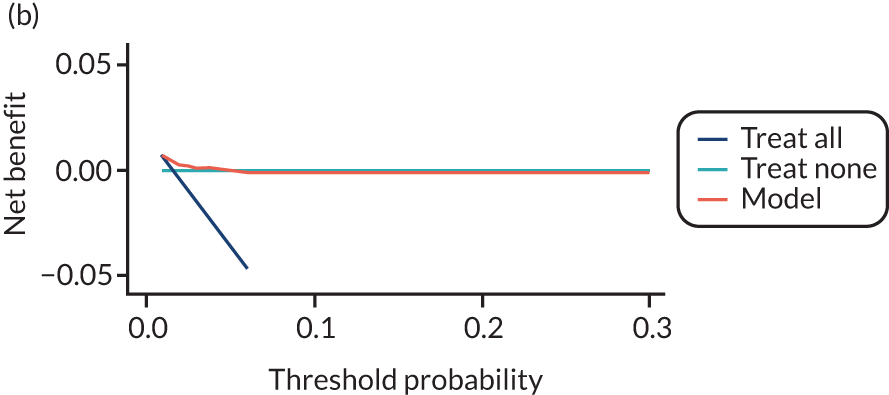
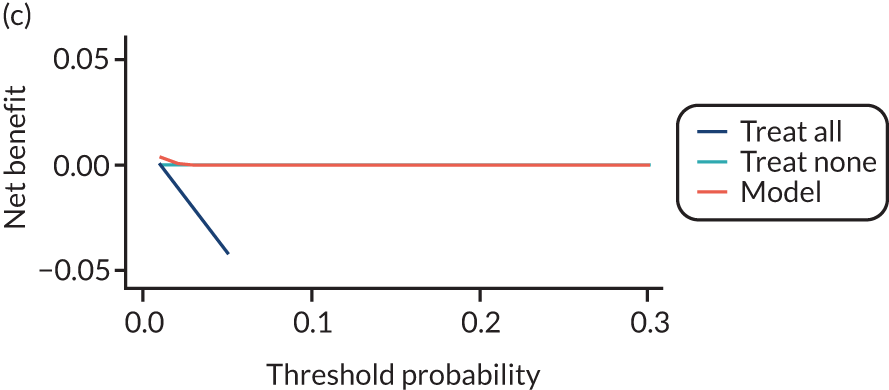
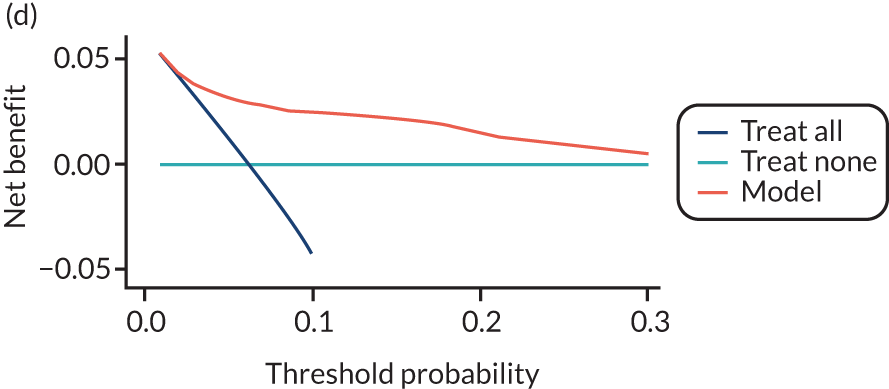
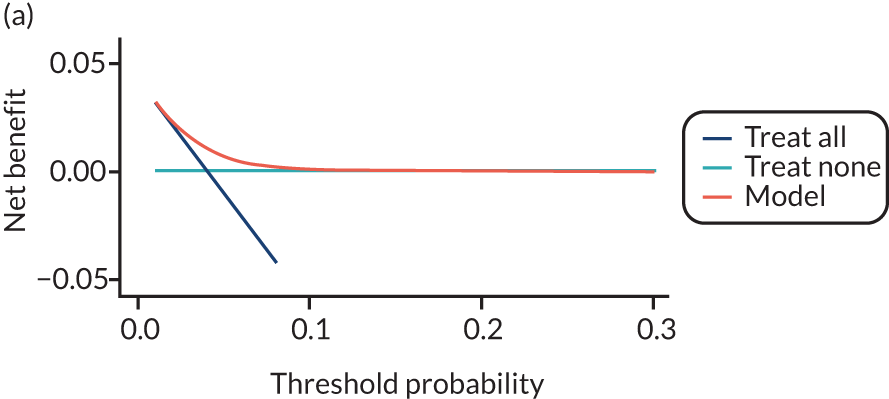
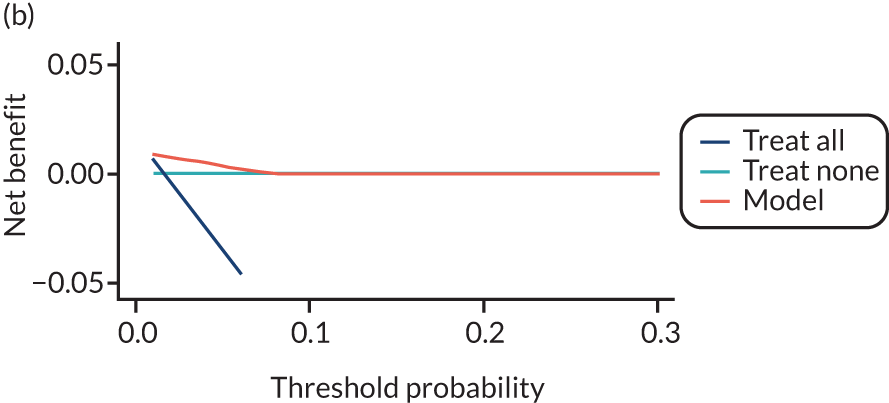
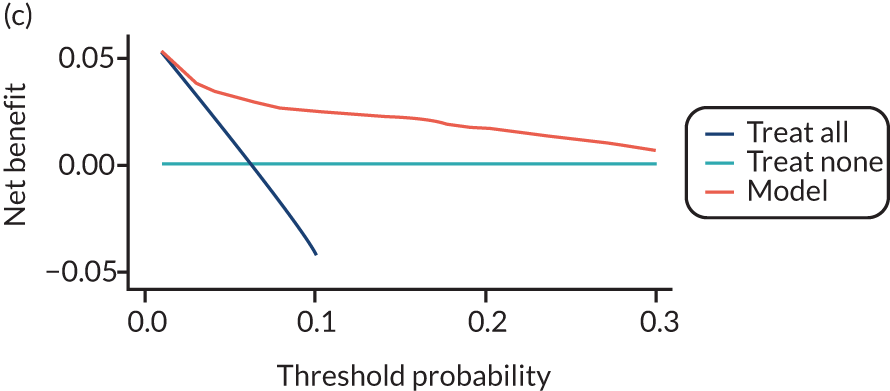
For each pre-eclampsia outcome (early, late or any onset), we compared prediction models using decision curve analysis in the data sets used most frequently in the external validation of the prediction models, enabling within-data set comparison of the models. 89,90 Decision curves show the net benefit (i.e. the benefit versus the harm) over a range of threshold probabilities (i.e. for treating women with a predicted risk above the threshold value) and can be compared with treat-all and treat-none strategies. For a probability threshold (pt), the net benefit is calculated as:
where ‘true positives’ and ‘false positives’ represent the numbers of individuals with a predicted probability ≥ pt who have and do not have the outcome of interest, respectively, and N is the total sample size. 89,90 Probability thresholds of between 5% and 20% will be clinically meaningful for making decisions about preventative interventions such as aspirin, including commencement of high-dose (150 mg) aspirin in the first trimester. Therefore, the model with the greatest net benefit for a particular threshold is considered to have the greatest clinical value.
Development and validation of pre-eclampsia prediction models
To develop new pre-eclampsia prediction models, non-UK data sets were considered in addition to the UK-only data sets. Prospective cohorts and trials were considered for inclusion in the development set. We excluded case–control studies as they cannot be used to estimate the baseline risk (intercept).
The number and proportion of missing values for each potential predictor and outcome were summarised by data set. Predictors were considered to be systematically missing for a data set if they were not recorded for any individuals or were recorded for very few individuals (< 10%) in that data set. No data sets included all potential predictors of interest; therefore, it was necessary to use a subset of predictors thought to be most predictive and of most interest.
A prioritised list of predictors was drawn up based on consensus among clinical experts in the collaborative group (see Prioritisation of predictors). To select data sets for development of a new prediction model (including maternal characteristics and clinical examination variables), it was necessary to compromise between the number of data sets included and the potential predictors that could be considered for inclusion in the models. The aim was to do this in such a way as to maximise both. We undertook the following process:
-
Ranked the prioritised predictors from the most to least relevant based on the scores from the clinical consensus meeting.
-
Excluded predictors that were rarely recorded across data sets (predictors recorded in data sets that total ≤ 5% of all events).
-
Summarised the number of data sets, total sample size and number of events included if all remaining predictors were included.
-
If no data sets or very few data sets included all remaining predictors, the lowest-ranking predictor from the set of predictors was dropped.
-
Repeated steps 3 and 4 until a reasonable number of data sets, sample size and number of events were achieved and, ideally, when excluding further predictors would not mean a significant gain in data sets included.
Subsets of the development data identified for models with clinical and maternal characteristics were used to develop models additionally including biochemical markers or ultrasound markers. These models built on the clinical models, and therefore they required the same clinical variables plus biochemical markers or ultrasound markers. When fitting the biochemical marker models, data sets were included only if both PlGF and sFlt-1 were recorded; however, each biochemical marker could be measured at either trimester 1 or trimester 2. These two biochemical markers were most commonly recorded together, so ensuring that both PlGF and sFlt-1 were recorded in the data sets meant that the relationship between biochemical markers (and trimester of measurement) could be better estimated and used in the imputation models, thereby reducing some uncertainty in the imputed values and reducing the risk of convergence issues.
Missing data
Multiple imputation was implemented using multivariable joint modelling to account for missing data and clustering of participants within data sets. This approach was selected (rather than imputation within data sets, as in External validation of existing pre-eclampsia models) to increase the number of data sets used for model development, as many of the potential predictors of interest were systematically missing (i.e. not recorded for anyone) in one or more data sets. The ‘jomo’91 package in R was designed to impute for multilevel (clustered) data and can therefore be used to impute for variables that are systematically missing in some IPPIC data sets, as well as for partially missing variables. 92 This package uses a Bayesian approach (Markov chain Monte Carlo sampling), so it is necessary to allow a burn-in for the chain to converge before sampling an imputed data set. Rather than repeating the whole process (including the burn-in) for each imputation, imputed data sets are sampled from the same chain by specifying the number of iterations to be left between the samples taken. Prior to running the full imputation, a dummy run was performed to check the chains for signs of non-convergence and to determine a suitable burn-in and sampling interval. Based on these checks, a burn-in of 20,000 was used for imputation of the data used to develop clinical characteristics models and was increased to 30,000 for data used to develop models additionally including biochemical markers or ultrasound markers. After the burn-in, imputations were sampled every 1000 iterations, until 25 imputed data sets had been sampled.
The imputation model included all potential predictors (for trimesters 1 and 2) and outcomes (early- and late-onset pre-eclampsia). Data were imputed separately (using different sets of data sets and predictors) for developing models including only maternal and clinical characteristics, and then for models additionally including biochemical markers or ultrasound markers.
For models with maternal and clinical characteristics, data were imputed for SBP and DBP rather than MAP, as MAP simply combines SBP and DBP and, therefore, imputing them separately would provide more flexibility in how they can be modelled subsequently. A preliminary complete-case analysis was performed to look for potential non-linear relationships between potential predictors and outcomes using multivariable fractional polynomial models. This led to BMI being considered on the original scale, as well as non-linearly using the natural logarithm transformation [ln(BMI)] and BMI–2. Data sets were imputed assuming each of these functions for BMI to enable non-linearity to be considered during model development. Biochemical markers and ultrasound markers were considered on their original scale and on the log-transformed scale (which was decided a priori), and therefore data sets were imputed separately for the transformed and the original biochemical markers or ultrasound markers.
After imputation, the distributions of values for variables were checked by plotting the mean ± SD for continuous variables against the imputation number (including the original complete data, imputation 0, for reference). For categorical variables, the proportions in each category were compared across imputations and with the original complete data. Methods for imputing for systematically missing predictors are still relatively new and therefore a cautious approach was taken. If the distribution of imputed values for a systematically missing predictor was unusual or extreme, then further examination was done to check the plausibility of the imputation. Alongside this, convergence of the Markov chain Monte Carlo samples was checked. If systematically missing predictor values could not reliably be imputed for a variable in a particular data set (e.g. adequate convergence was not achieved even after a long burn-in), then that data set was excluded from model development when that predictor would be considered (e.g. first-trimester BMI in a first-trimester prediction model).
Model development and validation
Prediction models were developed using random intercept logistic regression with backward elimination for variable selection. The random intercept was used to account for clustering of women within individual data sets. At each stage of the variable selection process, the same model (i.e. including the same predictors) was fitted to all imputations, and pooled Wald tests (using Rubin’s rules) were used for backwards elimination, with a p-value of > 0.157 (proxy for Akaike information criterion) for exclusion. 93,94 Models were developed separately for each pre-eclampsia outcome (any, early and late onset) using predictors recorded at trimester 1 and separately at trimester 2. Furthermore, for each outcome predicted at each trimester, three models were considered: using only clinical characteristics, using clinical characteristics plus biochemical markers, and using clinical characteristics plus ultrasound markers. Therefore, we aimed to develop 18 models in total (one for each combination of the three outcomes, two trimesters and three predictor sets). We were unable to develop models that included clinical characteristics, biochemical and ultrasound markers as no data sets included all relevant predictors of interest.
For each model developed, its predictive performance was assessed in an internal validation using study-specific and overall estimates of discrimination and calibration. After model development, the fitted model (with the average intercept) was applied back into each individual data set to obtain, for each participant, values of the LP and predicted probability of the pre-eclampsia outcome from the developed model. These were then used to calculate the performance statistics described in External validation of existing pre-eclampsia prediction models. For each data set, the ‘pool last’ approach was followed, whereby imputation-specific performance statistics were calculated and then pooled across imputations using Rubin’s rules and using a transformed scale where necessary (such as pooling logit C-statistics). 95 Calibration plots were also produced for data sets that had more than 100 events. The predicted probability of pre-eclampsia for each individual was obtained by pooling the imputation-specific estimates of the model’s LP and then applying the logit transformation. 82
Summarising study-specific performance after model development is recommended by Royston et al. 96 and Debray et al. ,47 and gives an indication of how the model will perform with new data from populations represented by the included studies. For each model developed, the data set-specific performance statistics were summarised across the data sets using a random-effects meta-analysis, in the same way as described in External validation of existing pre-eclampsia prediction models.
The performance statistics of models developed using different functional forms of BMI were compared (in terms of overall predictive performance and homogeneity of performance across data sets) after repeating the model development process in the imputed data sets for each functional form. The model that provided the best overall predictive performance across the different statistics was selected, thereby also selecting the functional form for BMI (if it remained in the model). The same strategy of comparing performance statistics was used for models with biochemical markers and ultrasound markers, to determine whether they should be modelled on their original scale or using a natural logarithm transformation.
For each developed model, to correct for optimism during model development (also known as overfitting), the predictor effects (beta estimates) were shrunk by multiplying each beta estimate by a global shrinkage factor. 97–99 The shrinkage factor was taken to be the summary calibration slope from the internal validation process (i.e. the pooled calibration slope from the meta-analysis of data set-specific calibration slope estimates). Bootstrapping was not practical computationally given the need to incorporate both non-linear trend examinations, backwards selection, and multiple imputation (including for systematically missing predictors). Following application of shrinkage, the model’s intercept was re-estimated to ensure that predictions were correct on average. This then provided the final model equation.
For each of the final models, decision curves were produced within each data set included in model development and validation. This shows the net benefit across different probability thresholds and compares the use of the model with treat-all and treat-none strategies.
Summarising the prognostic effect of individual predictors of pre-eclampsia
For each outcome (early-onset pre-eclampsia, late-onset pre-eclampsia and any-onset pre-eclampsia) and each candidate predictor (clinical, biochemical, and ultrasound marker) prioritised in Prioritisation of predictors, we separately performed an unadjusted two-stage IPD meta-analysis of the prognostic effect to obtain a summary estimate, 95% CI and 95% prediction interval for that predictor. The 95% prediction interval presents the heterogeneity on the same scale as the original outcome and estimates where the true effects are to be expected for similar exchangeable studies. 100 We used the two-stage approach because of the large numbers of studies.
The two-step approach first involves fitting a logistic regression model for each study to obtain the odds ratio (OR) for the prognostic effect, and then pooling the log ORs using a conventional random-effects meta-analysis. The random-effects model allows for heterogeneity between studies, and was estimated using restricted maximum likelihood. The 95% CI of the pooled effect was derived using the Hartung–Knapp approach. 101,102 Heterogeneity was summarised using the I2-statistic (which provides the proportion of total variability due to between-study heterogeneity) and 95% prediction intervals. 100 The trend across multiple categories and continuous variables was considered linear.
A pragmatic decision was made to perform all analyses on complete cases of singleton pregnancies on the IPPIC international data set only, that is, no statistical imputation method was carried out for missing outcome or predictor data. This was because of the length of time it would take to impute and perform pre- and post-imputation checks for the 78 data sets of the IPPIC international IPD, with different combinations of predictors in each data set, with some data sets as large as 600,000 pregnancies. The clustering of participants within data sets was accounted for by analysing each data set separately in the first stage. Clustering of pregnancies by women was not accounted for because of the small number of clusters of women who had been pregnant multiple times. Models were univariable and thus predictors were not adjusted for. Adjustment would have reduced statistical power owing to missing observations and would have distorted the combining of associations in the IPD second stage due to different availability of adjustable variables by data set. The analysis excluded all women with multifetal pregnancies (e.g. twins/triplets); however, we also explored the relationships between multiple birth as a predictor and all pre-eclampsia outcomes.
Chapter 4 Characteristics and quality of data sets included in the individual participant data meta-analysis
Study identification and individual participant data acquisition
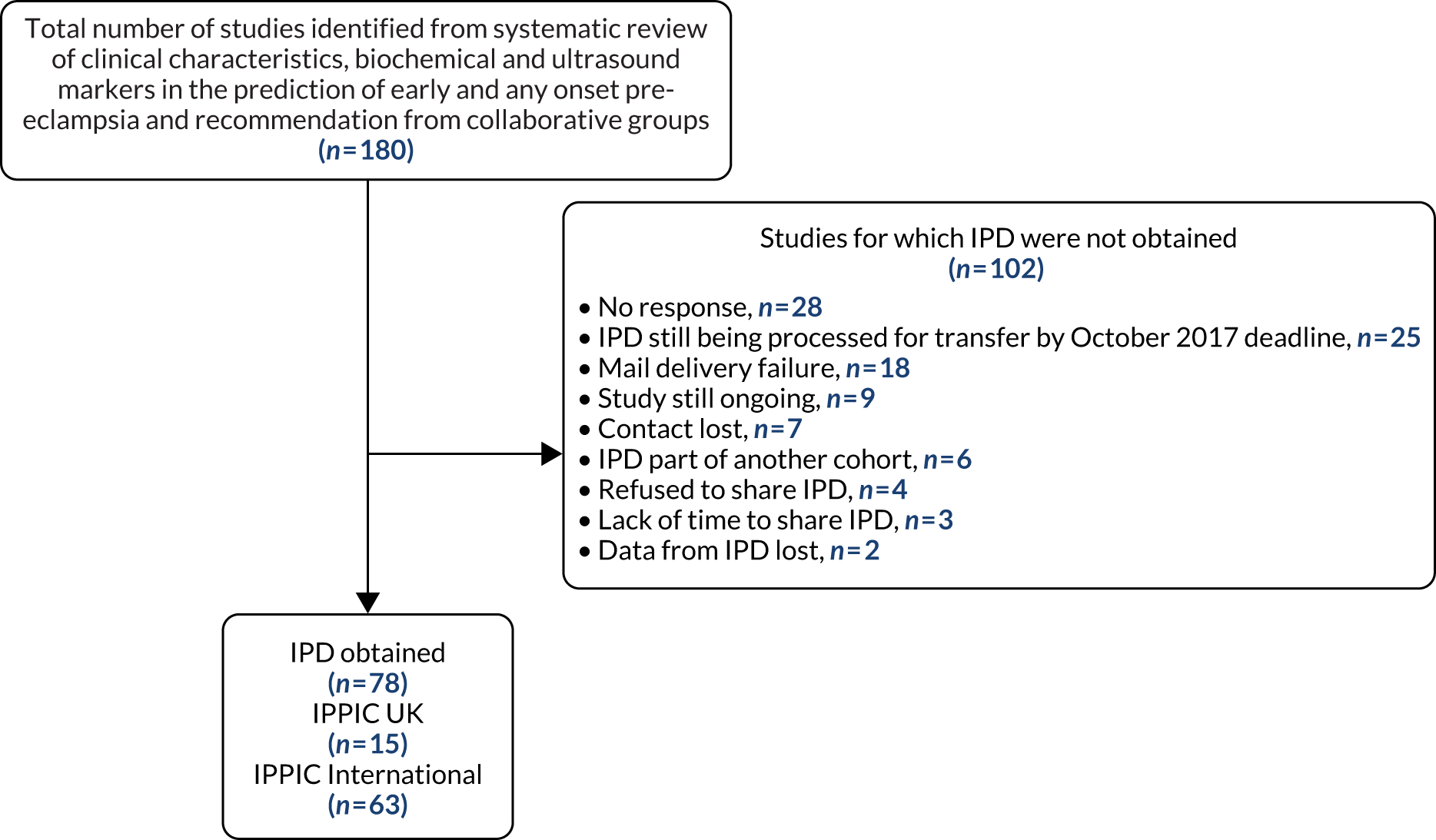
One hundred and twenty-five researchers from 73 teams in 25 countries had joined the IPPIC network by October 2017 and provided access to pseudonymised individual data for 3,674,684 pregnancies. 42,103–178 The most common reason for not obtaining the IPD was not receiving a response from the author to the request to share data (28/180) (Figure 2).
FIGURE 2.
Flow diagram of studies included in the IPD meta-analysis, showing reasons why IPD were not obtained.
Our search up to September 2014 of reviews that evaluated the performance of single or combined tests for predicting pre-eclampsia identified 73 citations. After evaluation of the abstracts, we included 62 published reviews evaluating one or more tests for predicting pre-eclampsia (Table 2). Clinical characteristics were studied in 32.3% (20/62) of published reviews, biochemical markers were studied in 59.7% (37/62) and ultrasound markers were studied in 8.1% (5/62).
| Systematic review (first author and year) | Number of primary studies | Number of women | Risk factors evaluated | Outcome reported |
|---|---|---|---|---|
| Maternal clinical characteristics | ||||
| Cnossen 2007179 | 36 | 1,699,073 | BMI | Any-onset pre-eclampsia |
| O’Brien 2003180 | 13 | 1,400,000 | Any-onset pre-eclampsia | |
| Wang 2013181 | 29 | 1,980,761 | Any-onset pre-eclampsia | |
| Duckitt 2005182 | 2 | 64,789 | Multiple clinical features | Any-onset pre-eclampsia |
| Alpoim 2013183 | 2 | 1875 | ABO blood group status | Early-onset pre-eclampsia |
| England 2007184 | 48 | N/A | Smoking | Early-onset pre-eclampsia |
| Rebelo 2013185 | 23 | 4265 | CRP, BMI | Any-onset pre-eclampsia |
| Duckitt 2005182 | 2 | 37,988 | Parity | Any-onset pre-eclampsia |
| Luo 2007186 | 26 | N/A | Any-onset pre-eclampsia | |
| Duckitt 2005182 | 2 | 65,314 | Age | Any-onset pre-eclampsia |
| Duckitt 2005182 | 2 | 907 | Blood pressure | Any-onset pre-eclampsia |
| Cnossen 2008187 | 34 | 60,599 | Any-onset pre-eclampsia | |
| Sgolastra 2013188 | 15 | 5023 | Periodontal disease | Any-onset pre-eclampsia |
| Kunnen 2010189 | 15 | N/A | Early-onset pre-eclampsia | |
| Morris 2012190 | 20 | 2978 | Proteinuria | Any-onset pre-eclampsia |
| Sanchez-Ramos 2013191 | 24 | 3186 | Early-onset pre-eclampsia | |
| Wolf 2014192 | 11 | 5411 | Leisure-time physical activity | Any-onset pre-eclampsia |
| Palmer 2013193 | 11 | N/A | Occupational exposures | Any-onset pre-eclampsia |
| Bonzini 2007194 | 9 | N/A | Any-onset pre-eclampsia | |
| Cnossen 2006195 | 5 | 572 | Uric acid | Any-onset pre-eclampsia |
| Uterine artery Doppler ultrasound | ||||
| Velauthar 201439 | 18 | 55,974 | First-trimester Doppler | Early-onset pre-eclampsia |
| Chien 2000196 | 27 | 12,994 | Any-trimester Doppler | Any-onset pre-eclampsia |
| Cnossen 200863 | 74 | 79,547 | Any-onset pre-eclampsia | |
| Kleinrouweler 2013197 | 8 | 6708 | Second-trimester Doppler | Early-onset pre-eclampsia |
| Pedrosa 2011198 | N/A | N/A | Doppler combined with other markers | Early-onset pre-eclampsia |
| Biochemical markers | ||||
| Kosmas 2003199 | 19 | 5145 | Factor V Leiden | Any-onset pre-eclampsia |
| Dudding 2008200 | 6 | 6755 | Any-onset pre-eclampsia | |
| Rodger 2010201 | 10 | 21,833 | Any-onset pre-eclampsia | |
| Xia 2012202 | 36 | 9203 | MTHFR gene C677T polymorphism | Any-onset pre-eclampsia |
| Kosmas 2004203 | 23 | 6213 | Any-onset pre-eclampsia | |
| Zusterzeel 2000204 | 4 | 579 | Any-onset pre-eclampsia | |
| Li 2014205 | 49 | 18,009 | Any-onset pre-eclampsia | |
| Wang 2013206 | 51 | 17,749 | Any-onset pre-eclampsia | |
| Widmer 2007207 | 10 | 1173 | sFlt-1 | Early-onset pre-eclampsia |
| Jacobs 2011208 | 11 | N/A | Early-onset pre-eclampsia | |
| Kleinrouweler 201265 | 19 | 6708 | Early-onset pre-eclampsia | |
| Widmer 2007207 | 14 | 2045 | PIGF | Early-onset pre-eclampsia |
| Kleinrouweler 201265 | 27 | N/A | Any-onset pre-eclampsia | |
| Huppertz 2013209 | 19 | 16,153 | PP13 | Early-onset pre-eclampsia |
| Schneuer 2012210 | 7 | 2989 | Early-onset pre-eclampsia | |
| Lau 2013211 | 41 | 1940 | TNF-alpha, IL-6 and IL-10 | Any-onset pre-eclampsia |
| Tabesh 2013212 | 8 | 2485 | Serum vitamin D | Any-onset pre-eclampsia |
| Morgan 2013213 | 12 | 5003 | PAI-1 promoter polymorphism | Any-onset pre-eclampsia |
| Dai 2013214 | 29 | 3228 | eNOS polymorphisms | Any-onset pre-eclampsia |
| Chen 2012215 | 18 | N/A | Any-onset pre-eclampsia | |
| Qi 2013216 | 33 | 10,671 | Any-onset pre-eclampsia | |
| Zhao 2013217 | 11 | 3088 | PAI-1 promoter polymorphism | Any-onset pre-eclampsia |
| Zhao 2012218 | 8 | 1995 | AGTR1 +1166A>C polymorphism | Any-onset pre-eclampsia |
| Zhong 2012219 | 11 | 1749 | ACE I/D polymorphism | Any-onset pre-eclampsia |
| Chen 2012215 | 30 | 8340 | Any-onset pre-eclampsia | |
| Ni 2012220 | 22 | 7534 | AGT M235T polymorphism | Any-onset pre-eclampsia |
| Kleinrouweler 201265 | 3 | N/A | VEGF | Any-onset pre-eclampsia |
| Hui 2012221 | 37 | 115,290 | Wide range of serum markers | Any-onset pre-eclampsia |
| Giguere 2011222 | 37 | N/A | 71 different markers | Early-onset pre-eclampsia |
| Abou-Nassar 2011223 | 28 | 5991 | Antiphospholipid antibodies | Any-onset pre-eclampsia |
| do Prado 2010224 | 12 | 7950 | Any-onset pre-eclampsia | |
| Gupta 2009225 | 17 | 745 | Lipid peroxidation | Any-onset pre-eclampsia |
| Bombell 2008226 | 16 | 2374 | TNF (–308A) polymorphism | Any-onset pre-eclampsia |
| Zafarmand 2008227 | 17 | 5275 | Angiotensinogen gene M235T polymorphism | Any-onset pre-eclampsia |
| Morris 200868 | 44 | 169,637 | Inhibin A, AFP and three others | Any-onset pre-eclampsia |
| Wiwanitkit 2006228 | 6 | 1690 | PAI-1 | Any-onset pre-eclampsia |
| Leeflang 200766 | 5 | 573 | FFN | Any-onset pre-eclampsia |
Characteristics of data sets in the IPPIC data repository
Seventy-eight data sets contributed data to the IPPIC data repository. 42,103–178 More than half of the data sets received (58%, 45/78) were prospective cohort studies; 15% (12/78) were randomised controlled trials and 17% (13/78) were large prospective registry data sets or birth cohorts. One data set was IPD made up of 31 RCTs. Most of the data sets were from participants in Europe (60%, 47/78), 18% (14/78) were from North America, 6% (5/78) were from South America, 5% (4/78) were from Asia and Australia, and one (1%) was from Africa. Three of the data sets provided included participants from multiple countries, such as Argentina, Colombia, Kenya, India, Peru, Thailand and New Zealand. Ninety-seven per cent (3,570,993) of the 3,674,684 pregnancies in the IPPIC repository were singleton pregnancies. Individual data set size ranged from 42 to 1,663,167 pregnancies, and the total number of reported pre-eclampsia outcomes in each data set ranged from 0 to 4252 for early-onset pre-eclampsia, from 0 to 38,305 for late-onset pre-eclampsia and from 3 to 42,608 for any-onset pre-eclampsia (see Appendix 4). About one-third of the data sets received were on women with high-risk pregnancies only (29%, 23/78), 14% (11/78) of the data sets were on women with low-risk pregnancies and more than half (55%, 43/78) of the data sets included women with pregnancies of any risk. Detailed study characteristics of all IPPIC data sets are provided in Appendix 5 and a summary of the missing data for prioritised predictors and each pre-eclampsia outcome is provided in Appendix 6.
Prioritisation of predictors of pre-eclampsia
In April 2017, the online survey was designed and run using smartsurvey.co.uk (see Appendix 7). Ninety-eight members of the IPPIC collaborative network who had agreed to share data by this date were sent an e-mail introducing the survey and explaining the participation requirements and survey objectives. Collaborators had 7 days within which to complete the online survey.
Fifty-four candidate predictor variables were identified (37 clinical characteristics, nine biochemical markers and eight ultrasound markers) and ranked by 33 (34%) IPPIC collaborators. Seventy per cent (23/33) of responders were from Europe, 12% were from both the American (4/33) and Asian (4/33) continents, and 6% (2/33) were from Africa. A consensus group made up of five clinical academics reviewed 13 candidate predictor variables ranked by the online survey participants as being ‘moderately important’. This included eight clinical characteristic variables, two biochemical markers and three ultrasound markers. Two each of the clinical characteristic variables and ultrasound markers reviewed by the consensus group (mode of conception, substance misuse in current pregnancy, umbilical artery pulsatility index and estimated fetal weight centile) were included following assessment by the group.
Overall, fewer than half (48%, 26/54) of all assessed predictors were ranked as being important, with 54% (20/37) of clinical characteristics, 33% (3/9) of biochemical markers and 38% (3/8) of ultrasound markers being prioritised as important in predicting pre-eclampsia (Table 3).
| Important | Unimportant |
|---|---|
| Clinical characteristics | |
| Previous any pre-eclampsia | Previous miscarriage |
| Chronic or pre-existing hypertension | History of early pregnancy bleeding in current pregnancy |
| SBP | Alcohol use |
| BMI | Diet in pregnancy |
| DBP | Physical activity |
| Parity | Interval between pregnancies |
| History of renal disease | Family history of cardiovascular disease |
| Multiple pregnancy | History of gestational diabetes |
| History of pre-existing diabetes | Gestational diabetes in current pregnancy |
| Age | Socioeconomic status |
| Previous autoimmune disease | Previous stillbirth |
| Family history of pre-eclampsia in first degree relative | Previous preterm delivery |
| MAP | Previous heritable thrombophilia |
| PCR | New partner |
| Urine dipstick | Height |
| Previous SGA | Weight |
| Smoking | 24-hour protein |
| Mode of conception | |
| Substance misuse in current pregnancy | |
| Ethnicity | |
| Ultrasound markers | |
| Uterine artery pulsatility index | CRL |
| Umbilical artery pulsatility index | Umbilical artery resistance index |
| Estimated fetal weight centile | Abdominal circumference |
| Notching on ultrasound scan | |
| Uterine artery resistance index | |
| Biochemical markers | |
| PlGF | CRP |
| sFlt-1 | Hypertriglyceridaemia |
| PAPP-A | Human chorionic gonadotropin |
| AFP | |
| PAI-1 polymorphism | |
| sEng | |
Quality of the IPPIC data sets
Risk-of-bias assessment using the PROBAST resulted in 77% (60/78) of the included IPD data sets being classified as having an overall low risk of bias, while 22% (17/78) were classified as having an unclear risk of bias. Only one data set (1%, 1/78) received an overall high risk of bias assessment. All of the included data sets had a low risk of bias in the domain of participant selection. For the domain of predictors, 94% (73/78) had a low risk of bias, while 1% (1/78) had a high and 5% (4/78) an unclear risk of bias assessment. The risk of bias in the outcome domain was unclear for 22% (17/78) of the included data sets and low in the rest (78%, 61/78). Detailed assessment of the risk of bias for the IPPIC data sets is presented in Appendix 8.
Characteristics of identified prediction models
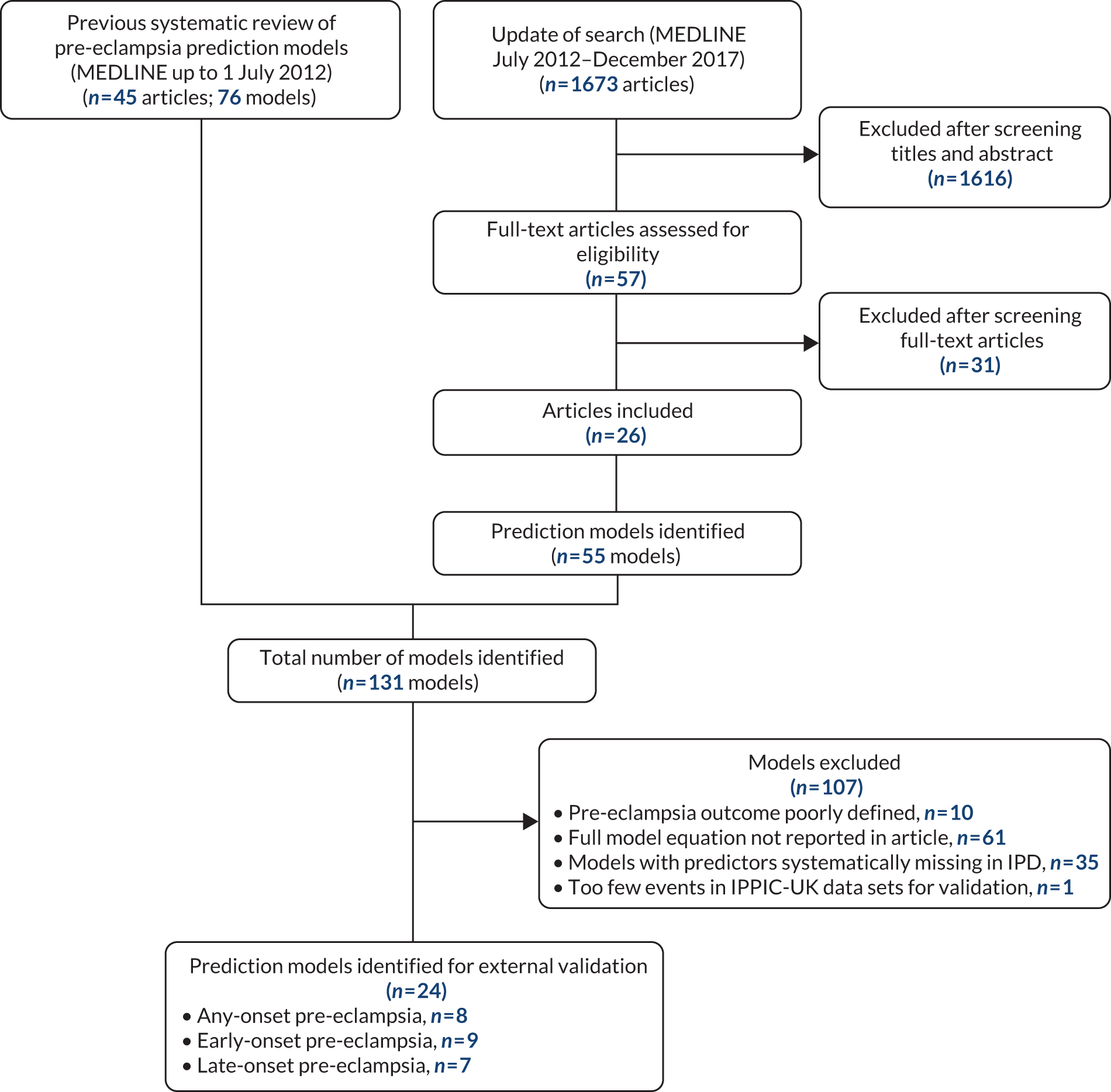
From our updated literature search (up to December 2017), we identified 131 models developed to predict pre-eclampsia. About half of these (53%, 70/131) reported the model equation in the publication, and only one-fifth of all models (18%, 24/131) from 12 publications met the inclusion criteria for the external validation of their predictive performance in the IPPIC-UK data sets. 115,128,147,229–237 The primary reasons for not including a model for external validation were the full prediction formula not being reported in the publication (47%, 61/131) and the absence of the predictor information in the IPPIC-UK data sets (27%, 35/131). Other reasons for not validating the models include pre-eclampsia being poorly defined in the study (8%, 10/131) and not enough events in the IPPIC-UK data set to validate the model (1%, 1/131). Figure 3 is the flow chart of prediction model selection for external validation, and Appendix 9 shows published pre-eclampsia prediction models reporting a model equation.
FIGURE 3.
Flow chart of pre-eclampsia prediction model selection for external validation in IPPIC-UK data set using IPD meta-analysis. Reproduced from Snell et al. 52 Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
Chapter 5 External validation of existing pre-eclampsia prediction models
Characteristics of included prediction models
We identified 24 prediction models that could be externally validated in the IPPIC-UK cohorts. Eight models predicted any-onset pre-eclampsia, nine models predicted early-onset pre-eclampsia and seven models predicted late-onset pre-eclampsia. About half of these models (13/24, 54%) were developed in unselected singleton pregnancies at any risk of pre-eclampsia. Two-thirds of the models included only clinical characteristics as predictors (15/24, 63%), one-fifth (5/24, 21%) included clinical characteristics and biochemical markers, and one-sixth (4/24, 17%) included clinical characteristics and ultrasound markers. It was not possible to validate any of the models that included all three predictor categories (clinical characteristics, biochemical markers and ultrasound markers).
The majority of models (22/24, 92%) were developed using binary logistic regression. The two models by Wright et al. 236 modelled the outcome of ‘gestational age at delivery with pre-eclampsia’ using competing risks models. The models by Wright et al. 236 used pre-eclampsia before 34 weeks to define early-onset pre-eclampsia. Over 80% of the models that we validated involved first-trimester predictors (88%, 21/24); only three included second-trimester predictors. Details of the validated models are given in Table 4.
| Model number | Authors, year | Predictor category | Prediction model equation for LPa |
|---|---|---|---|
| First-trimester any-onset pre-eclampsia models | |||
| 1 | Plasencia et al., 2007231 | Clinical characteristics | LP = –6.253 + 1.432 (if African Caribbean ethnicity) + 1.465 (if mixed ethnicity) + 0.084 (BMI) + 0.81 (if patient’s mother had PE) – 1.539 (if parous without previous PE) + 1.049 (if parous with previous PE) |
| 2 | Poon et al., 2008233 | Clinical characteristics | LP = –6.311 + 1.299 (if African Caribbean ethnicity) + 0.092 (BMI) + 0.855 (if woman’s mother had PE) – 1.481 (if parous without previous PE) + 0.933 (if parous with previous PE) |
| 3 | Wright et al., 2015236 | Clinical characteristics | Mean gestational age at delivery with PE = 54.3637 – 0.0206886 (age, years – 35, if age ≥ 35) + 0.11711 (height, cm – 164) – 2.6786 (if African Caribbean ethnicity) – 1.129 (if South Asian ethnicity) – 7.2897 (if chronic hypertension) – 3.0519 (if systemic lupus erythematosus or antiphospholipid syndrome) – 1.6327 (if conception by in vitro fertilisation) – 8.1667 (if parous with previous PE) + 0.0271988 (if parous with previous PE, previous gestation in weeks – 24)2 – 4.335 (if parous with no previous PE) – 4.15137651 (if parous with no previous PE, interval between pregnancies in years)–1 + 9.21473572 (if parous with no previous PE, interval between pregnancies in years)–0.5 – 0.0694096 (if no chronic hypertension, weight in kg – 69) – 1.7154 (if no chronic hypertension and family history of PE) – 3.3899 (if no chronic hypertension and diabetes mellitus type 1 or 2) |
| 4 | Baschat et al., 2014115 | Clinical characteristics and biochemical markers | LP = –8.72 + 0.157 (if nulliparous) + 0.341 (if history of hypertension) + 0.635 (if prior PE) + 0.064 (MAP) – 0.186 (PAPP-A, Ln MoM) |
| 5 | Goetzinger et al., 2010128 | Clinical characteristics and biochemical markers | LP = –3.25 + 0.51 (if PAPP-A < 10th percentile) + 0.93 (if BMI > 25) + 0.94 (if chronic hypertension) + 0.97 (if diabetes) + 0.61 (if African American ethnicity) |
| 6 | Odibo et al., 2011147 | Clinical characteristics and biochemical markers | LP = –3.389 – 0.716 (PAPP-A, MoM) + 0.05 (BMI) + 0.319 (if black ethnicity) + 1.57 (if history of chronic hypertension) |
| 7 | Odibo et al., 2011147 | Clinical characteristics and ultrasound markers | LP = –3.895 – 0.593 (mean uterine artery PI) + 0.944 (if pre-gestational diabetes) + 0.059 (BMI) + 1.532 (if history of chronic hypertension) |
| First-trimester early-onset pre-eclampsia models | |||
| 8 | Baschat et al., 2014115 | Clinical characteristics | LP = –5.803 + 0.302 (if diabetes) + 0.767 (if hypertension) + 0.00948 (MAP) |
| 9 | Crovetto et al., 2015229 | Clinical characteristics | LP = –5.177 + 2.383 (if black ethnicity) – 1.105 (if nulliparous) + 3.543 (if parous with previous PE) + 2.229 (if chronic hypertension) + 2.201 (if renal disease) |
| 10 | Kuc et al., 2013230 | Clinical characteristics | LP = –6.790 – 0.119 (maternal height, cm) + 4.8565 (maternal weight, Ln kg) + 1.845 (if nulliparous) + 0.086 (maternal age, years) + 1.353 (if smoker) |
| 11 | Plasencia et al., 2007231 | Clinical characteristics | LP = –6.431 + 1.680 (if African Caribbean ethnicity) + 1.889 (if mixed ethnicity) + 2.822 (if parous with previous PE) |
| 12 | Poon et al., 2010232 | Clinical characteristics | LP = –5.674 + 1.267 (if black ethnicity) + 2.193 (if history of chronic hypertension) – 1.184 (if parous without previous PE) + 1.362 (if parous with previous PE) + 1.537 (if conceived with ovulation induction) |
| 13 | Scazzocchio et al., 2013235 | Clinical characteristics | LP = –7.703 + 0.086 (BMI) + 1.708 (if chronic hypertension) + 4.033 (if renal disease) + 1.931 (if parous with previous PE) + 0.005 (if parous with no previous PE) |
| 14 | Wright et al., 2015236 | Clinical characteristics | Same as model 3 |
| 15 | Poon et al., 2009234 | Clinical characteristics and biochemical markers | LP = –6.413 – 3.612 (PAPP-A, Ln MoM) + 1.803 (if history of chronic hypertension) + 1.564 (if black ethnicity) – 1.005 (if parous without previous PE) + 1.491 (if parous with previous PE) |
| First-trimester late-onset pre-eclampsia models | |||
| 16 | Crovetto et al., 2015229 | Clinical characteristics | LP = –5.873 – 0.462 (if white ethnicity) + 0.109 (BMI) – 0.825 (if nulliparous) + 2.726 (if parous with previous PE) + 1.956 (if chronic hypertension) – 0.575 (if smoker) |
| 17 | Kuc et al., 2013230 | Clinical characteristics | LP = –14.374 + 2.300 (maternal weight, Ln kg) + 1.303 (if nulliparous) + 0.068 (maternal age, years) |
| 18 | Plasencia et al., 2007231 | Clinical characteristics | LP = –6.585 + 1.368 (if African Caribbean ethnicity) + 1.311 (if mixed ethnicity) + 0.091 (BMI) + 0.960 (if woman’s mother had PE) – 1.663 (if parous without previous PE) |
| 19 | Poon et al., 2010232 | Clinical characteristics | LP = –7.860 + 0.034 (maternal age, years) + 0.096 (BMI) + 1.089 (if black ethnicity) + 0.980 (if Indian or Pakistani ethnicity) + 1.196 (if mixed ethnicity) + 1.070 (if woman’s mother had PE) – 1.413 (if parous without previous PE) + 0.780 (if parous with previous PE) |
| 20 | Scazzocchio et al., 2013235 | Clinical characteristics | LP = 6.135 + 2.124 (if previous PE) + 1.571 (if chronic hypertension) + 0.958 (if diabetes) + 1.416 (if thrombophilic condition) – 0.487 (if multiparous) + 0.093 (BMI) |
| 21 | Poon et al., 2009234 | Clinical characteristics and biochemical markers | LP = –6.652 – 0.884 (PAPP-A, Ln MoM) + 1.127 (if family history of PE) + 1.222 (if black ethnicity) + 0.936 (if Indian or Pakistani ethnicity) + 1.335 (if mixed ethnicity) + 0.084 (BMI) – 1.255 (if parous without previous PE) + 0.818 (if parous with previous PE) |
| Second-trimester any-onset pre-eclampsia models | |||
| 22 | Yu et al., 2005238 | Clinical characteristics and ultrasound markers | LP = 1.8552 + 5.9228 (mean uterine artery PI)–2 – 14.4474 (mean uterine artery PI)–1 = – 0.5478 (if smoker) + 0.6719 (bilateral notch) + 0.0372 (age) + 0.4949 (if black ethnicity) + 1.5033 (if history of PE) – 1.2217 (if previous term live birth) + 0.0367 (T2 BMI) |
| Second-trimester early-onset pre-eclampsia models | |||
| 23 | Yu et al., 2005238 | Clinical characteristics and ultrasound markers | LP = –9.81223 + 2.10910 (mean uterine artery PI)3 – 1.79921 (mean uterine artery PI)3 + 1.059463 (if bilateral notch) |
| Second-trimester late-onset pre-eclampsia models | |||
| 24 | Yu et al., 2005238 | Clinical characteristics and ultrasound markers | LP = 0.7901 + 5.1473 (mean uterine artery PI)–2 – 12.5152 (mean uterine artery PI)–1 – 0.5575 (if smoker) + 0.5333 (if bilateral notch) + 0.0328 (age) + 0.4958 (if black ethnicity) + 1.5109 (if history of PE) + 1.1556 (if previous term live birth) + 0.0378 (BMI) |
Characteristics of the IPPIC-UK validation cohorts
Of the 78 data sets in IPPIC repository, 15 (19%, 15/78) were UK data sets. About three-quarters (73%, 11/15) of IPPIC-UK cohorts contained relevant data needed for external validation. 42,103,108,114,121,122,137,149,150,161,163 Four data sets106,112,137,177 did not include the predictor needed to validate the published prediction models.
Four of the included IPPIC-UK studies were prospective cohorts,42,108,161 three were prospective registry data sets103,114,163 and four were cohorts (control arm) from randomised trials. 121,122,149,150 All studies reported on early-, late- and any-onset pre-eclampsia. About half of the included IPPIC-UK cohorts consisted of unselected pregnant women (6/11); four included high-risk women such as those with abnormal uterine artery Doppler measurement,121,137 history of pre-eclampsia or underlying medical conditions150 and maternal obesity;122,149 one42 included only low-risk nulliparous women, and one161 included all nulliparous singleton pregnancies.
All women in the Screening for Pregnancy Endpoints (SCOPE)42 and Pregnancy Outcome Prediction (POP)161 studies were nulliparous. The percentage of nulliparous women in the other data sets ranged from 43% to 65%. Among the nine data sets with multiparous women, five recorded previous pre-eclampsia with percentages ranging from 0%122 to 27%. 150 The mean age was similar across studies. The median BMI was higher in EMPOWaR,122 Poston et al. 2006150 and Poston et al. 2015149 than in other studies. Most data sets that recorded ethnicity predominantly consisted of white women, apart from Allen et al. 108 and Velauthar et al. 39 (47% and 50% Asian women, respectively). All data sets were considered to be at low risk of bias for quality of participant selection (11/11, 100%); 91% (10/11) were considered to be at low risk and 9% (1/11) were considered to be at unclear risk of bias for predictor reporting; and 45% (5/11) were considered to be at low risk and 55% (6/11) were considered to be at unclear risk of bias for outcome reporting (see Appendix 8).
Detailed study characteristics (see Appendix 5) and summary statistics for the data sets used for external validation, along with a summary of the missing data for each predictor and outcome, are provided in Appendices 10 and 11, respectively.
External validation and meta-analysis of predictive performance
We were able to externally validate each of the 24 published models in at least one and in up to eight data sets. Initially, we could use only one data set of nulliparous women (SCOPE UK42) to validate models 3 (Wright et al. 236) and 14 (Wright et al. 236) owing to a lack of information on ‘interval between pregnancies’ in data sets. However, to increase the sample size available for validation (and the number of events), we included nulliparous subgroups in other data sets for validating these models if all other predictors were recorded. Models 3 (Wright et al. 236) and 14 (Wright et al. 236) were ‘competing risk’ models and did not provide LP values. Therefore, where necessary (e.g. for the assessment of calibration), we used logit probabilities for validation and for comparison with the other models. We were able to validate only models (22–24)238 with second-trimester predictors (Yu et al. 238 for all three models) in the POP study. 161 We did not impute second-trimester predictors in the other data sets as these were not recorded or were missing for a large proportion of individuals.
A summary of the LP and predicted probability distributions for each model and validation data set is given in Appendix 12. Median predicted probabilities were generally low for models, which is expected for a rare outcome. However, the median predicted probability in validation data sets was higher for model 10 for early-onset pre-eclampsia and for model 17 for late-onset pre-eclampsia (Kuc et al. 230 for both). The median predicted probability was 0.034–0.367 across validation data sets for model 10 (Kuc et al. 230) and 0.063–0.310 across validation data sets for model 17 (Kuc et al. 230).
Performance of the models
The summary (average) performance statistics across all validation data sets for each model are given in Table 5 and data set-specific performance statistics in data sets with > 100 pre-eclampsia events are provided for each model in Table 6. The summary performance statistics are also shown graphically for all models in Figure 4. Direct comparison of the prediction models is difficult owing to different data sets (and numbers of individuals and outcomes) contributing to the validation of each prediction model (see Appendix 13).
| Model number | Type of predictor | Authors, year | Number of validation cohorts | Total events | Summary estimate of performance statistic (95% CI); measures of heterogeneity (I2, τ2 where possible to estimate) | ||
|---|---|---|---|---|---|---|---|
| C-statistica | Calibration slope | Calibration-in-the-large | |||||
| Any-onset pre-eclampsia | |||||||
| Trimester 1 models | |||||||
| 1 | Clinical | Plasencia et al., 2007231 | 3 | 102 | 0.686 (0.531 to 0.809); I2 = 1%, τ2 = 0.001 | 0.693 (–0.026 to 1.413); I2 = 45%, τ2 = 0.035 | 0.143 (–1.471 to 1.757); I2 = 91%, τ2 = 0.38 |
| 2 | Poon et al., 2008233 | 3 | 102 | 0.688 (0.533 to 0.810); I2 = 3%, τ2 = 0.002 | 0.715 (–0.032 to 1.462); I2 = 45%, τ2 = 0.037 | 0.002 (–1.653 to 1.657); I2 = 92%, τ2 = 0.402 | |
| 3 | Wright et al., 2015236 | 3 | 76 | 0.624 (0.481 to 0.748); I2 = 0%, τ2 = 0 | 0.642 (–0.182 to 1.467); I2 = 0%, τ2 = 0 | 0.954 (–1.127 to 3.034); I2 = 93%, τ2 = 0.640 | |
| 4 | Clinical and biochemical markers | Baschat et al., 2014115 | 2 | 287 | 0.708 (0.467 to 0.870); I2 = 0%, τ2 = 0 | 1.238 (0.000 to 2.475); I2 = 0%, τ2 = 0 | –0.427 (–14.405 to 13.551); I2 = 98%, τ2 = 2.382 |
| 5 | Goetzinger et al., 2010128 | 3 | 343 | 0.659 (0.303 to 0.896); I2 = 93%, τ2 = 0.315 | 1.124 (–0.595 to 2.843); I2 = 76%, τ2 = 0.356 | –0.965 (–3.041 to 1.111); I2 = 97%, τ2 = 0.667 | |
| 6 | Odibo et al., 2011147 | 3 | 1774 | 0.715 (0.506 to 0.860); I2 = 90%, τ2 = 0.101 | 1.163 (0.243 to 2.083); I2 = 93%, τ2 = 0.104 | –0.786 (–2.615 to 1.044); I2 = 99%, τ2 = 0.511 | |
| 7 | Clinical and ultrasound markers | Odibo et al., 2011147 | 1 | 28 | 0.526 (0.390 to 0.658) | 0.277 (–0.642 to 1.195) | –0.520 (–0.907 to –0.134) |
| Trimester 2 models | |||||||
| 22 | Clinical and ultrasound markers | Yu et al., 2005238 | 1 | 273 | 0.610 (0.574 to 0.645) | 0.075 (0.007 to 0.144) | NE |
| Early-onset pre-eclampsia | |||||||
| Trimester 1 models | |||||||
| 8 | Clinical | Baschat et al., 2014115 | 5 | 204 | 0.675 (0.617 to 0.728); I2 = 0%, τ2 = 0 | 2.041 (0.560 to 3.522); I2 = 69%, τ2 = 0.692 | –0.102 (–1.699 to 1.494); I2 = 97%, τ2 = 1.535 |
| 9 | Crovetto et al., 2015229 | 3b | 21 | 0.575 (0.208 to 0.875); I2 = 69%, τ2 = 0.288 | 0.642 (–4.006 to 5.291); I2 = 81%, τ2 = 0.217 | –0.576 (–4.967 to 3.814); I2 = 95%, τ2 = 2.925 | |
| 10 | Kuc et al., 2013230 | 6 | 1449 | 0.661 (0.613 to 0.706); I2 = 32%, τ2 = 0.011 | 0.423 (0.294 to 0.552); I2 = 33%, τ2 = 0.004 | –4.330 (–5.411 to –3.250); I2 = 99%, τ2 = 0.946 | |
| 11 | Plasencia et al., 2007231 | 4b | 27 | 0.491 (0.429 to 0.553); I2 = 38%, τ2 = 0.005 | 0.513 (–2.050 to 3.076); I2 = 0%, τ2 = 0 | 0.472 (–0.797 to 1.740); I2 = 74%, τ2 = 0.452 | |
| 12 | Poon et al., 2010232 | 3 | 21 | 0.636 (0.308 to 0.873); I2 = 34%, τ2 = 0.105 | 0.991 (0.022 to 1.959); I2 = 0%, τ2 = 0 | –1.091 (–4.885 to 2.702); I2 = 93%, τ2 = 2.175 | |
| 13 | Scazzocchio et al., 2013235 | 3 | 21 | 0.743 (0.374 to 0.933); I2 = 14%, τ2 = 0.057 | 0.751 (0.144 to 1.358); I2 = 0%, τ2 = 0 | –0.699 (–3.885 to 2.486); I2 = 90%, τ2 = 1.481 | |
| 14 | Wright et al., 2015236 | 2 | 9 | 0.742 (0.040 to 0.995); I2 = 0%, τ2 = 0 | 0.919 (–4.378 to 6.216); I2 = 0%, τ2 = 0 | 0.282 (–14.337 to 14.900); I2 = 90%, τ2 = 2.395 | |
| 15 | Clinical and biochemical markers | Poon et al., 2009234 | 1 | 10 | 0.741 (0.507 to 0.888) | 0.452 (0.210 to 0.693) | –2.671 (–3.350 to –1.991) |
| Trimester 2 models | |||||||
| 23 | Clinical and ultrasound markers | Yu et al., 2005238 | 1 | 10 | 0.908 (0.826 to 0.954) | 0.557 (0.293 to 0.821) | 2.473 (1.716 to 3.229) |
| Late-onset pre-eclampsia | |||||||
| Trimester 1 models | |||||||
| 16 | Clinical | Crovetto et al., 2015229 | 5 | 384 | 0.634 (0.461 to 0.778); I2 = 87%, τ2 = 0.264 | 0.558 (–0.008 to 1.125); I2 = 92%, τ2 = 0.179 | –0.048 (–1.647 to 1.551); I2 = 98%, τ2 = 1.615 |
| 17 | Kuc et al., 2013230 | 8 | 5716 | 0.623 (0.572 to 0.671); I2 = 87%, τ2 = 0.025 | 0.657 (0.496 to 0.818); I2 = 60%, τ2 = 0.007 | –1.911 (–2.235 to –1.586); I2 = 98%, τ2 = 0.124 | |
| 18 | Plasencia et al., 2007231 | 3 | 90 | 0.672 (0.539 to 0.782); I2 = 0%, τ2 = 0 | 0.612 (0.042 to 1.182); I2 = 14%, τ2 = 0.008 | 0.202 (–1.113 to 1.517); I2 = 85%, τ2 = 0.234 | |
| 19 | Poon et al., 2010232 | 3 | 90 | 0.647 (0.475 to 0.787); I2 = 25%, τ2 = 0.020 | 0.565 (0.081 to 1.050); I2 = 0%, τ2 = 0 | 0.121 (–1.594 to 1.837); I2 = 91%, τ2 = 0.430 | |
| 20 | Scazzocchio et al., 2013235 | 1 | 26 | 0.597 (0.478 to 0.705) | 0.562 (–0.168 to 1.291) | 0.524 (0.128 to 0.920) | |
| 21 | Clinical and biochemical markers | Poon et al., 2009234 | 1 | 13 | 0.684 (0.550 to 0.792) | 0.799 (0.257 to 1.341) | –0.349 (–0.902 to 0.205) |
| Trimester 2 models | |||||||
| 24 | Clinical and ultrasound markers | Yu et al., 2005238 | 1 | 263 | 0.607 (0.570 to 0.642) | 0.077 (0.005 to 0.148) | NE |
| Model number | Authors, year | Type of predictor | Validation cohort | Number of women | Eventsa (%) | Performance statistic (95% CI) | ||
|---|---|---|---|---|---|---|---|---|
| C-statistic | Calibration slope | Calibration-in-the-large | ||||||
| Trimester 1 any-onset pre-eclampsia models | ||||||||
| 4 | Baschat et al., 2014115 | Clinical and biochemical | POP161 | 4212 | 273 (6.5) | 0.704 (0.670 to 0.737) | 1.237 (1.034 to 1.439) | 0.658 (0.533 to 0.782) |
| 5 | Goetzinger et al., 2010128 | POP161 | 4212 | 273 (6.5) | 0.764 (0.730 to 0.795) | 1.706 (1.499 to 1.913) | –0.070 (–0.195 to 0.054) | |
| 6 | Odibo et al., 2011147 | St George’s163 | 54,635 | 1487 (2.7) | 0.672 (0.654 to 0.690) | 0.962 (0.885 to 1.039) | –0.897 (–0.950 to –0.845) | |
| POP161 | 4212 | 273 (6.5) | 0.779 (0.744 to 0.812) | 1.490 (1.329 to 1.651) | –0.033 (–0.159 to 0.094) | |||
| Trimester 1 early-onset pre-eclampsia models | ||||||||
| 8 | Baschat et al., 2014115 | Clinical | Poston et al. 2006150 | 2422 | 144 (6.0) | 0.672 (0.626 to 0.716) | 1.281 (0.898 to 1.664) | 1.797 (1.628 to 1.967) |
| 10 | Kuc et al., 2013230 | St George’s163 | 54,635 | 151 (0.3) | 0.635 (0.587 to 0.679) | 0.343 (0.227 to 0.460) | –4.510 (–4.674 to –4.347) | |
| AMND114 | 136,635 | 1237 (0.9) | 0.681 (0.665 to 0.696) | 0.470 (0.430 to 0.511) | –3.387 (–3.445 to –3.330) | |||
| Trimester 1 late-onset pre-eclampsia models | ||||||||
| 16 | Crovetto et al., 2015229 | Clinical | POP161 | 4212 | 263 (6.2) | 0.781 (0.745 to 0.813) | 1.248 (1.120 to 1.376) | 1.309 (1.177 to 1.441) |
| 17 | Kuc et al., 2013230 | ALSPAC103 | 14,344 | 266 (1.9) | 0.657 (0.616 to 0.696) | 0.761 (0.550 to 0.973) | –1.574 (–1.699 to –1.448) | |
| St George’s163 | 54,635 | 1336 (2.4) | 0.636 (0.621 to 0.651) | 0.632 (0.560 to 0.704) | –1.970 (–2.025 to –1.915) | |||
| AMND114 | 136,635 | 3733 (2.7) | 0.844 (0.640 to 0.943) | 0.746 (0.449 to 1.042) | –1.439 (–2.092 to –0.787) | |||
| POP161 | 4212 | 263 (6.2) | 0.599 (0.561 to 0.636) | 0.673 (0.452 to 0.894) | –1.487 (–1.613 to –1.361) | |||
| Trimester 2 any-onset pre-eclampsia models | ||||||||
| 22 | Yu et al., 2005238 | Clinical and ultrasound | POP161 | 4212 | 273 (6.5) | 0.610 (0.574 to 0.645) | 0.075 (0.007 to 0.144) | NE |
| Trimester 2 late-onset pre-eclampsia models | ||||||||
| 24 | Yu et al., 2005238 | Clinical and ultrasound | POP161 | 4212 | 263 (6.2) | 0.607 (0.570 to 0.642) | 0.077 (0.005 to 0.148) | NE |
FIGURE 4.
Plot of the summary meta-analysis estimates and CIs of the C-statistic (pooled across IPPIC-UK validation data sets) for each prediction model. (a) Any-onset pre-eclampsia; (b) early-onset pre-eclampsia; and (c) late-onset pre-eclampsia. This figure contains information from several sources. 115,128,147,229–236,238 C, clinical characteristics only; C + B, clinical characteristics and biochemical markers; C + U, clinical characteristics and ultrasound markers.
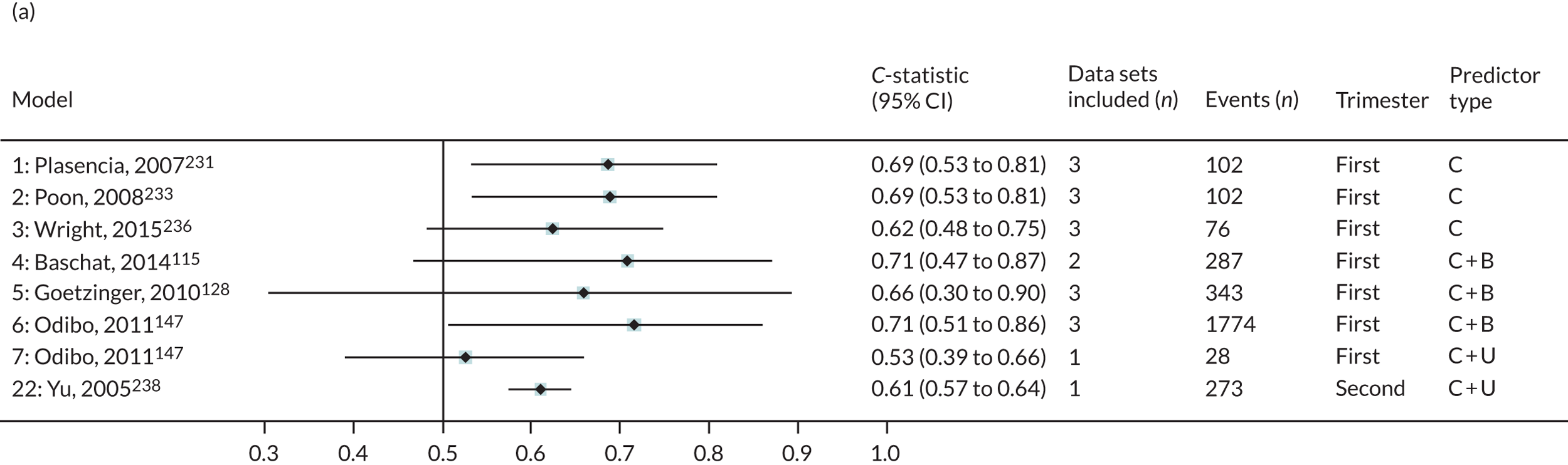
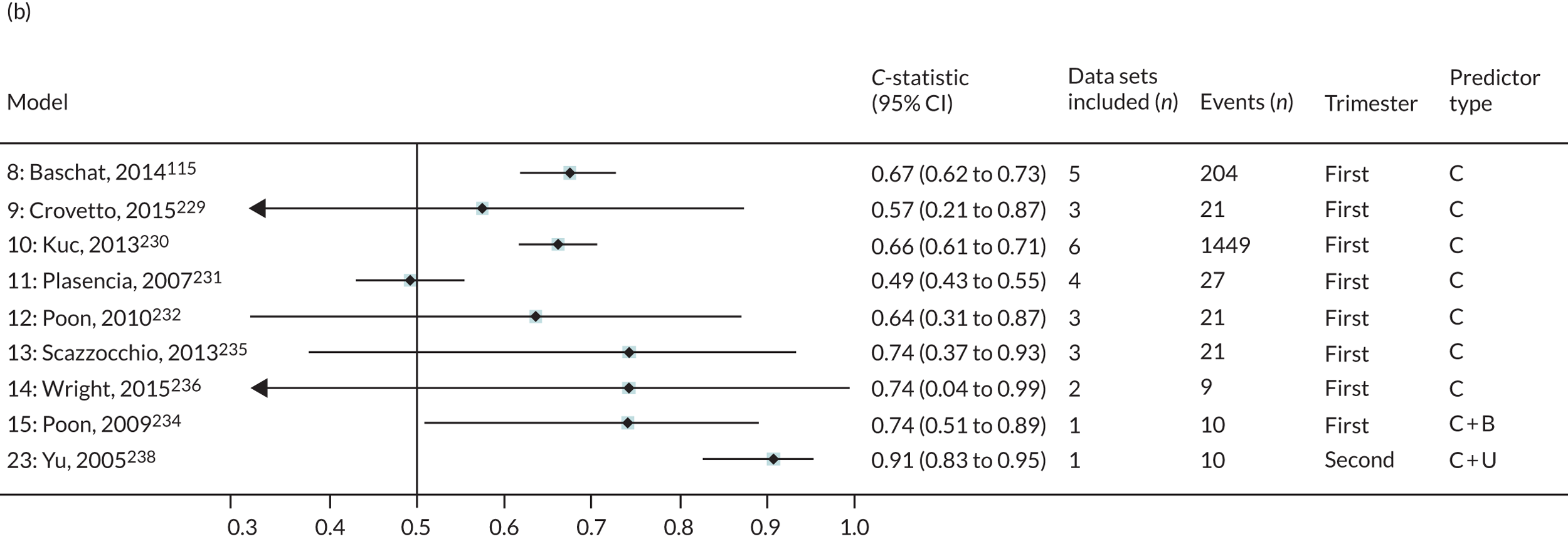
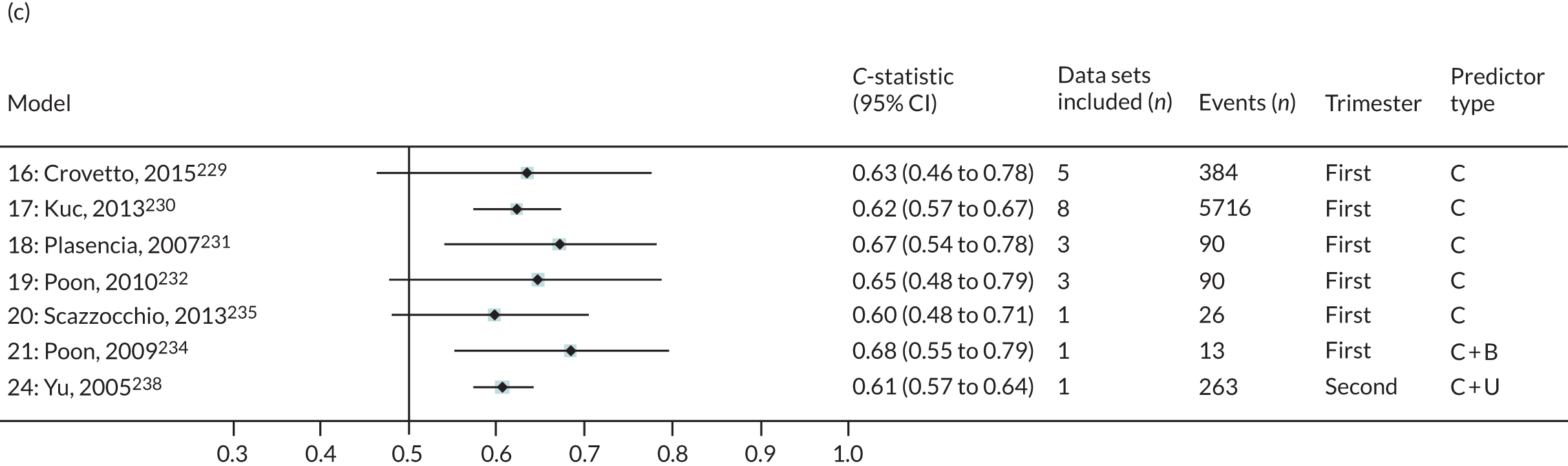
Any-onset pre-eclampsia
First-trimester clinical characteristics only models
Two (Plasencia et al. , model 1;231 Poon et al. , model 2233) of the three first-trimester clinical characteristics models were validated in three cohorts with total pre-eclampsia events > 100. The summary C-statistics were low (< 0.7), and the summary calibration slopes were < 1 for both models.
First-trimester clinical and biochemical models
There was a sufficient number of pre-eclampsia events in total (> 100) for all three clinical and biochemical first-trimester models (Baschat et al. , model 4;115 Goetzinger et al. , model 5;128 Odibo et al. , model 6147). Of these, only two models had summary C-statistics > 0.7; but only one model’s (Odibo et al. , model 6147) C-statistic had a lower limit of the 95% CI > 0.50. Their summary calibration slopes were > 1, indicating potential underfitting with predictions that do not span a wide enough range of probabilities compared with what is observed in the validation data sets. One model (Odibo et al. , model 7147) including first-trimester clinical and ultrasound predictors could be validated only in a data set with a very small number of events (i.e. 28 pre-eclampsia outcomes).
When validated in individual cohorts that had at least 100 pre-eclampsia events, promising discrimination was observed for all three models with C-statistics of 0.70 (Baschat et al. , model 4115), 0.76 (Goetzinger et al. , model 5128) and 0.78 (Odibo et al. , model 6147) in the POP study161 cohort of singleton nulliparous women; the corresponding calibration slopes were 1.24, 1.71 and 1.49, indicating underfitting of risks (range of predictions too narrow). Models 5 (Goetzinger et al. 128) and 6 (Odibo et al. 147) systematically predicted risks that were too high in all of the validation data sets (calibration-in-the-large < 0).
Second-trimester models
Only one model (Yu et al. , model 22238), which comprised clinical and ultrasound predictors, could be validated. Both discrimination (C-statistic 0.61) and calibration slope were poor (0.075).
Early-onset pre-eclampsia
First-trimester clinical models
Of the seven models, only two (Baschat et al. , model 8;115 Kuc et al. , model 10230) could be validated in data sets with adequate total numbers of pre-eclampsia events (> 100). Both had low discrimination, with summary C-statistics < 0.7; the calibration was poor, with calibration slopes of 2.04 and 0.42, respectively.
The performance of the two models (Baschat et al. , model 8;115 Kuc et al. , model 10230) in individual cohorts with event size > 100 was low, with C-statistics ranging from 0.63 to 0.68; the calibration slope estimate was either too high (1.28) or too low (0.34, 0.47), respectively. Model 10230 systematically predicted too high in all their validation data sets (calibration-in-the-large < 0).
First-trimester clinical and biochemical models
The one model (Poon et al. , model 15234) with clinical and biochemical first-trimester predictors had an insufficient number of events (n = 10) to provide adequate validation.
Second-trimester models
One model (Yu et al. , model 23238) comprising clinical and ultrasound markers had an insufficient number of events (n = 10) to provide adequate validation.
Late-onset pre-eclampsia
First-trimester clinical models
Two of the five models (Crovetto et al. , model 16;229 Kuc et al. , model 17230) were validated across cohorts with total event sizes above 100 and had low summary C-statistics (< 0.7). The summary calibration slope ranged from 0.56 to 0.80 for all five models. CIs were generally wide, with only a few data sets for most model validations and small numbers of pre-eclampsia outcomes per meta-analysis (see Figure 4).
When the performance of the two first-trimester clinical models was assessed within individual cohorts with pre-eclampsia event numbers above 100, model 16 (Crovetto et al. 229) showed promising discrimination (C-statistic 0.78) and a calibration slope of 1.25 in the POP study. The other model (Kuc et al. ,230 model 17) showed low discrimination and poor calibration (C-statistic 0.60, calibration slope 0.67) in the POP cohort, low discrimination in St George’s and ALSPAC cohorts (C-statistic 0.64 and 0.66, respectively), and high discrimination (C-statistic 0.84) with a calibration slope of 0.75 in the Aberdeen Maternity and Neonatal Databank (AMND) cohort. 103,114,161,163
First-trimester clinical and biochemical models
The one model (Poon et al. , model 21234) with clinical and biochemical markers had an insufficient number of pre-eclampsia events (n = 13) to provide adequate validation.
Second-trimester models
The only model (Yu et al. , model 24238) with second-trimester clinical and ultrasound markers was validated in a single data set (263 events) and showed low C-statistic (0.61) and poor calibration (calibration slope 0.08).
Heterogeneity
Where it was possible to estimate it, heterogeneity across studies varied from small (e.g. models 1, 2 and 3 had I2 ≤ 3%, τ2 ≤ 0.002) to large (e.g. models 5 and 6 had I2 ≥ 90%, τ2 ≥ 0.1) for the C-statistic (on the logit scale), and from moderate to large in the calibration slope for two-thirds of all models (17/24). All models validated in multiple IPD data sets had high levels of heterogeneity in calibration-in-the-large performance, with the I2 often > 90% (see Table 5).
For the majority of models (67%, 16/24), the summary calibration slope was ≤ 0.7. Although this could be a result of chance for some models (95% CIs include 1), it is likely to suggest overfitting in the model development (as the ideal value is 1, and values < 1 indicate predictions that are too extreme) (Figure 5). The exceptions to this were models 4, 5 and 6 (for any-onset pre-eclampsia) and model 8 (for early-onset pre-eclampsia).
FIGURE 5.
Plot of the summary meta-analysis estimates and CIs of calibration (pooled across IPPIC-UK validation data sets) for each prediction model. (a) Any-onset pre-eclampsia; (b) early-onset pre-eclampsia; and (c) late-onset pre-eclampsia. This figure contains information from several sources. 115,128,147,229–236,238 C, clinical characteristics only; C + B, clinical characteristics and biochemical markers; C + U, clinical characteristics and ultrasound markers.
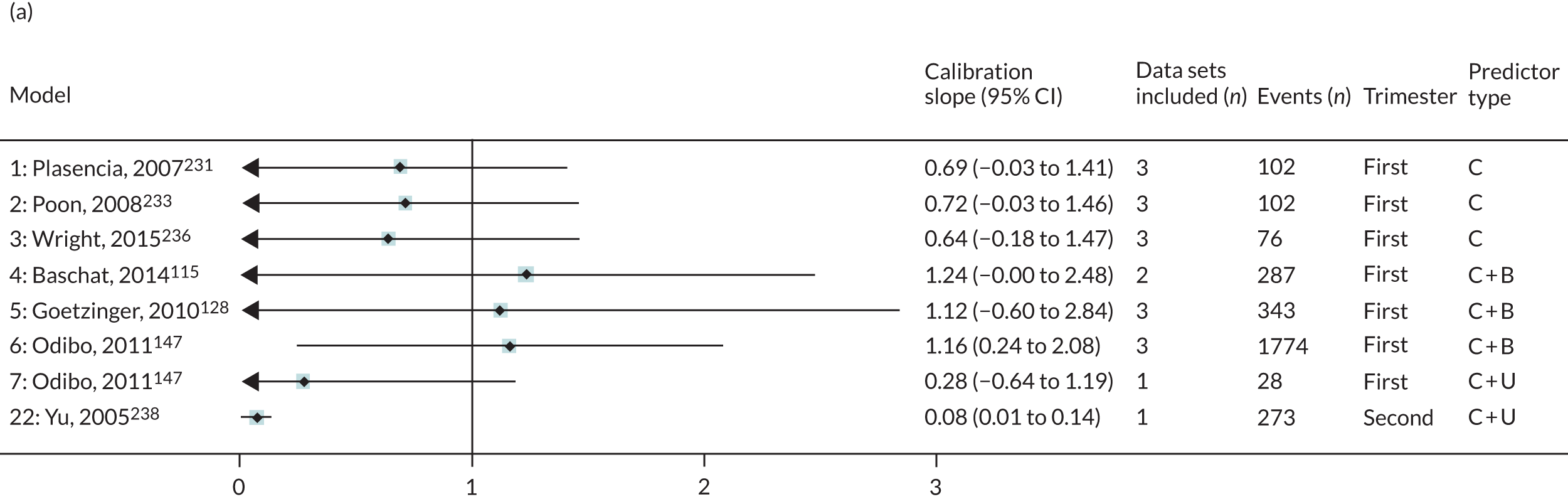
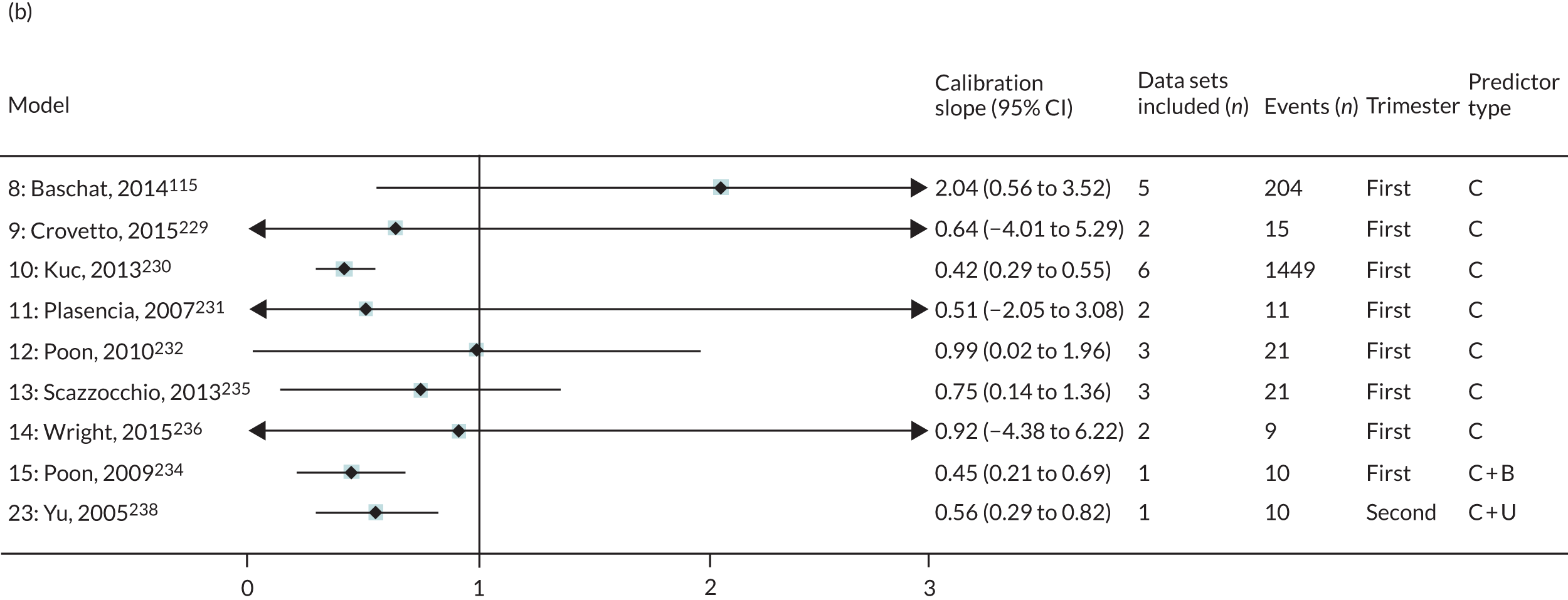
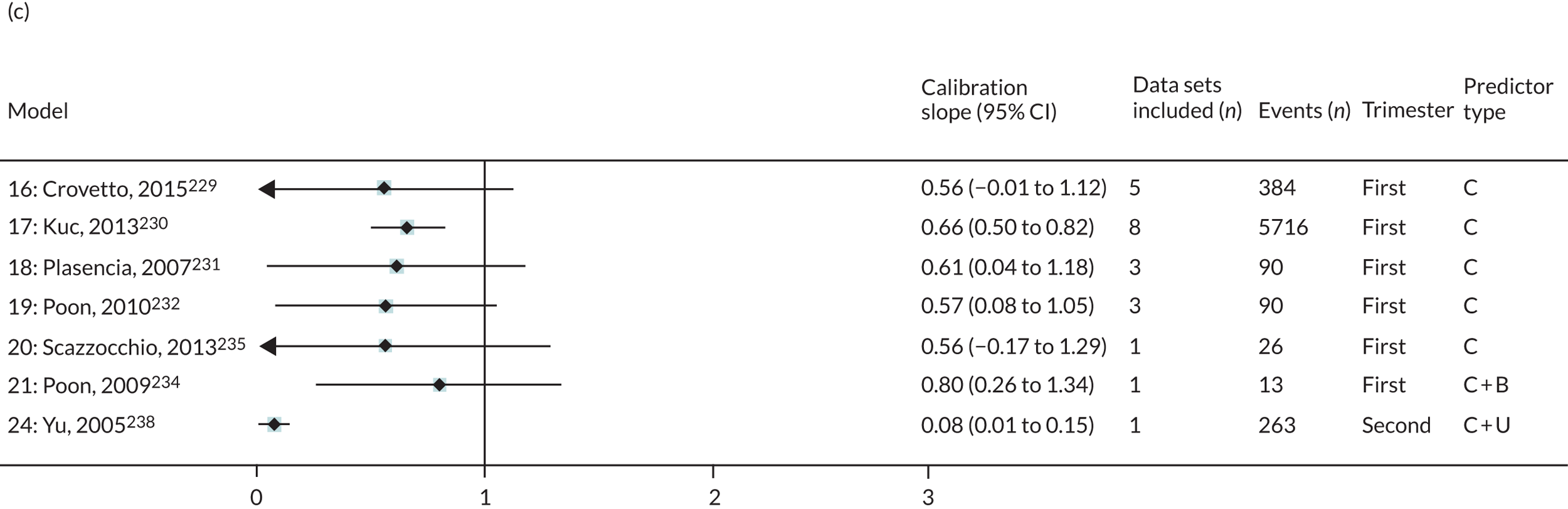
Many of the models did not have summary calibration-in-the-large values close to zero, and, even if the average is near zero, this could be due to it performing poorly in the individual validation data sets and averaging out near zero, such as for models 2, 8 and 16.
The predictive performance is likely to be optimistic in small development studies with few pre-eclampsia events, and predictor effects will be too large. If not corrected for (e.g. by penalisation/shrinkage methods), this will result in predictions that are too extreme when validated (hence calibration slope < 1); in other words, the predictor effects will not be as strong as they were thought to be at model development.
By indicating whether predictions are systematically too high or too low, calibration-in-the-large may suggest the need for recalibration of the model intercept. There was very large uncertainty in the summary estimates of calibration-in-the-large for most models, which reflects often small numbers of events and large heterogeneity across the individual data sets contributing towards the validation (Figure 6).
FIGURE 6.
Plot of the summary meta-analysis estimates and CIs of calibration-in-the-large (pooled across IPPIC-UK validation datasets) for each prediction model. (a) Any-onset pre-eclampsia; (b) early-onset pre-eclampsia; and (c) late-onset pre-eclampsia. This figure contains information from several sources. 115,128,147,229–236,238 C, clinical characteristics only; C + B, clinical characteristics and biochemical markers; C + U, clinical characteristics and ultrasound markers.
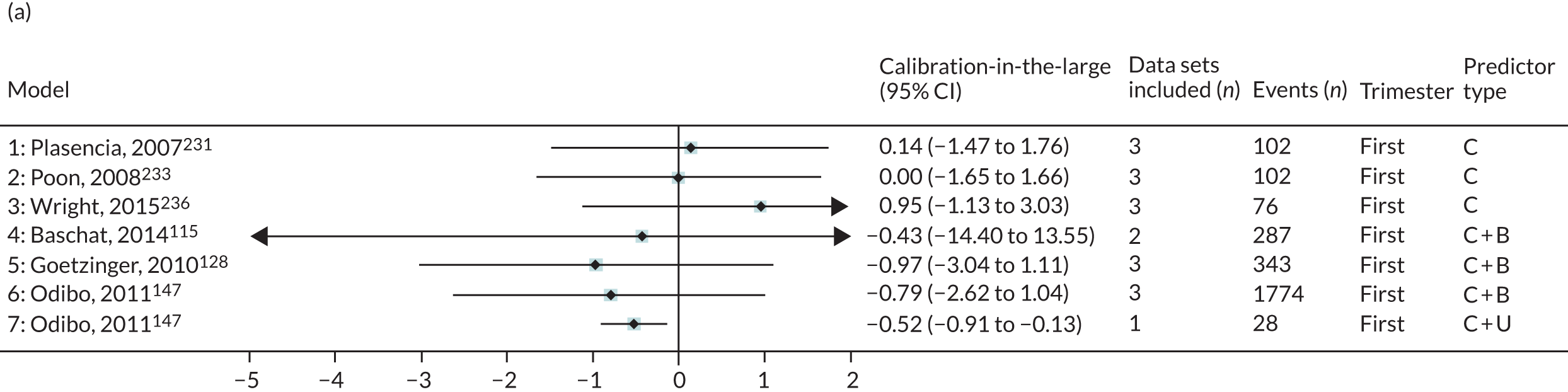
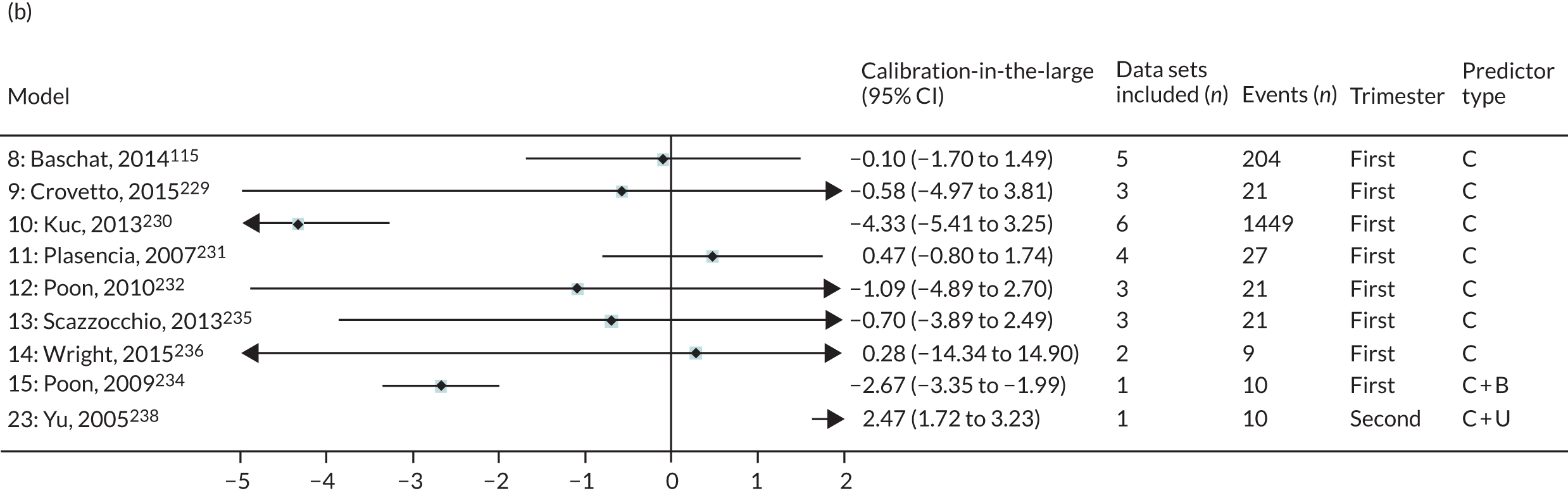
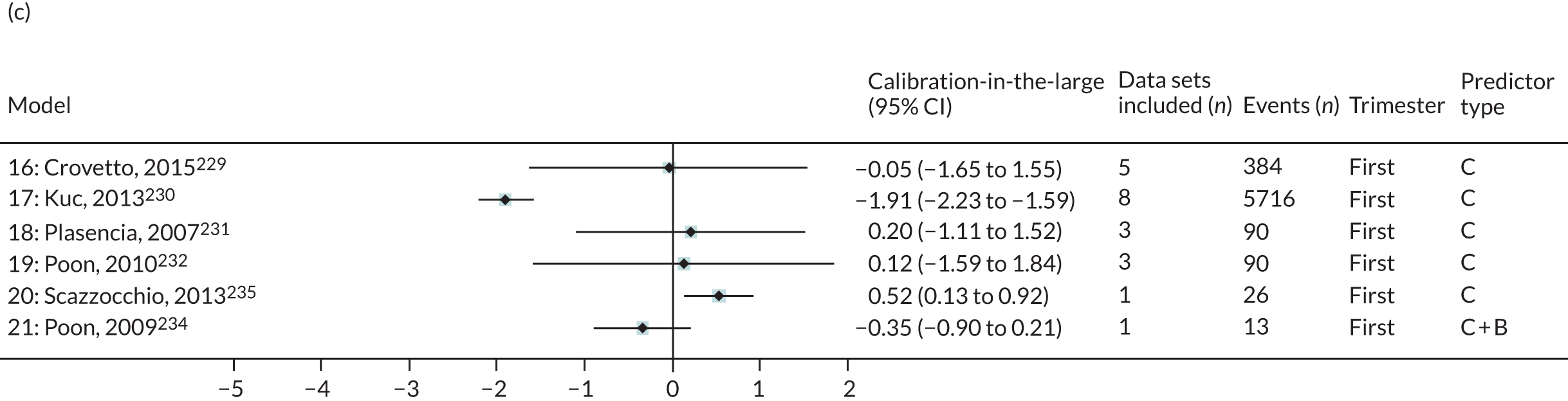
To illustrate the general concern about poor calibration, calibration plots are presented for models validated in data sets with > 100 events (Figures 7–9). These clearly show the extent of miscalibration, with predicted outcome risk from the models being far greater than the observed probabilities across the entire range of predicted risk in most cases. For any-onset pre-eclampsia, the exceptions to this are model 4 (Baschat et al. , 2014115) and model 5 (Goetzinger et al. , 2010128), when validated in POP (see Figure 7). 161 Model 4 and model 5 predictions are too low for those at greatest risk. For early-onset pre-eclampsia, model 8 (Baschat et al. , 2014115), validated in Poston 2006150 (see Figure 8), shows that predictions were all very low (all predicted probabilities < 3%) but the observed frequency of events was higher (up to around 20%). Model 10 (Kuc et al. , 2013230), validated in the St George’s cohort,163 predicted an average of near 50% for the highest risk group; however, their observed risk was still very close to zero. For late-onset pre-eclampsia, model 16 (Crovetto et al. , 2015229) underpredicted risk, particularly in those at highest risk in POP,161 and model 17 (Kuc et al. , 2013230) overpredicted risk in both St George’s163 and AMND114 (see Figure 9).
FIGURE 7.
Calibration plots for models predicting any-onset pre-eclampsia using first-trimester clinical characteristics and biochemical markers in data sets with > 100 outcome events. (a) Model 4115 in POP;161 (b) model 5128 in POP;161 (c) model 6147 in St George’s;163 and (d) model 6147 in POP. 161 Reproduced from Snell et al. 52 Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
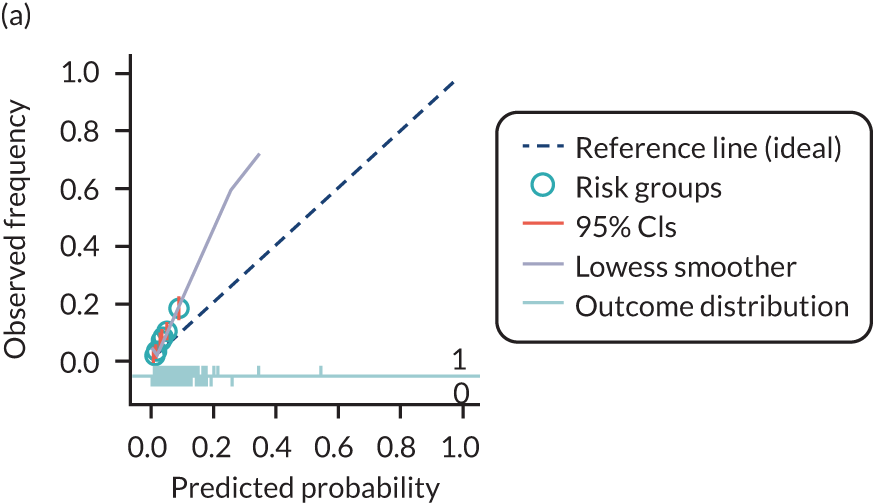
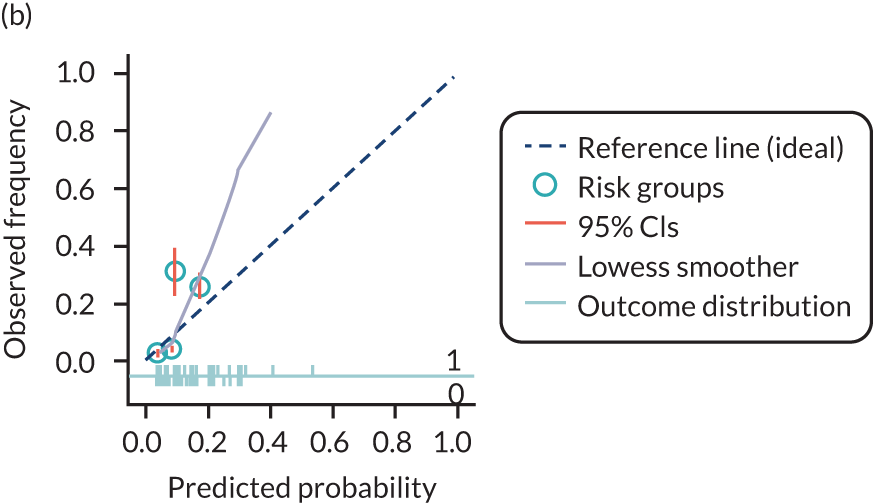
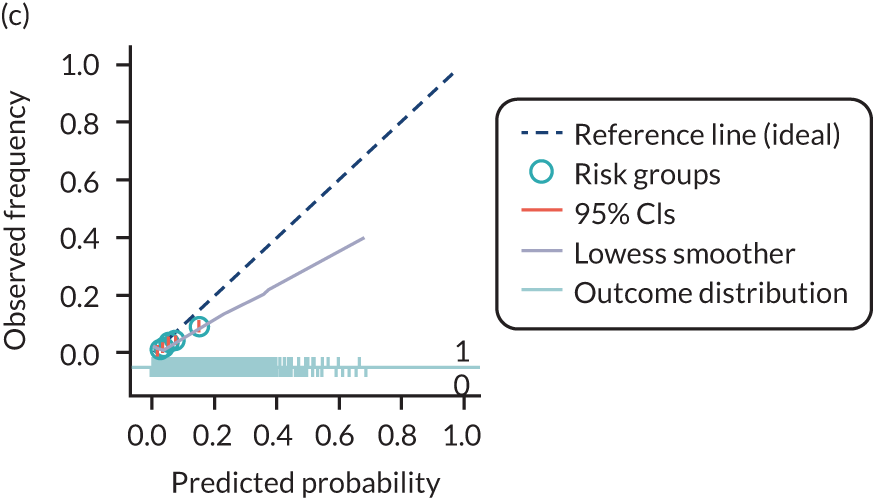
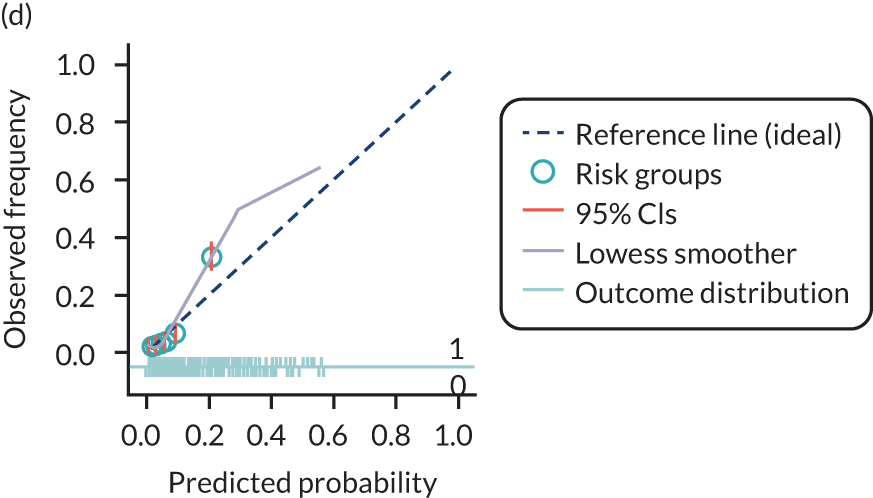
FIGURE 9.
Calibration plots for models predicting late-onset pre-eclampsia using first-trimester clinical characteristics marker in data sets with > 100 outcome events. (a) Model 16229 in POP;161 (b) model 17230 in ALSPAC;103 (c) model 17230 in St George’s;163 (d) model 17230 in AMND;114 and (e) model 17230 in POP. 161
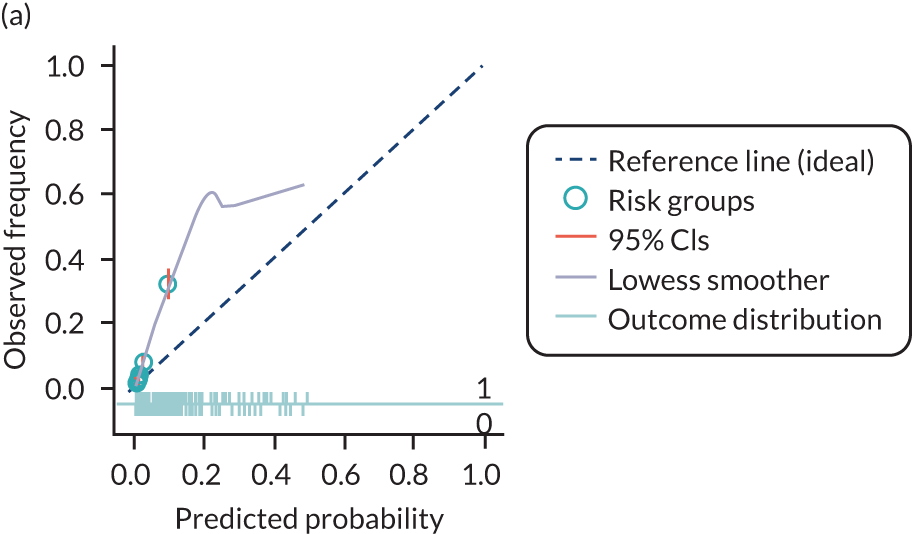
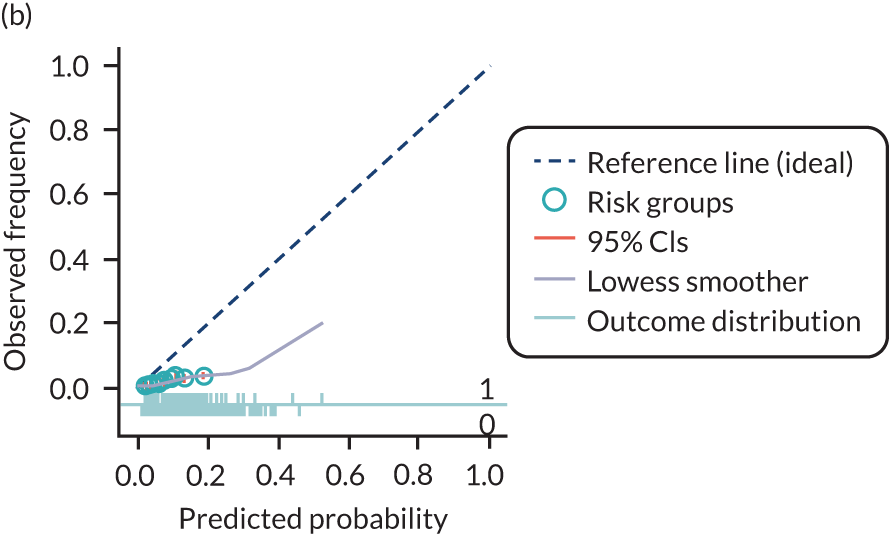
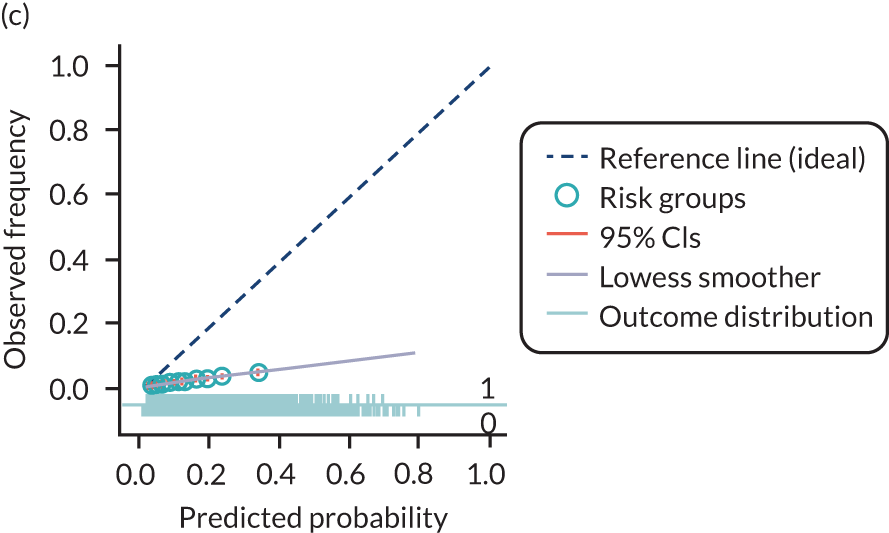
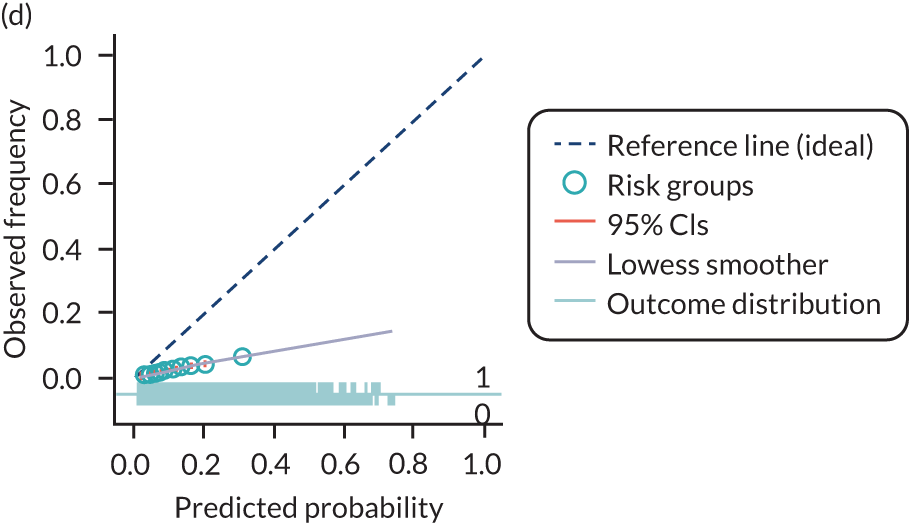
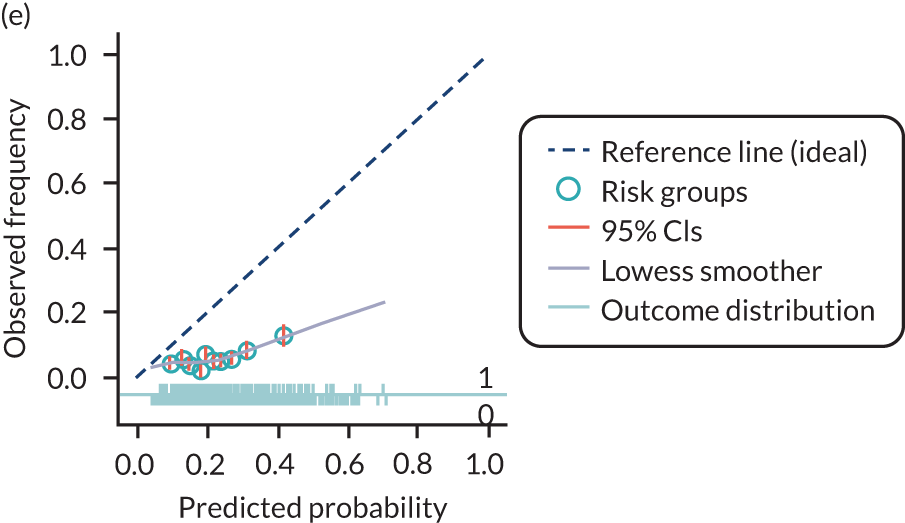
Decision curve analysis
Comparisons of models for any-, early- and late-onset pre-eclampsia using decision curve analysis were carried out in the SCOPE,42 Allen et al. ,108 Poston et al. 2015149 and POP161 data sets. Net benefit values are often best multiplied by 1000 to reveal the extra number of women who would be correctly treated per 1000 women who used the model, with none treated incorrectly.
Any-onset pre-eclampsia
Models validated in cohorts with > 100 events
The Goetzinger et al. 128 and Odibo et al. 147 models (models 5 and 6) showed a positive net benefit between relevant predicted probability thresholds of 4% to 20% in the POP data set161 comprising any nulliparous women with a singleton pregnancy. The Baschat et al. 115 model (model 4) also showed a net benefit from 5% to 20% in the POP data set. 161 These models, however, showed a net harm across thresholds in the Allen et al. 108 and Poston et al. 2015149 data sets (Figure 10).
FIGURE 10.
Decision curves for prediction models of any-onset pre-eclampsia in the (a) SCOPE UK,42 (b) Allen et al. ,108 (c) Poston et al. 2015149 and (d) POP161 data sets. Reproduced from Snell et al. 52 Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
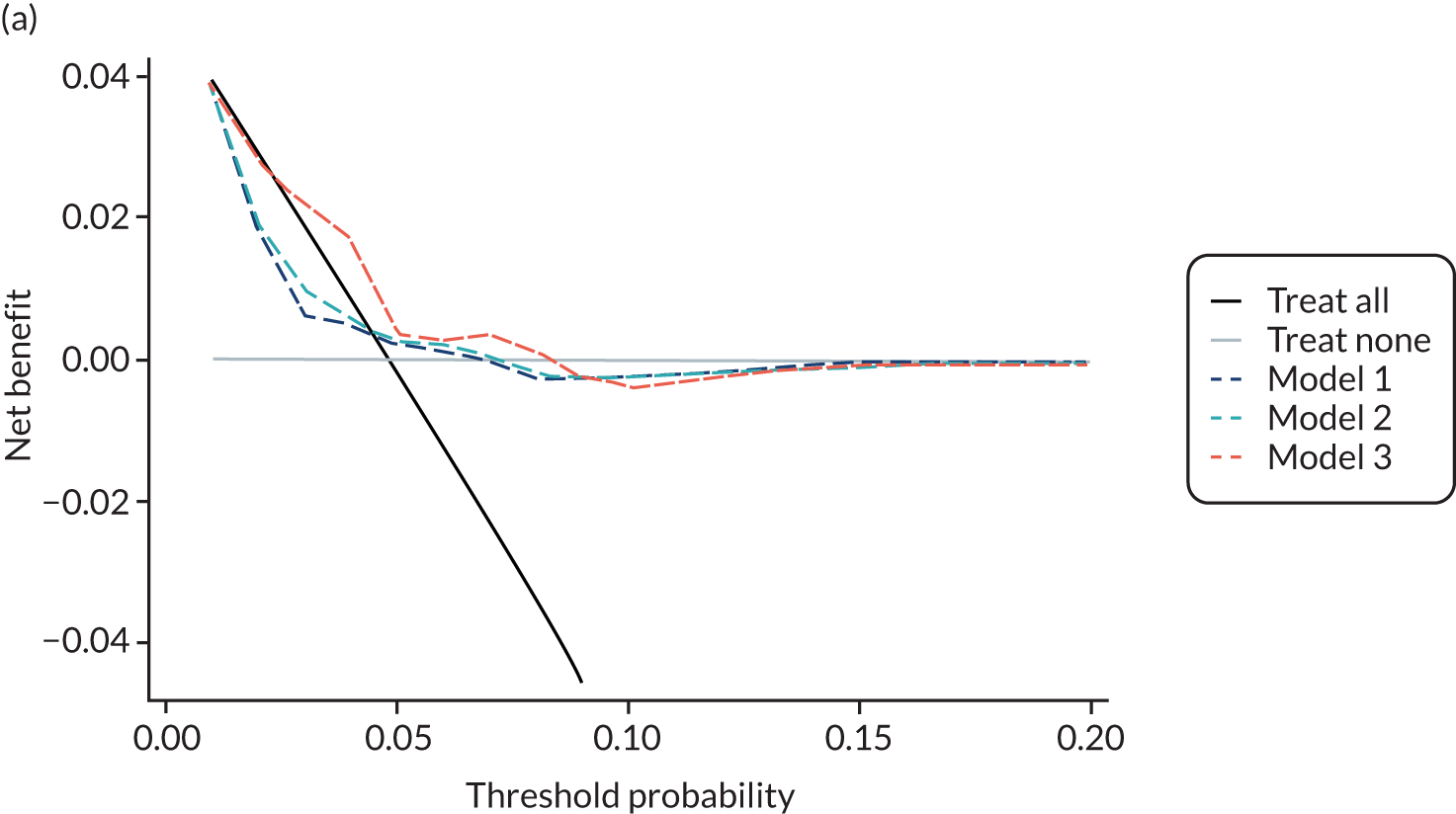
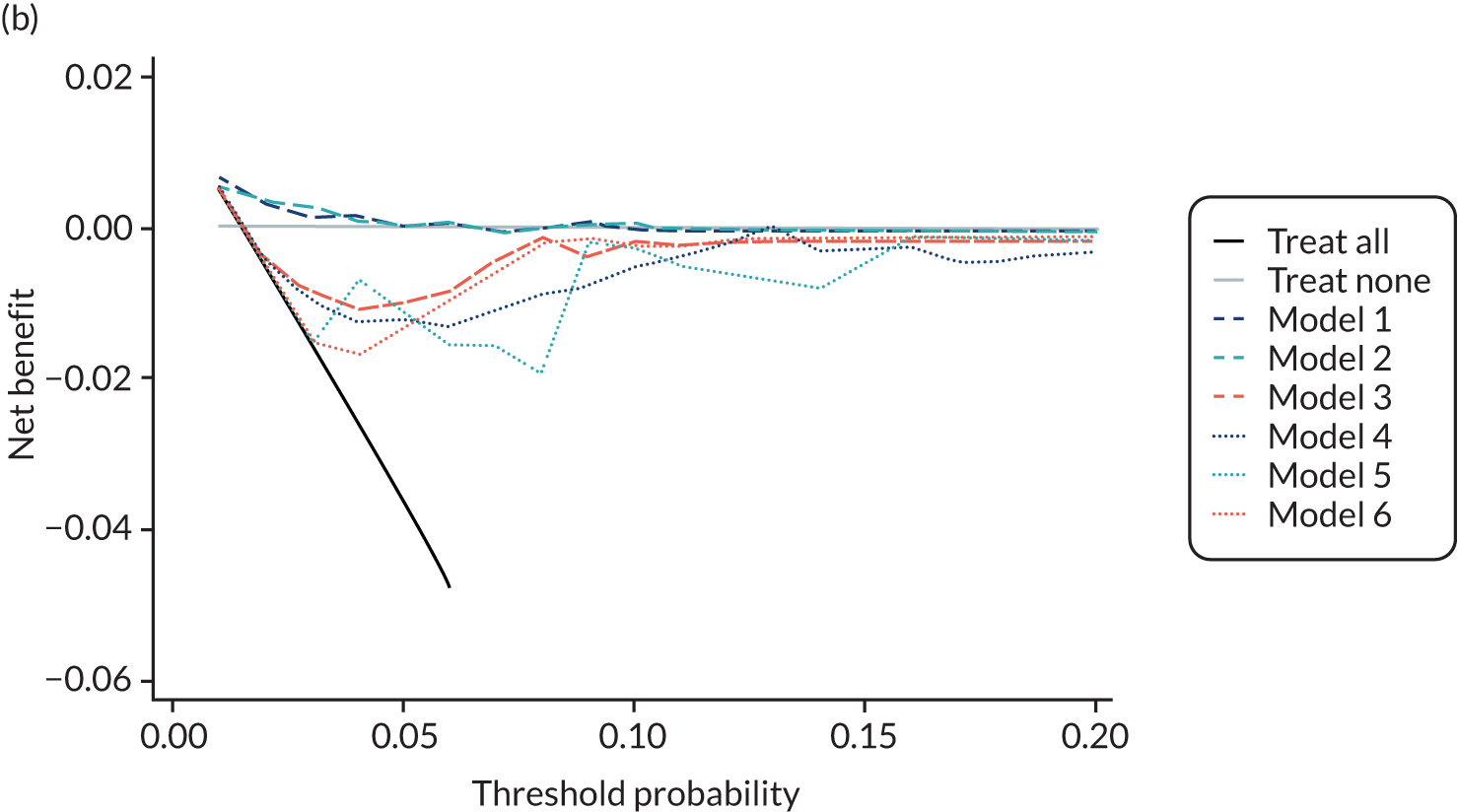
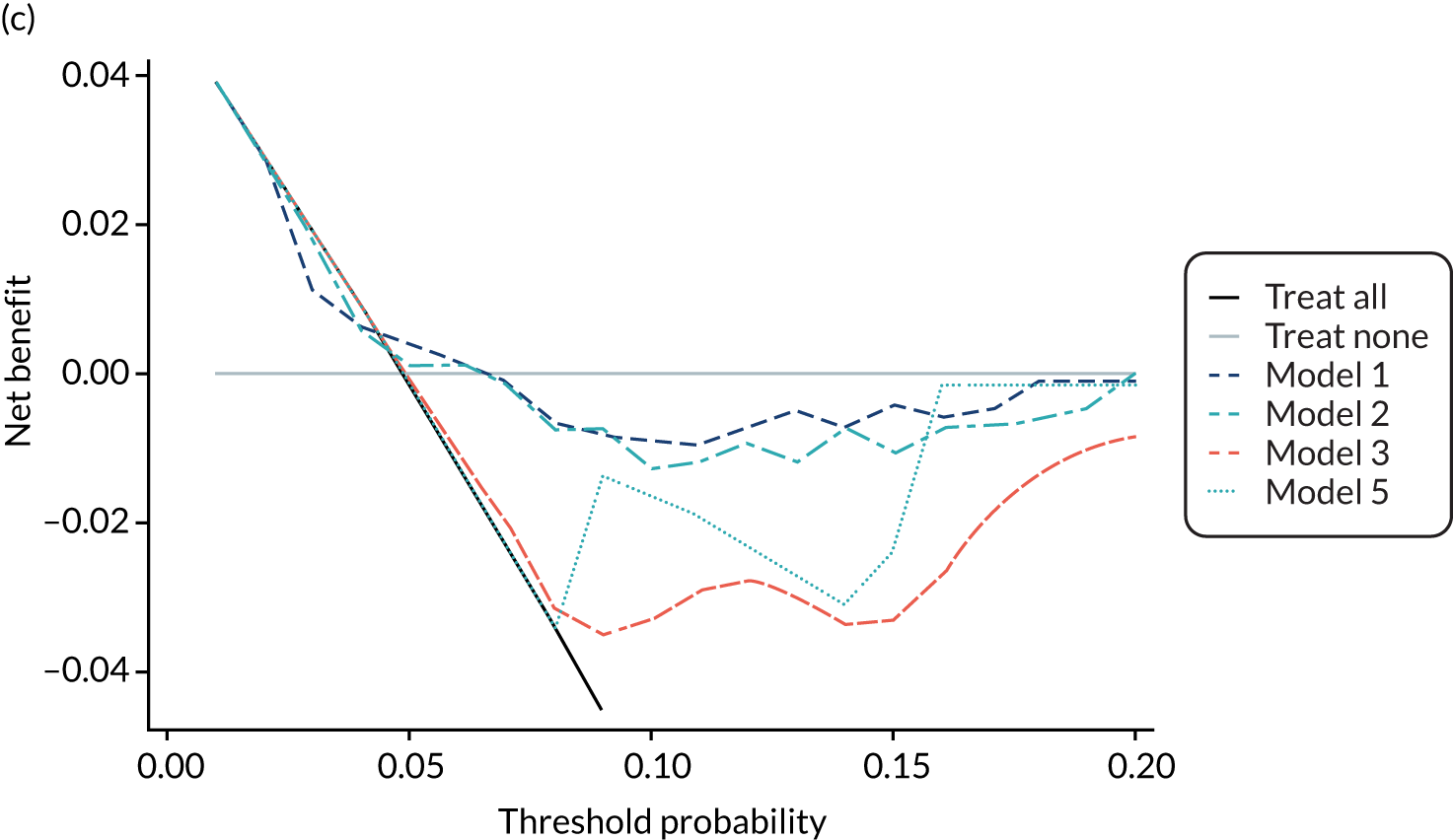
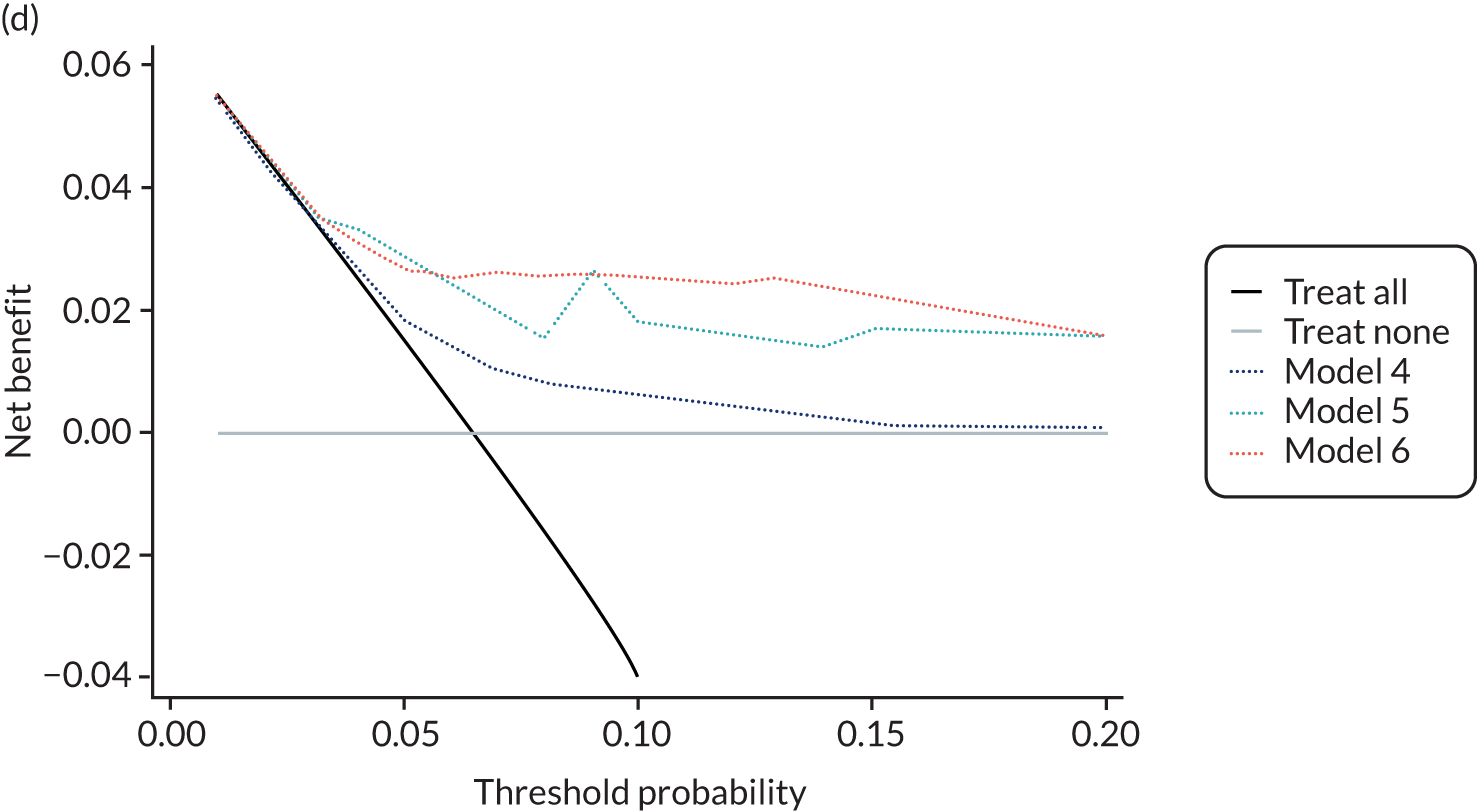
Early-onset pre-eclampsia
Decision curves for early-onset pre-eclampsia are given in Appendix 14. These did not show any net benefit, with some models showing potential harm compared with a treat-none strategy. This is likely to be because of how rare the outcome is and how few events there were in the data sets used for validation.
Late-onset pre-eclampsia
Models validated in cohorts with > 100 events
For late-onset pre-eclampsia, the Crovetto et al. 229 model (model 16) performed better than the Kuc et al. 2013230 model (model 17) in POP,161 with net benefit at thresholds of 4–20%; however, this model did not demonstrate any net benefit over a treat-all or a treat-none strategy in the other three studies (Figure 11).
FIGURE 11.
Decision curves for prediction models of late-onset pre-eclampsia in (a) SCOPE UK,42 (b) Allen et al. ,108 (c) Poston et al. 2015149 and (d) POP161 data sets. Net benefit values on the y-axis are often best multiplied by 1000 to reveal the extra number of women who would be correctly treated per 1000 women who used the model, with none treated incorrectly. Reproduced from Snell et al. 52 Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
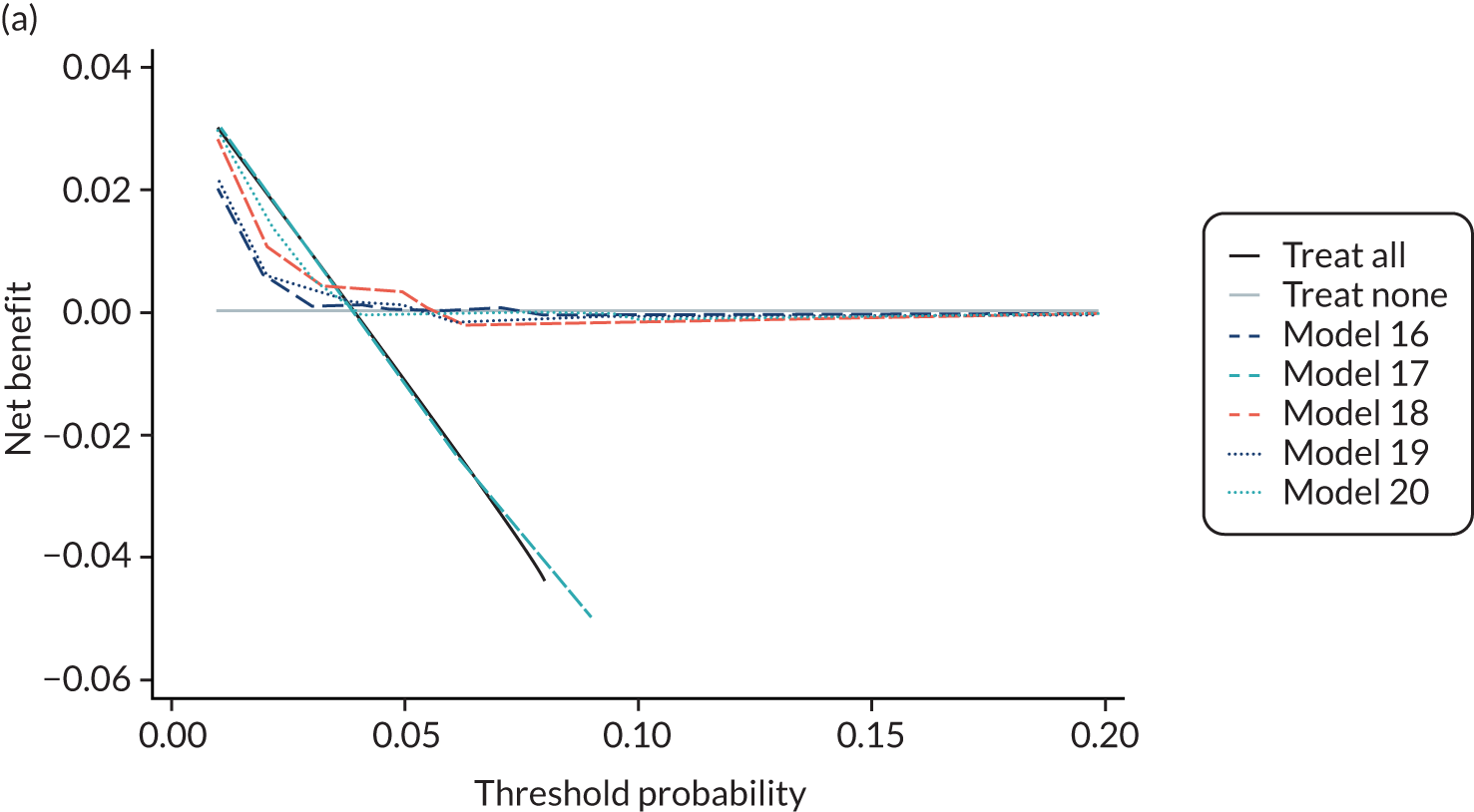
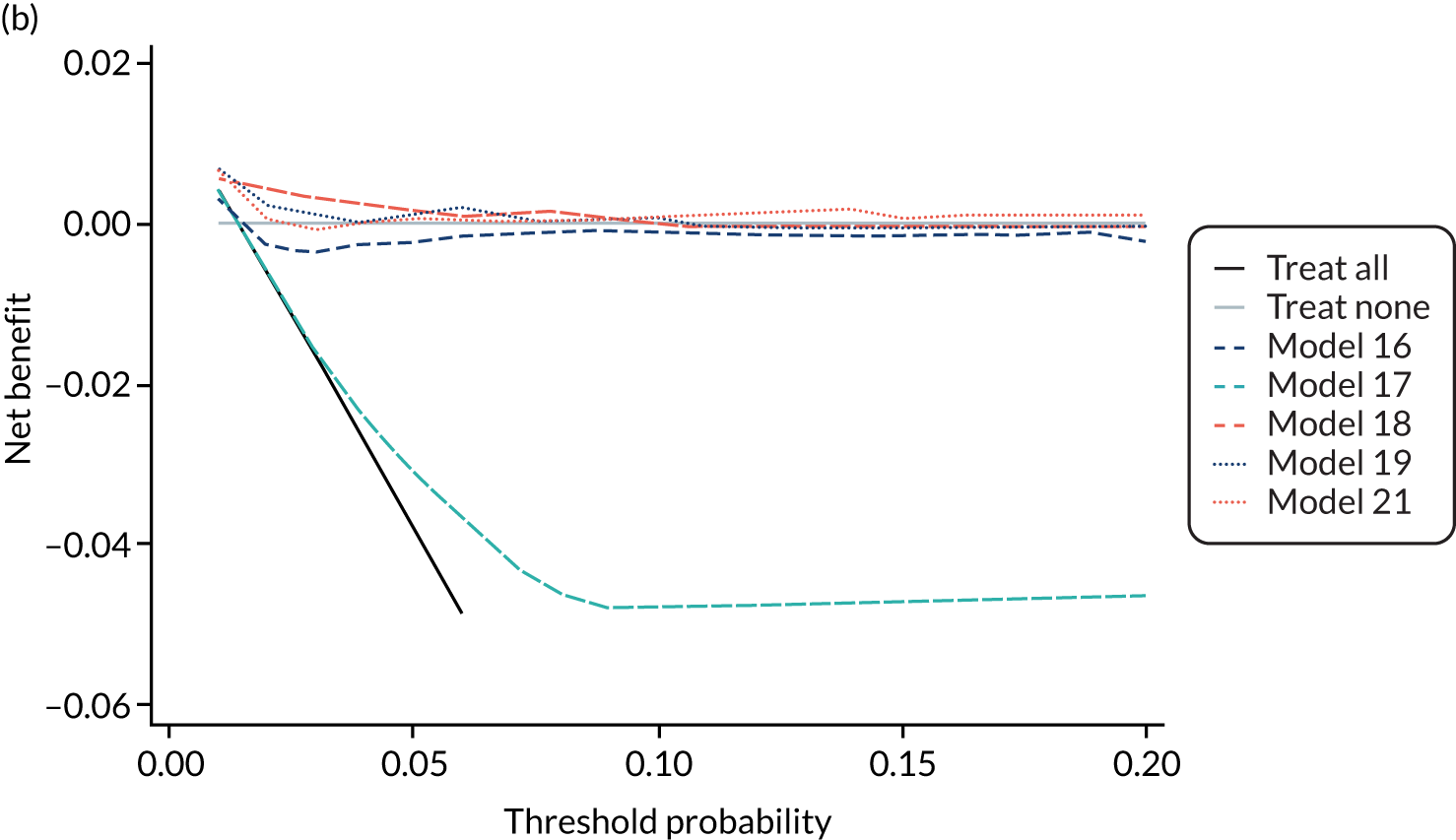
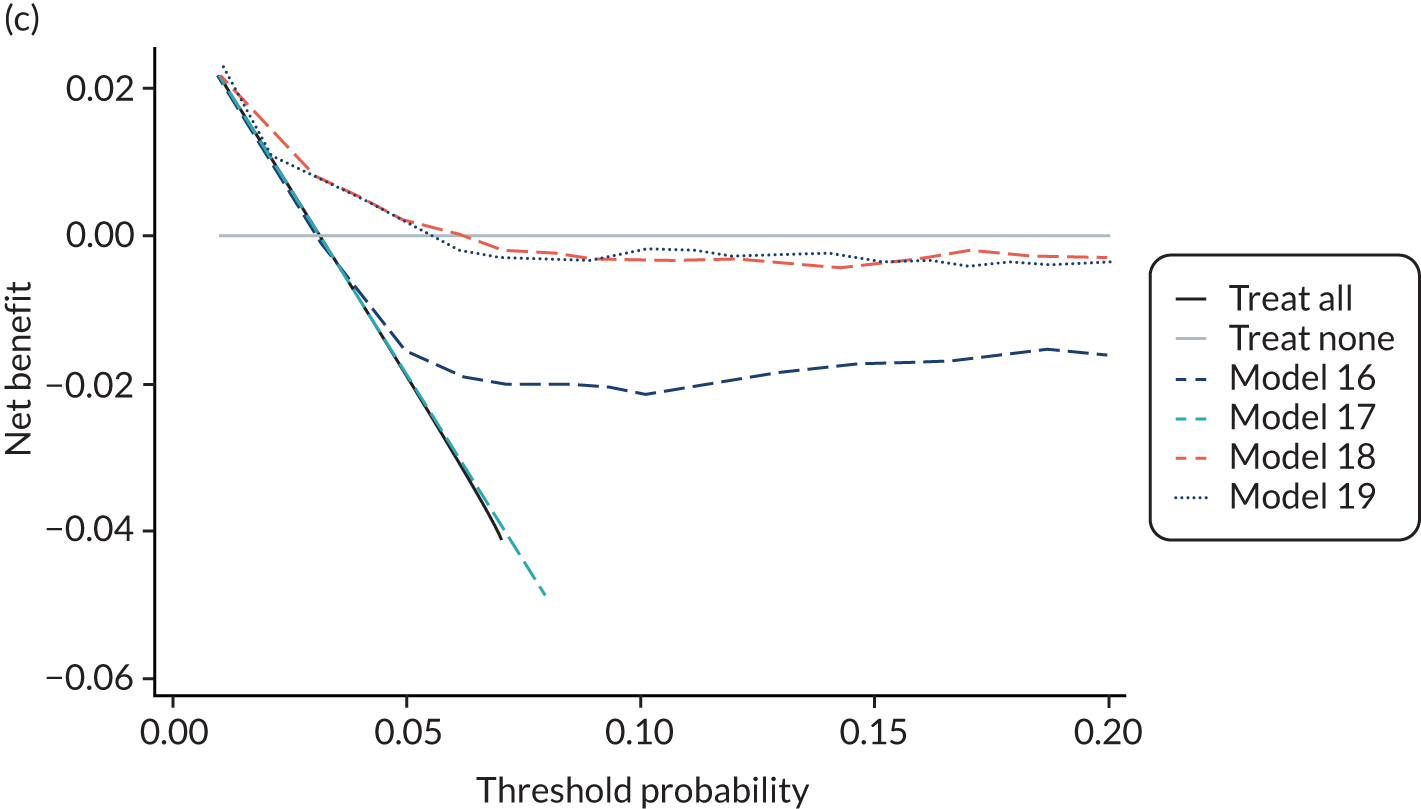
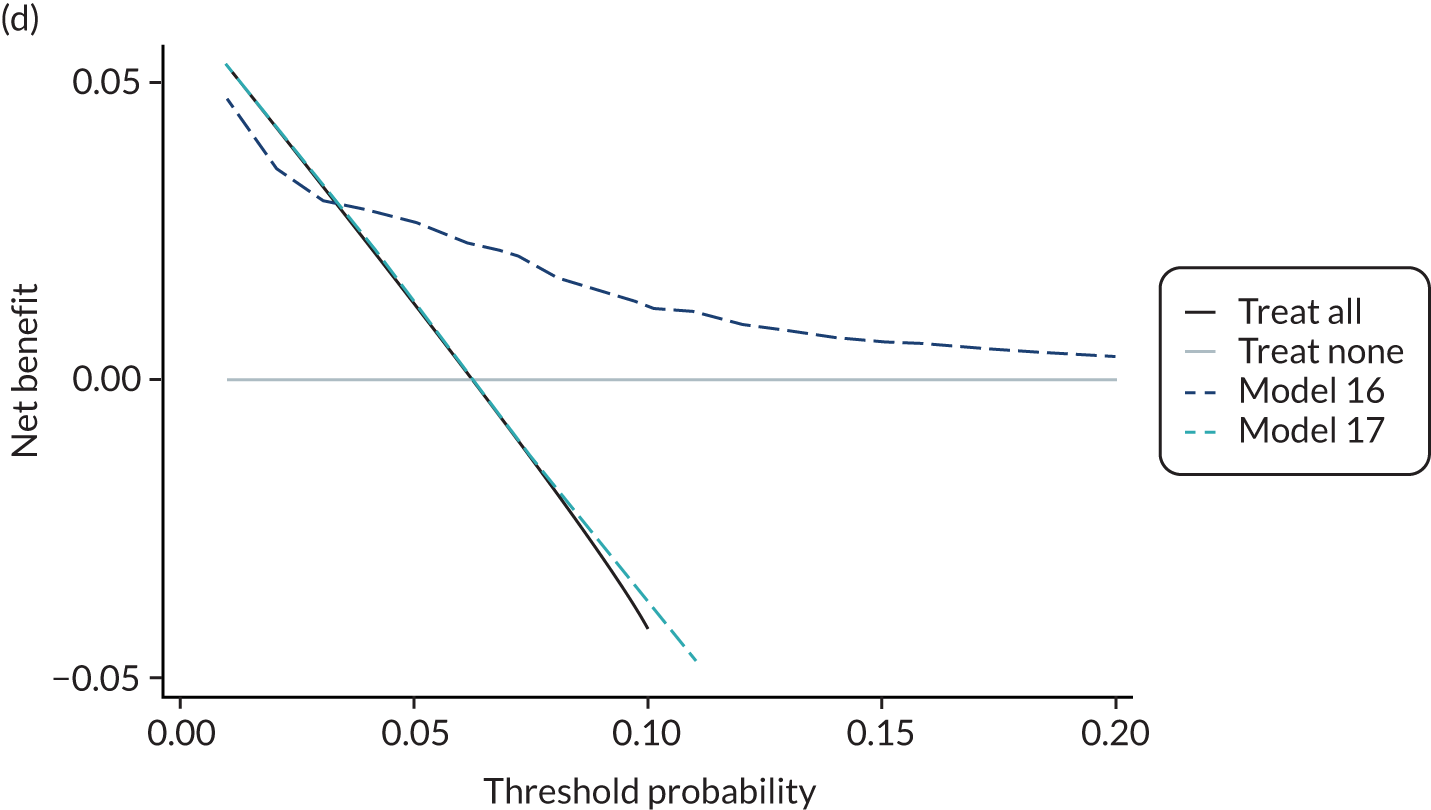
Summary
In summary, although some existing models showed a reasonable ability to discriminate between women who had a pre-eclampsia outcome and those who did not, calibration performance was, overall, poor across the data sets, with large heterogeneity in the calibration performance across different IPPIC data sets. Many of the models demonstrated likely overfitting at model development, with predictions that were too extreme compared with the observed risk in the data sets (calibration slope < 1). A model prediction could also be systematically too low or too high depending on the data set used to validate it (calibration-in-the-large ≠ 0). The models were validated in a rather heterogeneous group of data sets that had different eligibility criteria. These findings suggest that the differences between women in the data sets are not adequately captured by the set of predictors included in the models. There was also little difference in predictive performance when biochemical markers or ultrasound markers were combined with maternal and clinical characteristics, compared with models with only maternal and clinical characteristics. An important limitation to this work is that it was possible to externally validate only 24 out of the 70 existing models that reported model equations.
None of the existing models validated performed well enough to warrant recalibration. Owing to the large heterogeneity in predictive performance of these existing models across data sets, simple recalibration strategies were unlikely to improve the overall performance or reduce heterogeneity in performance. Therefore, in Chapter 6 we go on to develop and internally validate new prediction models for pre-eclampsia outcomes using the combined IPPIC data to ascertain whether or not this can address the shortcomings of existing models.
Chapter 6 Development and validation of pre-eclampsia prediction models
In this chapter, we describe the results of developing new prediction models using the IPPIC data sets. The methods for this chapter are detailed in Chapter 3, Development and validation of pre-eclampsia prediction models. We aimed to develop 18 models, one for each combination of the three outcomes (any-onset, early-onset and late-onset pre-eclampsia), two trimesters (predictors measured at trimester 1 or at trimester 2) and three predictor sets (clinical characteristics only, clinical characteristics plus biochemical markers, and clinical characteristics plus ultrasound markers).
Summary of international data sets and predictor availability
A total of 78 data sets were included in the IPPIC project. As explained in Chapter 3, the available data sets did not record all of the variables of interest, or the same combination of variables as other data sets. The timing of measurements also differed (i.e. trimester 1, trimester 2 or both). Potential predictors deemed ‘important’ based on the clinical consensus (see Table 3) were ranked according to the mean of their scores (ranging from 1, not important, to 5, very important; Table 7).
| Rank | Variable | Mean of scores | Data sets with variable, n (%) | Pre-eclampsia, n (% of all) |
|---|---|---|---|---|
| Maternal clinical characteristics | ||||
| 1 | Previous any PE | 4.91 | 55 (69) | 33,583 (33) |
| 2 | Chronic or pre-existing hypertension | 4.67 | 70 (86) | 90,084 (89) |
| 3 | SBP (first or second trimester) | 4.48 | 41 (51) | 12,879 (13) |
| 4 | BMI (first or second trimester) | 4.45 | 52 (65) | 34,453 (34) |
| 5 | DBP (first or second trimester) | 4.45 | 41 (51) | 12,879 (13) |
| 6 | Parity | 4.30 | 52 (65) | 41,020 (41) |
| 13 | MAP (first or second trimester) | 4.27 | 44 (55) | 13,018 (13) |
| 7 | History of renal disease | 4.24 | 50 (63) | 84,595 (84) |
| 8 | Multiple pregnancy | 4.12 | 60 (75) | 87,961 (87) |
| 9 | History of pre-existing diabetes | 4.12 | 63 (79) | 93,588 (93) |
| 10 | Age | 3.94 | 73 (91) | 97,724 (97) |
| 14 | PCR (first or second trimester) | 3.91 | 1 (1) | 278 (< 1) |
| 12 | Family history of PE in first-degree relative | 3.85 | 14 (18) | 995 (1) |
| 11 | Previous autoimmune disease | 3.82 | 43 (54) | 37,664 (37) |
| 17 | Previous SGA | 3.79 | 18 (23) | 4064 (4) |
| 15 | Urine dipstick (first or second trimester) | 3.73 | 12 (15) | 2209 (2) |
| 16 | Ethnicity | 3.61 | 57 (71) | 72,248 (72) |
| 18 | Smoking | 3.06 | 57 (71) | 94,252 (94) |
| 19 | Mode of conception | 2.97 | 23 (29) | 31,096 (31) |
| 20 | Substance misuse in current pregnancy | 2.48 | 17 (21) | 13,844 (14) |
| Biochemical markers | ||||
| 1 | PlGF (first or second trimester) | 4.21 | 19 (24) | 3453 (3) |
| 2 | sFlt1 (first or second trimester) | 3.94 | 12 (15) | 3175 (3) |
| 3 | PAPP-A (first or second trimester) | 3.15 | 9 (11) | 871 (< 1) |
| Ultrasound markers | ||||
| 1 | Uterine artery PI (first or second trimester) | 4.03 | 20 (25) | 2999 (3) |
| 2 | Estimated fetal weight centile (first or second trimester) | 3.33 | 4 (5) | 591 (< 1) |
| 3 | Umbilical artery PI (first or second trimester) | 3.24 | 8 (10) | 887 (< 1) |
None of the individual IPPIC data sets had all of the clinical characteristics of interest, and therefore some variables had to be excluded to form the model development sets. We first excluded variables that were not recorded in many data sets or for which few data were available across the data sets with that variable (removed if the proportion of events in data sets with the variable accounted for < 5% of events across all data sets). Protein–creatinine ratio (PCR), urine dipstick, family history of pre-eclampsia and previous small for gestational age fetus were all excluded. We also excluded multiple pregnancy as we aimed to develop models applicable to women with singleton pregnancies. Variables were then removed according to their ranking (lowest-ranking first) until we had a reasonable number of data sets for model development. Substance misuse, mode of conception, smoking and ethnicity were removed. Twelve data sets had all of the remaining clinical variables of interest, and these are summarised in Table 8.
| Maternal characteristics and outcomes | SCOPE42 (N = 5628) | Allen108 (N = 1045) | Poston 2015149 (N = 1554) | Baschat115 (N = 1704) | Antsaklis110 (N = 3328) | WHO175 (N = 7273) | NICH LR159 (N = 3097) | POUCH131 (N = 3019) | van Kuijk 2014166 (N = 230) | STORK G135 (N = 812) | Vinter173 (N = 304) | POP161 (N = 4212) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Maternal age (years), mean (SD), range | 28.6 (5.5), 14–45 | 29.9 (5.1), 15–48 | 30.5 (5.5), 16–45 | 30.2 (6.5), 18–50 | 30.9 (4.8), 14–47 | 22.5 (5.8), 11–51 | 20.6 (4.4), 15–39 | 26.4 (5.8), 15–47 | 30.8 (4.8), 19–40 | 29.8 (4.8), 19–45 | 29.2 (4.2), 19–43 | 29.9 (5.1), 16–48 |
| BMI (kg/m2), median [IQR], range | 24.2a [21.9–27.5], 15.4 to 58.5 | 23.6 [21.1–26.8], 14.8 to 51.1 | 35.1a [32.8–38.5], 30.0 to 66.0 | 26.7 [23.1–32.2], 15.9 to 72.9 | 22.7 [20.6–25.7], 14.5 to 50.1 | 22.2 [20.1–24.9], 13.1 to 50.5 | 22.7a [20.4–25.7], 13.4 to 51.2 | 27.7a [24.3–32.9], 15.1 to 66.3 | 24.1 [21.7–28.4], 18.0 to 38.3 | 24.2 [21.4–27.0], 16.6 to 43.0 | 33.6 [31.7–36.7], 28.8 to 46.0 | 24.1 [21.8–27.3], 14.7 to 54.7 |
| Nulliparous, n (%) | 5628 (100) | 584 (56) | 674 (43) | 736 (43) | 3328 (100) | 6710 (92) | 3097 (100) | 1293 (43) | 0 (0) | 377 (46) | 160 (53) | 4212 (100) |
| Previous pre-eclampsia, n (%) | 0 (0) | 17 (2) | 69 (4) | 95 (6) | 0 (0) | 460 (6) | 0 (0) | 106 (4) | 209 (92) | 0 (0) | 1 (< 1) | 0 (0) |
| History of hypertension | 24 (< 1) | 10 (< 1) | 0 (0) | 162 (10) | 2372 (71) | 234 (3) | 0 (0) | 92 (3) | 79 (34) | 13 (2) | 0 (0) | 220 (5) |
| History of renal disease | 0 (0) | 3 (< 1) | 0 (0) | 4 (< 1) | 0 (0) | 0 (0) | 0 (0) | 94 (9) | 0 (0) | 5 (< 1) | 0 (0) | 41 (< 1) |
| History of diabetes | 0 (0) | 11 (1) | 0 (0) | 81 (5) | 28 (< 1) | 59 (< 1) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 16 (< 1) |
| Outcome, n (%) | ||||||||||||
| Any-onset pre-eclampsia | 278 (5) | 14 (1) | 54 (4) | 106 (6) | 32 (1) | 141 (2) | 156 (5) | 44 (3) | 43 (20) | 46 (6) | 19 (6) | 273 (6) |
| Early-onset pre-eclampsia | 44 (< 1) | 1 (< 1) | 5 (< 1) | 21 (1) | 13 (< 1) | 18 (< 1) | 12 (< 1) | 12 (< 1) | 15 (7) | 1 (< 1) | 2 (< 1) | 10 (< 1) |
| Late-onset pre-eclampsia | 234 (4) | 13 (1) | 49 (3) | 85 (5) | 19 (< 1) | 123 (2) | 144 (5) | 32 (2) | 28 (13) | 45 (6) | 17 (6) | 263 (6) |
Biochemical and ultrasound markers were recorded in very few data sets (see Table 7). To develop models including either biochemical or ultrasound markers in addition to clinical characteristics, only subsets of the 12 data sets could be used. Four data sets included biochemical markers and six data sets included ultrasound markers. Estimated fetal weight centile was excluded as a potential predictor as none of the data sets recorded this variable.
Missingness and multiple imputation
Data sets were included if they recorded either first-trimester or second-trimester measurements for the potential predictors of interest; therefore, many variables were systematically missing (not recorded or recorded for < 10% of individuals) in a data set and some were partially missing (missing for some individuals) in a data set. Table 9 summarises the missingness for variables and outcomes in the development data sets.
| Variable | Missing or not recorded, n (%) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Scope42 (N = 5628) | Allen et al.108 (N = 1045) | Poston et al. 2015149 (N = 1554) | Baschat et al.115 (N = 1704) | Antsaklis et al.110 (N = 3328) | WHO175 (N = 7273) | NICH LR159 (N = 3097) | POUCH131 (N = 3019) | van Kuijk et al. 2014166 (N = 230) | STORK G135 (N = 812) | Vinter et al.173 (N = 304) | POP161 (N = 4212) | |
| Maternal clinical characteristics | ||||||||||||
| Previous PE | 0 (0) | 0 (0) | 8 (1) | 0 (0) | 0 (0) | 1 (0) | 0 (0) | 1 (0) | 2 (1) | 0 (0) | 0 (0) | 0 (0) |
| History of hypertension | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (0) | 0 (0) | 1 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Trimester 1 SBP | 5490 (98) | 5 (0) | 1536 (99) | 0 (0) | 2906 (87) | 2581 (35) | 3020 (98) | 2454 (81) | 127 (55) | 421 (52) | 0 (0) | 279 (7) |
| Trimester 2 SBP | 7 (0) | 1040 (100) | 9 (1) | 1704 (100) | 3321 (100) | 1022 (14) | 87 (3) | 1653 (55) | 183 (80) | 254 (31) | 304 (100) | 4076 (97) |
| Trimester 1 BMI | 5490 (98) | 5 (0) | 1536 (99) | 0 (0) | 480 (14) | 2585 (36) | 3024 (98) | 3019 (100) | 127 (55) | 414 (51) | 0 (0) | 152 (4) |
| Trimester 2 BMI | 7 (0) | 1040 (100) | 0 (0) | 1704 (100) | 2966 (89) | 1078 (15) | 177 (6) | 1 (0) | 183 (80) | 245 (30) | 304 (100) | 57 (1) |
| Trimester 1 DBP | 5490 (98) | 5 (0) | 1536 (99) | 0 (0) | 2913 (88) | 2581 (35) | 3020 (98) | 2454 (81) | 127 (55) | 421 (52) | 0 (0) | 279 (7) |
| Trimester 2 DBP | 7 (0) | 1040 (100) | 9 (1) | 1704 (100) | 3321 (100) | 1023 (14) | 87 (3) | 1653 (55) | 183 (80) | 254 (31) | 304 (100) | 4076 (97) |
| Nulliparous | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Trimester 1 MAP | 5490 (98) | 5 (0) | 1536 (99) | 0 (0) | 2913 (88) | 2581 (35) | 3020 (98) | 2454 (81) | 127 (55) | 421 (52) | 0 (0) | 280 (7) |
| Trimester 2 MAP | 7 (0) | 1040 (100) | 9 (1) | 1704 (100) | 3321 (100) | 1023 (14) | 87 (3) | 1653 (55) | 183 (80) | 254 (31) | 304 (100) | 4076 (97) |
| History of renal disease | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1921 (64) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Multiple pregnancy | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 231 (3) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| History of diabetes | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Age | 0 (0) | 1 (0) | 0 (0) | 0 (0) | 11 (0) | 1 (0) | 99 (3) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Trimester 1 PCR | 5612 (100) | 1045 (100) | 1554 (100) | 1704 (100) | 3328 (100) | 7273 (100) | 3097 (100) | 3019 (100) | 230 (100) | 812 (100) | 304 (100) | 4212 (100) |
| Trimester 2 PCR | 4464 (79) | 1045 (100) | 1549 (100) | 1704 (100) | 3328 (100) | 7273 (100) | 3097 (100) | 3019 (100) | 230 (100) | 812 (100) | 304 (100) | 4212 (100) |
| Family history of PE | 0 (0) | 6 (1) | 61 (4) | 1704 (100) | 3328 (100) | 7273 (100) | 3097 (100) | 3019 (100) | 230 (100) | 812 (100) | 304 (100) | 4212 (100) |
| History of autoimmune disease | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 2 (0) | 0 (0) | 2458 (81) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Previous SGA | 5628 (100) | 0 (0) | 1554 (100) | 1704 (100) | 3328 (100) | 7273 (100) | 3097 (100) | 3019 (100) | 0 (0) | 0 (0) | 304 (100) | 4212 (100) |
| Trimester 1 urine dipstick | 5490 (98) | 1045 (100) | 1554 (100) | 1704 (100) | 3328 (100) | 2584 (36) | 3020 (98) | 3019 (100) | 230 (100) | 812 (100) | 304 (100) | 4212 (100) |
| Trimester 2 urine dipstick | 7 (0) | 1045 (100) | 404 (26) | 1704 (100) | 3328 (100) | 1079 (15) | 87 (3) | 0 (0) | 230 (100) | 812 (100) | 304 (100) | 4212 (100) |
| Ethnicity | 0 (0) | 2 (0) | 0 (0) | 0 (0) | 7 (0) | 6 (0) | 0 (0) | 0 (0) | 230 (100) | 0 (0) | 0 (0) | 0 (0) |
| Smoking | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 6 (0) | 2 (0) | 6 (0) | 6 (0) | 230 (100) | 0 (0) | 0 (0) | 0 (0) |
| Spontaneous conception | 0 (0) | 0 (0) | 4 (0) | 0 (0) | 3328 (100) | 7273 (100) | 3097 (100) | 3019 (100) | 20 (9) | 0 (0) | 304 (100) | 0 (0) |
| Substance misuse | 0 (0) | 0 (0) | 1554 (100) | 0 (0) | 3328 (100) | 7273 (100) | 3097 (100) | 7 (0) | 230 (100) | 812 (100) | 304 (100) | 4212 (100) |
| Biochemical markers | ||||||||||||
| Trimester 1 PlGF | 5628 (100) | 5 (0) | 1554 (100) | 704 (41) | 3328 (100) | 4842 (67) | 3097 (100) | 3019 (100) | 230 (100) | 812 (100) | 304 (100) | 135 (3) |
| Trimester 2 PlGF | 51 (1) | 966 (92) | 445 (29) | 1704 (100) | 3328 (100) | 4026 (55) | 3097 (100) | 1715 (57) | 230 (100) | 812 (100) | 304 (100) | 191 (5) |
| Trimester 1 sFlt-1 | 5628 (100) | 1045 (100) | 1554 (100) | 1704 (100) | 3328 (100) | 4989 (69) | 3097 (100) | 3019 (100) | 230 (100) | 812 (100) | 304 (100) | 134 (3) |
| Trimester 2 sFlt-1 | 48 (1) | 1045 (100) | 1554 (100) | 1704 (100) | 3328 (100) | 4125 (57) | 3097 (100) | 1712 (57) | 230 (100) | 812 (100) | 304 (100) | 191 (5) |
| Trimester 1 PAPP-A | 5628 (100) | 119 (11) | 1554 (100) | 704 (41) | 3328 (100) | 7273 (100) | 3097 (100) | 3019 (100) | 230 (100) | 812 (100) | 304 (100) | 134 (3) |
| Trimester 2 PAPP-A | 48 (1) | 1040 (100) | 1554 (100) | 1704 (100) | 3328 (100) | 7273 (100) | 3097 (100) | 3019 (100) | 230 (100) | 812 (100) | 304 (100) | 4212 (100) |
| Ultrasound markers | ||||||||||||
| Trimester 1 uterine artery PI | 5628 (100) | 5 (0) | 1554 (100) | 266 (16) | 2895 (87) | 7273 (100) | 3097 (100) | 3019 (100) | 230 (100) | 812 (100) | 304 (100) | 4212 (100) |
| Trimester 2 uterine artery PI | 5628 (100) | 1040 (100) | 1554 (100) | 1704 (100) | 3204 (96) | 7273 (100) | 3097 (100) | 3019 (100) | 230 (100) | 812 (100) | 304 (100) | 133 (3) |
| Trimester 1 estimated fetal weight | 5628 (100) | 1045 (100) | 1554 (100) | 1704 (100) | 3328 (100) | 7273 (100) | 3097 (100) | 3019 (100) | 230 (100) | 812 (100) | 304 (100) | 4212 (100) |
| Trimester 2 estimated fetal weight | 5628 (100) | 1045 (100) | 1554 (100) | 1704 (100) | 3328 (100) | 7273 (100) | 3097 (100) | 3019 (100) | 230 (100) | 144 (18) | 304 (100) | 43 (1) |
| Trimester 1 umbilical artery PI | 5628 (100) | 1045 (100) | 1554 (100) | 1704 (100) | 3328 (100) | 7273 (100) | 3097 (100) | 3019 (100) | 230 (100) | 812 (100) | 304 (100) | 4212 (100) |
| Trimester 2 umbilical artery PI | 3102 (55) | 1045 (100) | 1554 (100) | 1704 (100) | 3328 (100) | 7273 (100) | 3097 (100) | 3019 (100) | 230 (100) | 187 (23) | 304 (100) | 70 (2) |
| Outcomes | ||||||||||||
| Any-onset PE | 5 (0) | 0 (0) | 47 (3) | 0 (0) | 985 (30) | 0 (0) | 150 (5) | 1648 (55) | 13 (6) | 0 (0) | 0 (0) | 5 (0) |
| Early-onset PE | 5 (0) | 0 (0) | 47 (3) | 0 (0) | 1007 (30) | 0 (0) | 150 (5) | 1648 (55) | 13 (6) | 0 (0) | 0 (0) | 5 (0) |
| Late-onset PE | 5 (0) | 0 (0) | 47 (3) | 0 (0) | 1007 (30) | 0 (0) | 150 (5) | 1648 (55) | 13 (6) | 0 (0) | 0 (0) | 5 (0) |
When checking convergence following imputation of systematically missing predictors (as described in Chapter 3, Development and validation of pre-eclampsia prediction models), values for first-trimester BMI were deemed poorly imputed for POUCH131 (i.e. convergence was not achieved despite the large burn-in, and extreme values were imputed), so the data set was excluded when developing models using first-trimester clinical characteristics. Three studies108,115,173 were excluded from model development using second-trimester clinical characteristics, owing to poor imputation of blood pressures and BMI.
For model development including biochemical markers, all four data sets were included for the first-trimester models, and POUCH131 was excluded for the second-trimester models. For model development including ultrasound markers, two data sets135,161 were excluded for first-trimester models, and three data sets108,135,149 were excluded for second-trimester models. The relevant imputation checking plots are provided in Appendix 15 along with an explanation of why these studies were excluded. A summary of the overall sample size and number of events contributing to model development for each combination of trimester of measurement (first or second) and predictor set (clinical characteristics, clinical characteristics plus biochemical markers, clinical characteristics plus ultrasound markers) is given in Table 10.
| Model development including | Number of studies | Total sample size | Any-onset pre-eclampsia | Early-onset pre-eclampsia | Late-onset pre-eclampsia |
|---|---|---|---|---|---|
| First-trimester clinical characteristics | 11 | 29,187 | 1187 | 149 | 1039 |
| Second-trimester clinical characteristics | 9 | 29,153 | 1144 | 152 | 993 |
| First-trimester clinical characteristics and biochemical markers | 4 | 20,132 | 795 | 103 | 692 |
| Second-trimester clinical characteristics and biochemical markers | 3 | 17,117 | 692 | 72 | 620 |
| First-trimester clinical characteristics and ultrasound markers | 4 | 11,705 | 443 | 84 | 359 |
| Second-trimester clinical characteristics and ultrasound markers | 3 | 13,168 | 596 | 72 | 524 |
Models including clinical characteristics only
The model development process was performed for data sets imputed with each form of BMI, namely BMI, ln(BMI) and BMI–2. The resulting models and performance statistics for all clinical characteristic models predicting any-onset, early-onset and late-onset pre-eclampsia are presented in Appendix 16. As an example of how the functional form was decided for BMI, let us consider the model including first-trimester clinical characteristics for any-onset pre-eclampsia. The model that included BMI rather than ln(BMI) or BMI–2 had an overall calibration slope closest to 1 (with least heterogeneity across data sets), calibration-in-the large closest to 0 (with least heterogeneity across data sets), and the C-statistic with least heterogeneity across data sets (C-statistic estimates were very similar across the three models). Therefore, the model with BMI was selected. This process was repeated for models developed using clinical characteristics for all three pre-eclampsia outcomes using first-trimester measurements for potential predictors, and then separately for models using second-trimester measurements.
A summary of the predictors retained for each model after variable selection is given in Table 11. For all but one model, BMI was best modelled linearly if it was retained. However, for early-onset pre-eclampsia (for which there are fewer events), first-trimester BMI was modelled non-linearly using (BMI/10)–2. The relationship between BMI and risk (log-odds) of early pre-eclampsia is shown in Figure 12. Autoimmune disease was only retained in the model including second-trimester predictors for early-onset pre-eclampsia. Nulliparity was dropped from the models for early-onset pre-eclampsia and diabetes was dropped from the first-trimester model for late-onset pre-eclampsia and all of the second-trimester models. DBP was not retained in the first-trimester models for any-onset or late-onset pre-eclampsia but was retained in the model for early-onset pre-eclampsia and all second-trimester models.
| Variable | Prediction from first-trimester predictors | Prediction from second-trimester predictors | ||||
|---|---|---|---|---|---|---|
| Any-onset PE | Early-onset PE | Late-onset PE | Any-onset PE | Early-onset PE | Late-onset PE | |
| Age | ✓ | ✓ | ✓ | ✓ | ✓ | |
| SBP | ✓ | ✓ | ✓ | ✓ | ✓ | |
| DBP | ✓ | ✓ | ✓ | ✓ | ||
| BMI | ✓ linear | ✓ (–2) | ✓ linear | ✓ linear | ✓ linear | |
| Nulliparity | ✓ | ✓ | ✓ | ✓ | ||
| Previous PE | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Renal disease | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Hypertension | ✓ | ✓ | ✓ | ✓ | ✓ | |
| Diabetes | ✓ | ✓ | Omitteda | |||
| Autoimmune disease | ✓ | |||||
FIGURE 12.
Relationship between first-trimester BMI and risk of early-onset pre-eclampsia when using (BMI/10)–2 transformation.
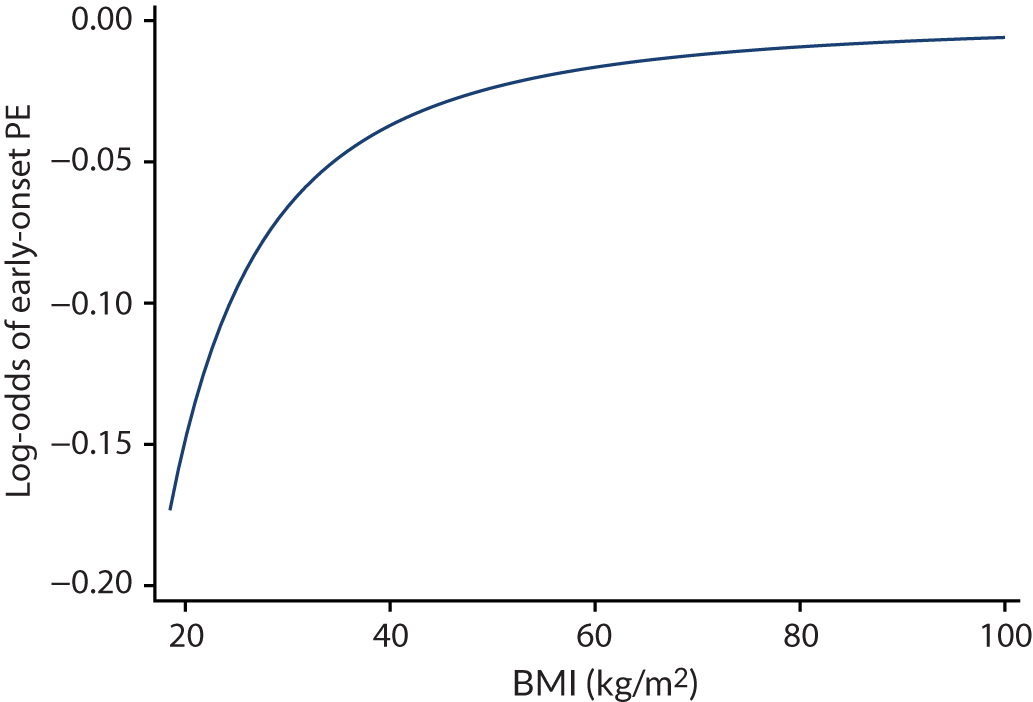
The parameter estimates for models with first-trimester predictors and second-trimester predictors are given in Tables 12 and 13, respectively. These are the developed models before adjustment for optimism due to overfitting. Increasing values of BMI, previous pre-eclampsia, history of hypertension, renal disease and diabetes were all associated with an increased risk of the pre-eclampsia outcomes. Increasing age was associated with a decrease in risk of pre-eclampsia. When retained in the models, increasing values of SBP and DBP were associated with an increase in risk for any-onset and late-onset pre-eclampsia, although second-trimester SBP was negatively associated with risk for early-onset pre-eclampsia.
| Variable | Any-onset PE | Early-onset PE | Late-onset PE | ||||||
|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | Beta | p-value | OR (95% CI) | Beta | p-value | OR (95% CI) | Beta | p-value | |
| Age | 0.984 (0.972 to 0.995) | –0.016 | 0.006 | 0.965 (0.934 to 0.996) | –0.036 | 0.985 (0.973 to 0.997) | –0.015 | 0.018 | |
| SBP | 1.017 (1.010 to 1.025) | 0.017 | < 0.001 | 0.983 (0.959 to 1.006) | –0.018 | 0.089 | 1.016 (1.008 to 1.025) | 0.016 | < 0.001 |
| DBP | 1.078 (1.032 to 1.126) | 0.075 | |||||||
| aBMI or (BMI/10)–2 | 1.041 (1.027 to 1.056) | 0.040 | < 0.001 | 0.553 (0.270 to 1.133) | –0.593 | 0.080 | 1.039 (1.024 to 1.055) | 0.039 | < 0.001 |
| Nulliparous | 2.759 (2.073 to 3.673) | 1.015 | < 0.001 | 3.089 (2.268 to 4.206) | 1.128 | < 0.001 | |||
| Previous PE | 3.952 (2.692 to 5.802) | 1.374 | < 0.001 | 5.710 (3.087 to 10.565) | 1.742 | < 0.001 | 3.368 (2.204 to 5.145) | 1.214 | < 0.001 |
| Renal disease | 5.163 (2.665 to 10.002) | 1.641 | < 0.001 | 9.749 (2.534 to 37.512) | 2.277 | < 0.001 | 3.891 (1.919 to 7.892) | 1.359 | < 0.001 |
| Hypertension | 5.555 (4.384 to 7.039) | 1.715 | < 0.001 | 1.730 (1.005 to 2.976) | 0.548 | 0.028 | 6.265 (4.895 to 8.018) | 1.835 | < 0.001 |
| Diabetes | 1.479 (0.883 to 2.477) | 0.391 | 0.137 | 5.567 (2.449 to 12.653) | 1.717 | 0.003 | |||
| Intercept (average) | –6.868 | –7.924 | –7.052 | ||||||
| Variable | Any-onset PE | Early-onset PE | Late-onset PE | ||||||
|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | Beta | p-value | OR (95% CI) | Beta | p-value | OR (95% CI) | Beta | p-value | |
| Age | 0.983 (0.97 to 0.996) | –0.017 | 0.013 | 0.985 (0.972 to 0.999) | –0.015 | 0.034 | |||
| SBP | 1.011 (1.003 to 1.020) | 0.011 | 0.010 | 1.011 (1.002 to 1.020) | 0.011 | 0.012 | |||
| DBP | 1.021 (1.007 to 1.034) | 0.020 | 0.003 | 1.060 (1.018 to 1.104) | 0.059 | 0.006 | 1.013 (1.001 to 1.026) | 0.013 | 0.035 |
| BMI | 1.065 (1.052 to 1.079) | 0.063 | < 0.001 | 1.072 (1.059 to 1.086) | 0.070 | < 0.001 | |||
| Nulliparous | 2.697 (1.905 to 3.820) | 0.992 | < 0.001 | 3.592 (2.469 to 5.225) | 1.279 | < 0.001 | |||
| Previous PE | 4.271 (2.794 to 6.528) | 1.452 | < 0.001 | 8.497 (4.579 to 15.767) | 2.140 | < 0.001 | 3.500 (2.144 to 5.713) | 1.253 | < 0.001 |
| Renal disease | 2.721 (1.323 to 5.596) | 1.001 | 0.007 | 3.861 (1.074 to 13.878) | 1.351 | 0.039 | 2.257 (1.101 to 4.625) | 0.814 | 0.027 |
| Hypertension | 6.253 (4.859 to 8.047) | 1.833 | < 0.001 | 7.74 (6.020 to 9.953) | 2.046 | < 0.001 | |||
| Autoimmune disease | 2.817 (0.766 to 10.359) | 1.036 | 0.118 | ||||||
| Intercept (average) | –8.260 | –9.641 | –8.475 | ||||||
Following internal validation, the predictive performance of the models is summarised in Table 14. The average (pooled) C-statistic for the models was close to 0.7, with considerable heterogeneity in the C-statistic across individual data sets. C-statistics were slightly higher for second-trimester models than for first-trimester models. The calibration slope was generally around 0.9 for models of any-onset and late-onset pre-eclampsia but was greater than 1 for early-onset pre-eclampsia (1.001 and 1.105). Again, there was large heterogeneity in the calibration slope across data sets for most models. Average calibration-in-the-large was close to zero but, again, there was large heterogeneity across data sets, suggesting that the baseline risk differs across the individual data sets and is not being fully captured by the predictors included in the models.
| Number of | C-statistic | Calibration slope | Calibration-in-the-large | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Data sets | Participants | Events | Estimate (95% CI) | τ2 (for logit C-statistic) | I2 (for logit C-statistic) | Estimate (95% CI) | τ2 | I 2 | Estimate (95% CI) | τ2 | I 2 |
| Any-onset pre-eclampsia | |||||||||||
| First-trimester models | |||||||||||
| 11 | 29,187 | 1187 | 0.677 (0.612 to 0.736) | 0.139 | 83.9 | 0.915 (0.603 to 1.226) | 0.155 | 86.4 | 0.012 (–0.630 to 0.654) | 0.881 | 98.7 |
| Second-trimester models | |||||||||||
| 9 | 29,153 | 1144 | 0.703 (0.648 to 0.753) | 0.076 | 79.3 | 0.969 (0.664 to 1.273) | 0.129 | 88.3 | 0.011 (–0.772 to 0.795) | 1.003 | 99.0 |
| Early-onset pre-eclampsia | |||||||||||
| First-trimester models | |||||||||||
| 11 | 29,187 | 149 | 0.719 (0.590 to 0.820) | 0.260 | 49.1 | 1.001 (0.795 to 1.206) | 0.000 | 0.0 | 0.090 (–0.377 to 0.557) | 0.318 | 77.9 |
| Second-trimester models | |||||||||||
| 9 | 29,153 | 152 | 0.723 (0.601 to 0.819) | 0.223 | 56.4 | 1.105 (0.868 to 1.341) | 0.000 | 0.0 | 0.037 (–0.465 to 0.539) | 0.287 | 76.4 |
| Late-onset pre-eclampsia | |||||||||||
| First-trimester models | |||||||||||
| 11 | 29,187 | 1039 | 0.677 (0.615 to 0.733) | 0.116 | 79.7 | 0.919 (0.621 to 1.217) | 0.131 | 82.0 | 0.016 (–0.705 to 0.736) | 1.105 | 98.8 |
| Second-trimester models | |||||||||||
| 9 | 29,153 | 993 | 0.705 (0.649 to 0.756) | 0.076 | 78.4 | 0.930 (0.656 to 1.204) | 0.096 | 84.2 | 0.014 (–0.897 to 0.925) | 1.358 | 99.1 |
To illustrate the heterogeneity in predictive performance across individual data sets, Appendix 17 provides the forest plots of predictive performance measures for the second-trimester any-onset pre-eclampsia model. It is evident that there is large variability around the average values; for example, the observed calibration slope varies from 0.45 to 1.57 across data sets, and CIs often do not overlap.
Models including clinical characteristics and biochemical markers
Next, we examined whether or not biochemical markers should be included in the prediction models, in addition to the clinical characteristics identified for each outcome in the previous section (see Models including clinical characteristics only). Therefore, the clinical characteristics were forced into the models and only the biochemical markers were eligible for removal in the backwards elimination process. First-trimester BMI was previously modelled as (BMI/10)–2 for early pre-eclampsia; however, this transformation was not selected for any of the other clinical characteristic models and therefore may be a result of overfitting in data for which we have the fewest events. Therefore, for comparability and consistency across models, we used BMI rather than (BMI/10)–2 for early pre-eclampsia in the clinical plus biochemical marker models.
In the same way as for BMI in the clinical characteristics models, we considered non-normality for the biochemical markers by developing prediction models with biochemical markers on their original scale and compared this with models developed using natural logarithm-transformed biochemical marker values. Comparisons of biochemical marker and ln(biochemical marker) models are given in Appendix 18. For the first-trimester model for early pre-eclampsia, the model with better predictive performance came from the data with log-transformed biochemical markers; however, all biochemical markers were dropped from the model. Therefore, in this case, we selected the model that retained a biochemical marker in the model (see Table 16).
Table 15 shows which biochemical markers were included in each model and if they were log-transformed. PAPP-A was not retained in any of the models, whereas sFlt-1 was retained in all first-trimester prediction models and the second-trimester model for early pre-eclampsia. PlGF was retained in all models except the first-trimester model for early pre-eclampsia. Models included the original biochemical marker values apart from the second-trimester model for early pre-eclampsia, which used ln(biochemical marker) values. For all but the second-trimester model for early pre-eclampsia, biochemical markers were negatively associated with the pre-eclampsia outcomes, so risk decreased with increasing biochemical marker values (Tables 16 and 17).
| Variable | Prediction from first-trimester predictors | Prediction from second-trimester predictors | ||||
|---|---|---|---|---|---|---|
| Any-onset PE | Early-onset PE | Late-onset PE | Any-onset PE | Early-onset PE | Late-onset PE | |
| sFlt-1 | ✓ | ✓ | ✓ | ✓ (ln) | ||
| PAPP-A | ||||||
| PlGF | ✓ | ✓ | ✓ | ✓ (ln) | ✓ | |
| Variable | Any-onset PE | Early-onset PE | Late-onset PE | ||||||
|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | Beta | p-value | OR (95% CI) | Beta | p-value | OR (95% CI) | Beta | p-value | |
| Age | 0.977 (0.962 to 0.992) | –0.023 | 0.003 | 0.995 (0.954 to 1.039) | –0.005 | 0.831 | 0.976 (0.961 to 0.991) | –0.025 | 0.002 |
| SBP | 1.016 (1.004 to 1.028) | 0.016 | 0.011 | 1.055 (1.026 to 1.085) | 0.053 | < 0.001 | 1.009 (0.998 to 1.020) | 0.009 | 0.113 |
| DBP | 0.996 (0.960 to 1.033) | –0.004 | 0.836 | ||||||
| BMI | 1.042 (1.009 to 1.077) | 0.041 | 0.015 | 0.993 (0.933 to 1.057) | –0.007 | 0.831 | 1.052 (1.024 to 1.080) | 0.051 | < 0.001 |
| Nulliparous | 3.113 (1.816 to 5.337) | 1.136 | < 0.001 | 4.093 (2.258 to 7.418) | 1.409 | < 0.001 | |||
| Previous PE | 3.857 (2.252 to 6.607) | 1.350 | < 0.001 | 4.427 (2.002 to 9.789) | 1.488 | < 0.001 | 3.311 (1.745 to 6.283) | 1.197 | < 0.001 |
| Renal disease | 2.603 (1.376 to 4.925) | 0.957 | 0.004 | 4.397 (1.494 to 12.942) | 1.481 | 0.007 | 2.135 (1.088 to 4.190) | 0.758 | 0.028 |
| Hypertension | 9.403 (7.113 to 12.430) | 2.241 | < 0.001 | 2.300 (1.034 to 5.117) | 0.833 | 0.041 | 10.739 (8.039 to 14.346) | 2.374 | < 0.001 |
| Diabetes | 1.021 (0.401 to 2.605) | 0.021 | 0.965 | ||||||
| sFlt-1 | 0.9998 (0.9996 to 0.9999) | –2.42 × 10–4 | 0.012 | 0.9995 (0.9989 to 1.0001) | –5.15 × 10–4 | 0.079 | 0.9998 (0.9996 to 1.0000) | –2 × 10–4 | 0.026 |
| PlGF | 0.995 (0.992 to 0.998) | –0.005 | 0.001 | 0.994 (0.991 to 0.997) | –0.006 | 0.001 | |||
| Intercept (average) | –5.893 | –10.206 | –5.787 | ||||||
| Variable | Any-onset PE | Early-onset PE | Late-onset PE | ||||||
|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | Beta | p-value | OR (95% CI) | Beta | p-value | OR (95% CI) | Beta | p-value | |
| Age | 0.981 (0.966 to 0.996) | –0.019 | 0.012 | 0.980 (0.965 to 0.996) | –0.020 | 0.012 | |||
| SBP | 1.019 (1.009 to 1.030) | 0.019 | < 0.001 | 1.018 (1.008 to 1.029) | 0.018 | 0.001 | |||
| DBP | 0.991 (0.979 to 1.004) | –0.009 | 0.167 | 1.068 (1.039 to 1.096) | 0.065 | < 0.001 | 0.986 (0.973 to 0.999) | –0.014 | 0.031 |
| BMI | 1.072 (1.057 to 1.088) | 0.069 | < 0.001 | 1.076 (1.060 to 1.093) | 0.073 | < 0.001 | |||
| Nulliparous | 8.156 (3.534 to 18.820) | 2.099 | < 0.001 | 12.673 (4.777 to 33.622) | 2.539 | < 0.001 | |||
| Previous PE | 6.237 (2.755 to 14.123) | 1.831 | < 0.001 | 10.594 (4.615 to 24.318) | 2.360 | < 0.001 | 5.664 (2.223 to 14.431) | 1.734 | < 0.001 |
| Renal disease | 4.150 (1.838 to 9.368) | 1.423 | 0.001 | 23.285 (2.613 to 207.474) | 3.148 | 0.005 | 3.680 (1.588 to 8.524) | 1.303 | 0.002 |
| Hypertension | 11.034 (8.334 to 14.608) | 2.401 | < 0.001 | 11.380 (8.531 to 15.180) | 2.432 | < 0.001 | |||
| Autoimmune disease | 3.169 (0.397 to 25.315) | 1.154 | 0.277 | ||||||
| sFlt-1 or In(sFlt-1 + 0.5)a | 1.581 (1.100 to 2.271) | 0.458 | 0.013 | ||||||
| PlGF or In(PlGF + 0.5)a | 0.997 (0.996 to 0.998) | –0.003 | < 0.001 | 0.372 (0.277 to 0.500) | –0.988 | < 0.001 | 0.997 (0.996 to 0.998) | –0.003 | < 0.001 |
| Intercept (average) | –7.895 | –9.343 | –8.054 | ||||||
Table 18 shows the average predictive performance for the models, obtained through internal validation using meta-analysis of the data set-specific performance statistics. Models with the clinical characteristics identified in Models including clinical characteristics only were also refitted in the same data so that it was possible to compare the predictive performance of models with and models without the biochemical markers in the same data sets. For all models, the average predictive performance improved with the addition of biochemical markers, and, for most models and performance statistics, heterogeneity across data sets was reduced (lower I2 and τ2 values). The average calibration slope was generally < 1 (between 0.857 and 0.961), except for the early-onset pre-eclampsia models, which had calibration slopes of 1.038 and 1.079 for the first- and second-trimester models, respectively.
| Model | Number of | C-statistic | Calibration slope | Calibration-in-the-large | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Data sets | Participants | Events | Estimate (95% CI) | τ2 (for logit C-statistic) | I2 (for logit C-statistic) | Estimate (95% CI) | τ2 | I 2 | Estimate (95% CI) | τ2 | I 2 | |
| First-trimester predictors for any pre-eclampsia (C) | 4 | 20,132 | 795 | 0.675 (0.488 to 0.819) | 0.212 | 92.1 | 0.870 (0.258 to 1.481) | 0.123 | 89.3 | –0.009 (–0.924 to 0.907) | 0.318 | 97.8 |
| First-trimester predictors for any pre-eclampsia (C + B) | 0.696 (0.516 to 0.832) | 0.205 | 90.7 | 0.901 (0.355 to 1.447) | 0.094 | 86.7 | –0.004 (–0.939 to 0.931) | 0.333 | 97.9 | |||
| First-trimester predictors for early pre-eclampsia (C) | 4 | 20,132 | 103 | 0.749 (0.582 to 0.865) | 0.066 | 28.6 | 1.047 (0.694 to 1.401) | 0 | 0 | 0.025 (–1.175 to 1.226) | 0.507 | 90.2 |
| First-trimester predictors for early pre-eclampsia (C + B) | 0.762 (0.576 to 0.883) | 0.107 | 37.0 | 1.038 (0.666 to 1.410) | 0 | 0 | 0.036 (–1.269 to 1.342) | 0.610 | 91.7 | |||
| First-trimester predictors for late pre-eclampsia (C) | 4 | 20,132 | 692 | 0.665 (0.476 to 0.814) | 0.222 | 93.1 | 0.809 (0.213 to 1.404) | 0.117 | 87.7 | –0.010 (–0.901 to 0.881) | 0.295 | 97.3 |
| First-trimester predictors for late pre-eclampsia (C + B) | 0.688 (0.505 to 0.827) | 0.212 | 91.9 | 0.857 (0.338 to 1.376) | 0.085 | 84.4 | –0.005 (–0.932 to 0.922) | 0.322 | 97.5 | |||
| Second-trimester predictors for any pre-eclampsia (C) | 3 | 17,117 | 692 | 0.715 (0.488 to 0.868) | 0.141 | 93.9 | 0.917 (0.194 to 1.641) | 0.079 | 93.0 | 0.002 (–1.573 to 1.577) | 0.395 | 98.7 |
| Second-trimester predictors for any pre-eclampsia (C + B) | 0.754 (0.526 to 0.894) | 0.151 | 92.4 | 0.961 (0.358 to 1.564) | 0.052 | 88.7 | 0.004 (–1.284 to 1.293) | 0.262 | 98.0 | |||
| Second-trimester predictors for early pre-eclampsia (C) | 3 | 17,117 | 72 | 0.709 (0.392 to 0.902) | 0.111 | 40.1 | 1.045 (0.508 to 1.582) | 0 | 0 | 0.007 (–2.158 to 2.173) | 0.700 | 93.0 |
| Second-trimester predictors for early pre-eclampsia (C + B) | 0.830 (0.629 to 0.934) | 0.041 | 17.4 | 1.079 (0.567 to 1.591) | 0.011 | 26.0 | 0.099 (–0.972 to 1.169) | 0.107 | 66.4 | |||
| Second-trimester predictors for late pre-eclampsia (C) | 3 | 17,117 | 620 | 0.706 (0.437 to 0.881) | 0.194 | 95.0 | 0.884 (0.152 to 1.615) | 0.079 | 91.8 | 0.002 (–1.509 to 1.513) | 0.363 | 98.4 |
| Second-trimester predictors for late pre-eclampsia (C + B) | 0.746 (0.493 to 0.898) | 0.181 | 93.2 | 0.936 (0.355 to 1.517) | 0.047 | 86.5 | 0.004 (–1.295 to 1.303) | 0.266 | 97.8 | |||
Models including clinical characteristics and ultrasound markers
When considered in addition to clinical characteristics, ultrasound markers were not retained in any of the models fitted, whether using the original values or using the logarithm-transformed values. Therefore, the models reverted to the clinical characteristic models, which were reported in more detail and using more data sets in Models including clinical characteristics only.
Shrinkage and final models
Following model development and internal validation, shrinkage was applied to the beta coefficients and the final model equations are given in Table 19, along with the average performance statistics from meta-analysis across data sets for these models, including 95% CIs for the average performance and 95% prediction intervals for the performance of the model in a new but similar data set. Performance of the models in the individual data sets can be found in Appendix 19.
| Model number | Predictors | Model equation | Number of | Average statistic (95% CI) [95% prediction interval] | ||||
|---|---|---|---|---|---|---|---|---|
| Data sets | Participants | Events | C-statistic | Calibration slope | Calibration-in-the-large | |||
| Any-onset pre-eclampsia | ||||||||
| First-trimester models | ||||||||
| 1 | Clinical | Logit(p) = –6.5297 – 0.0151 × age + 0.0156 × SBP + 0.0370 × BMI + 0.9287 × nulliparous + 1.2574 × previous PE + 1.5019 × renal disease + 1.5689 × hypertension + 0.3578 × diabetes | 11 | 29,187 | 1187 | 0.677 (0.612 to 0.736) [0.462 to 0.836] | 1.000 (0.659 to 1.34) [–0.033 to 2.032] | 0.012 (–0.613 to 0.637) [–2.149 to 2.172] |
| 7 | Clinical and biochemical markers | Logit(p) = –5.6061 – 0.0208 × age + 0.0142 × SBP + 0.0373 × BMI + 1.0232 × nulliparous + 1.2163 × previous PE + 0.8620 × renal disease + 2.0191 × hypertension + 0.0191 × diabetes – 0.000218 × sFlt-1 – 0.00478 × PlGF | 4 | 20,132 | 795 | 0.696 (0.516 to 0.832) [0.202 to 0.954] | 1.000 (0.394 to 1.606) [–0.676 to 2.677] | –0.001 (–0.903 to 0.902) [–2.691 to 2.689] |
| Second-trimester models | ||||||||
|
Clinical | Logit(p) = –8.0909 – 0.0165 × age + 0.0108 × SBP + 0.0198 × DBP + 0.0614 × BMI + 0.9616 × nulliparous + 1.4068 × previous PE + 0.9700 × renal disease + 1.7763 × hypertension | 9 | 29,153 | 1144 | 0.703 (0.648 to 0.753) [0.541 to 0.827] | 0.999 (0.685 to 1.314) [0.065 to 1.934] | 0.011 (–0.762 to 0.784) [–2.456 to 2.478] |
| 10 | Clinical and biochemical markers | Logit(p) = –7.6942 – 0.0186 × age + 0.0185 × SBP – 0.0083 × DBP + 0.0668 × BMI + 2.0169 × nulliparous + 1.7592 × previous PE + 1.3675 × renal disease + 2.3073 × hypertension – 0.00262 × PlGF | 3 | 17,117 | 692 | 0.754 (0.526 to 0.894) [0.009 to 0.999] | 1.000 (0.373 to 1.628) [–2.537 to 4.537] | 0.001 (–1.284 to 1.287) [–7.520 to 7.522] |
| Early-onset pre-eclampsia | ||||||||
| First-trimester models | ||||||||
| 2 | Clinical | Logit(p) = –7.9161 – 0.0360 × age – 0.0176 × SBP + 0.0753 × DBP – 0.5937 × (BMI/10)–2 + 1.7440 × previous PE + 2.2795 × renal disease + 0.5486 × hypertension + 1.7185 × diabetes | 11 | 29,187 | 149 | 0.719 (0.590 to 0.820) [0.400 to 0.907] | 1.000 (0.795 to 1.205) [0.792 to 1.208] | 0.079 (–0.388 to 0.545) [–1.283 to 1.440] |
| 8 | Clinical and biochemical markers | Logit(p) = –10.3842 – 0.0048 × age + 0.0555 × SBP – 0.0040 × DBP – 0.0069 × BMI + 1.5442 × previous PE + 1.5372 × renal disease + 0.8644 × hypertension – 0.000535 × sFlt-1 | 4 | 20,132 | 103 | 0.762 (0.576 to 0.883) [0.341 to 0.952] | 1.000 (0.642 to 1.359) [0.516 to 1.485] | 0.009 (–1.319 to 1.338) [–3.858 to 3.877] |
| Second-trimester models | ||||||||
| 5 | Clinical | Logit(p) = –10.1805 + 0.0647 × DBP + 2.3643 × previous PE + 1.4929 × renal disease + 1.1445 × autoimmune disease | 9 | 29,153 | 152 | 0.723 (0.601 to 0.819) [0.418 to 0.905] | 1.000 (0.785 to 1.214) [0.780 to 1.219] | 0.058 (–0.464 to 0.580) [–1.369 to 1.485] |
| 11 | Clinical and biochemical markers | Logit(p) = –9.7075 + 0.0705 × DBP + 2.5467 × previous PE + 3.3965 × renal disease + 1.2447 × autoimmune disease + 0.4942 × ln(sFlt-1 + 0.5) – 1.0661 × ln(PlGF + 0.5) | 3 | 17,117 | 72 | 0.830 (0.629 to 0.934) [0.078 to 0.996] | 1.000 (0.526 to 1.475) [–0.879 to 2.879] | 0.119 (–1.176 to 1.414) [–6.600 to 6.838] |
| Late-onset pre-eclampsia | ||||||||
| First-trimester models | ||||||||
| 3 | Clinical | Logit(p) = –6.7280 – 0.0138 × age + 0.0150 × SBP + 0.0355 × BMI + 1.0363 × nulliparous + 1.1159 × previous PE + 1.2487 × renal disease + 1.6863 × hypertension | 11 | 29,187 | 1039 | 0.677 (0.615 to 0.733) [0.480 to 0.826] | 1.000 (0.675 to 1.324) [0.051 to 1.949] | 0.015 (–0.685 to 0.715) [–2.397 to 2.427] |
| 9 | Clinical and biochemical markers | Logit(p) = –5.4124 – 0.0210 × age + 0.0076 × SBP + 0.0434 × BMI + 1.2077 × nulliparous + 1.0260 × previous PE + 0.6499 × renal disease + 2.0344 × hypertension – 0.000171 × sFlt-1 – 0.0051 × PlGF | 4 | 20,132 | 692 | 0.689 (0.505 to 0.827) [0.190 to 0.954] | 1.000 (0.394 to 1.606) [–0.674 to 2.674] | 0.000 (–0.895 to 0.896) [–2.655 to 2.656] |
| Second-trimester models | ||||||||
| 6 | Clinical | Logit(p) = –8.0911 – 0.0137 × age + 0.0104 × SBP + 0.0124 × DBP + 0.0647 × BMI + 1.1891 × nulliparous + 1.1650 × previous PE + 0.7569 × renal disease + 1.9032 × hypertension | 9 | 29,153 | 993 | 0.705 (0.649 to 0.756) [0.542 to 0.829] | 1.000 (0.706 to 1.294) [0.156 to 1.843] | 0.012 (–0.872 to 0.896) [–2.809 to 2.833] |
| 12 | Clinical and biochemical markers | Logit(p) = –7.7252 – 0.0189 × age + 0.0171 × SBP – 0.0136 × DBP + 0.0687 × BMI + 2.3770 × nulliparous + 1.6232 × previous PE + 1.2194 × renal disease + 2.2762 × hypertension – 0.00245 × PlGF | 3 | 17,117 | 620 | 0.745 (0.493 to 0.898) [0.005 to 0.999] | 1.000 (0.379 to 1.621) [–2.464 to 4.464] | 0.001 (–1.307 to 1.309) [–7.651 to 7.654] |
After shrinkage and recalibration of the intercept, each model is, on average, perfectly calibrated across data sets. However, as was observed for existing models in Chapter 5, large heterogeneity remains in all of the performance statistics across data sets. The prediction intervals for potential performance in new settings are generally very wide. For example, the prediction interval for the calibration slope of model 4 ranges from 0.07 to 1.93. Therefore, although IPPIC models may predict well on average across populations, they may not be as accurate in particular populations. Figure 13 presents the calibration plots for model 1 (first-trimester clinical characteristics for the prediction of any pre-eclampsia) by data set. The model is fairly well calibrated for Baschat et al. ;115 however, the predictions are too high for pregnant women in the WHO cohort,175 but not high enough for those at high risk in the POP cohort. 161 Calibration plots for the other models (excluding for early pre-eclampsia, which had too few events in individual data sets) are given in Appendix 20. Heterogeneity in calibration performance could be reduced if, when applying the models in practice, model parameters (e.g. intercept) could be recalibrated to each population and setting. This would require local data for recalibration and model updating.
FIGURE 13.
Calibration plots for the final (shrunken) model predicting any-onset pre-eclampsia using first-trimester clinical characteristics, in data sets (with > 100 events) used in the development and validation of the model. (a) SCOPE;42 (b) Baschat;115 (c) WHO;175 (d) NICH LR;159 and (e) POP. 161
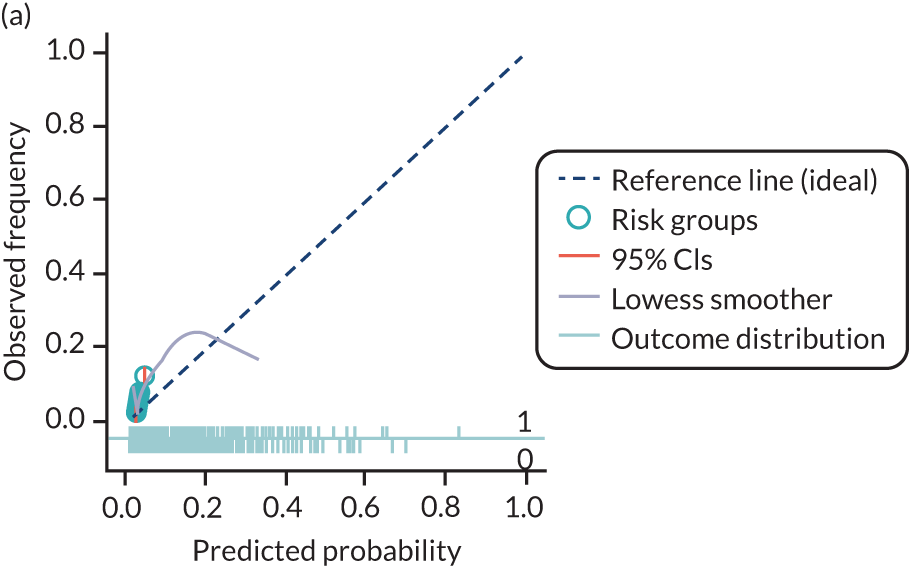
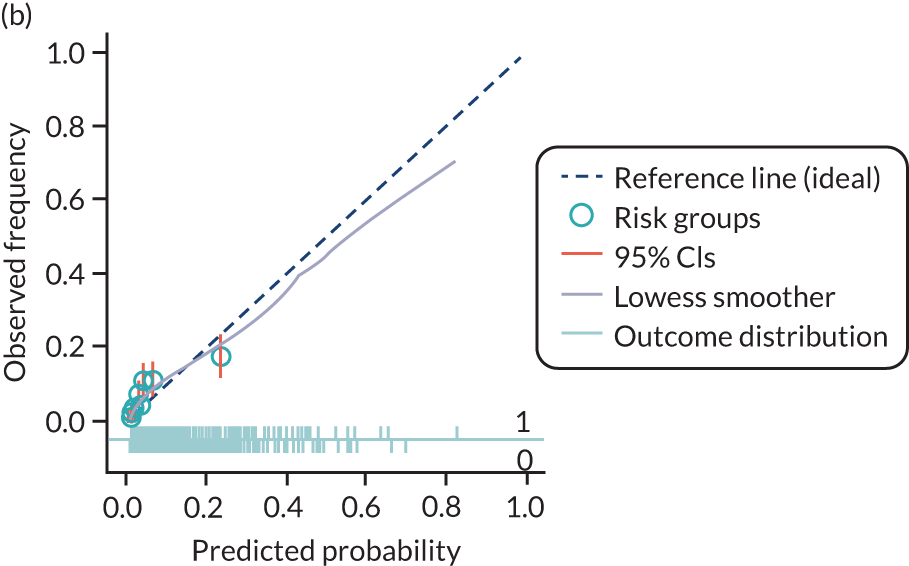
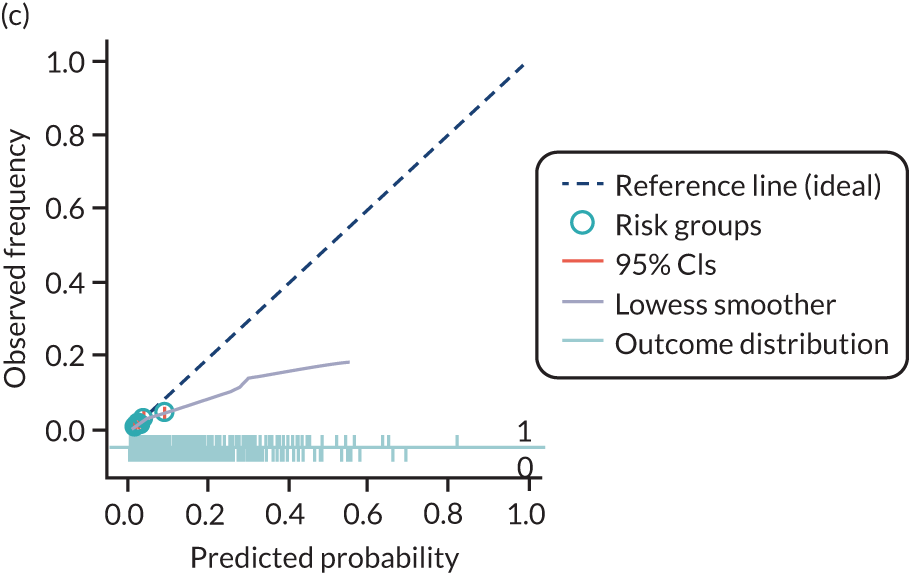
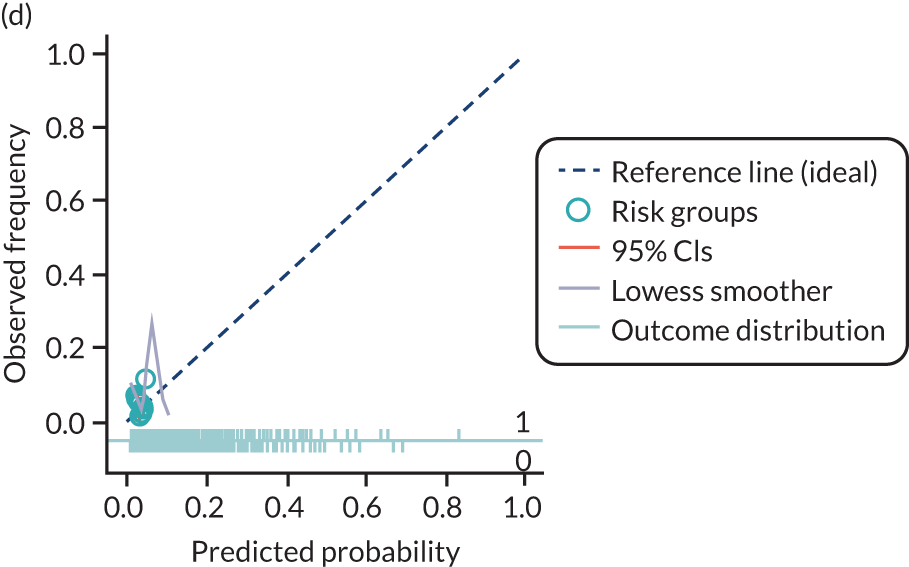
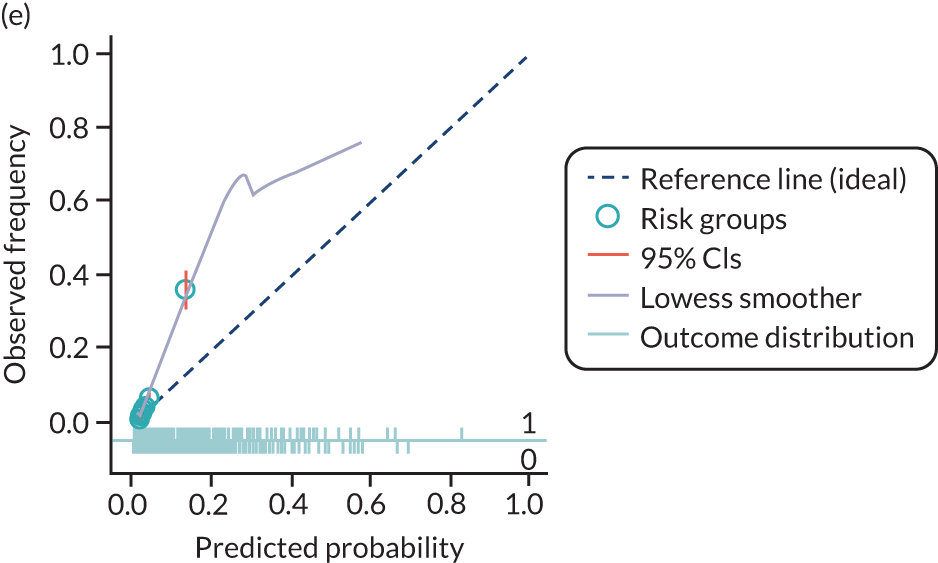
Decision curve analysis
Figures 14–17 are the decision curves in each data set for models predicting any pre-eclampsia. The decision curves show the net benefit or harm across different probability thresholds of the model and compared with the ‘treat-all’ and ‘treat-none’ strategies. Net benefit values are often best multiplied by 1000 to reveal the extra number of women who would be correctly treated per 1000 women who used the model, with none treated incorrectly.
FIGURE 14.
Decision curves for the final (shrunken) model predicting any pre-eclampsia using first-trimester clinical characteristics, in data sets used in the development and validation of the model. (a) SCOPE;42 (b) Allen et al. ;108 (c) Poston et al. 2015;149 (d) Baschat et al. ;115 (e) Antsaklis et al. ;110 (f) WHO;175 (g) NICH LR;159 (h) van Kuijk et al. 2014;166 (i) STORK G;135 (j) Vinter et al. ;173 and (k) POP. 161 Net benefit values are often best multiplied by 1000 to reveal the extra number of women who would be correctly treated per 1000 women who used the model, with none treated incorrectly.
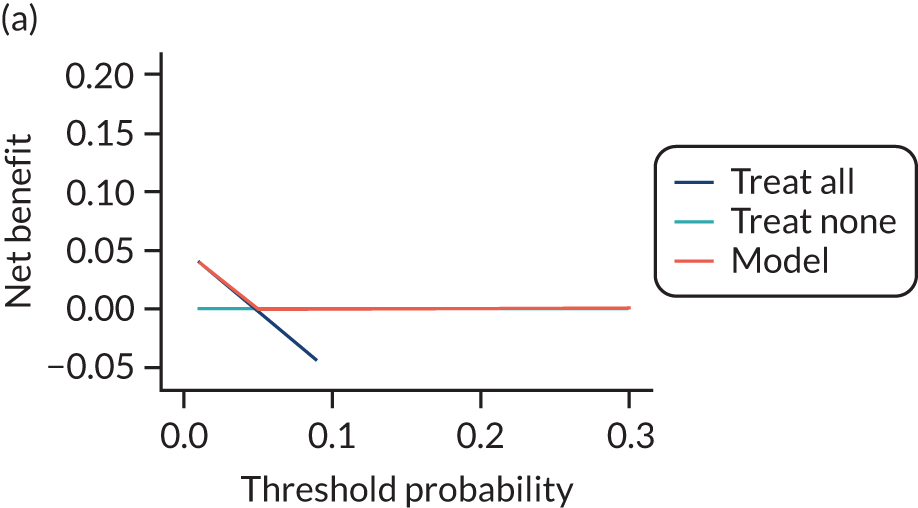
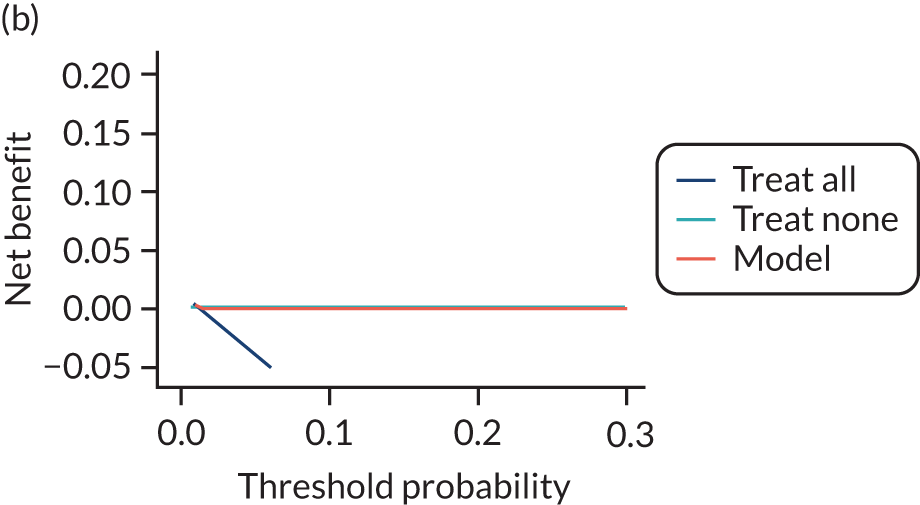
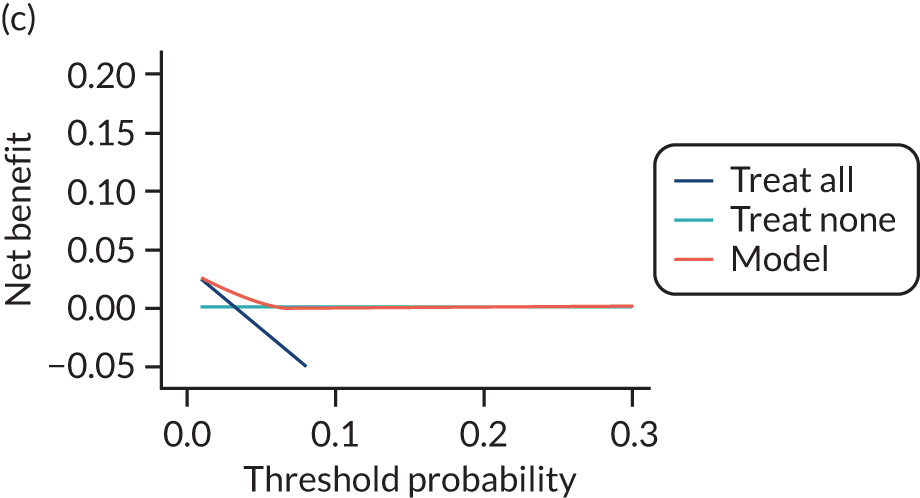
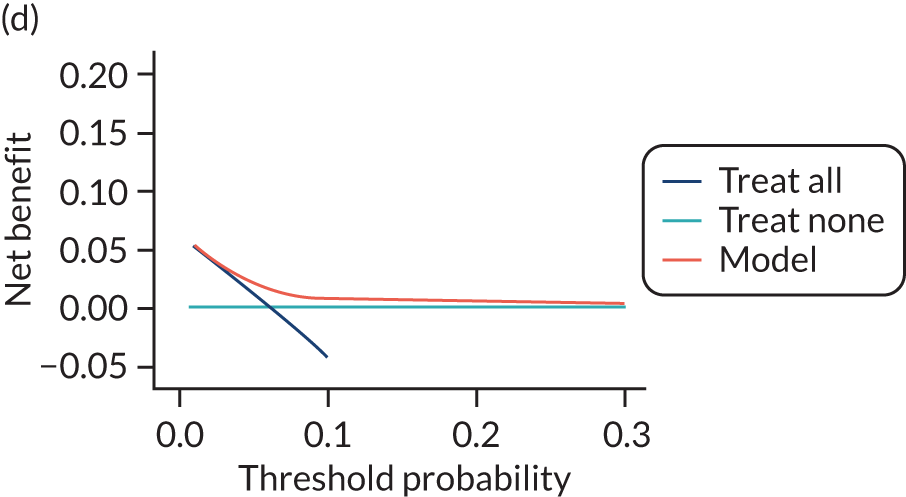
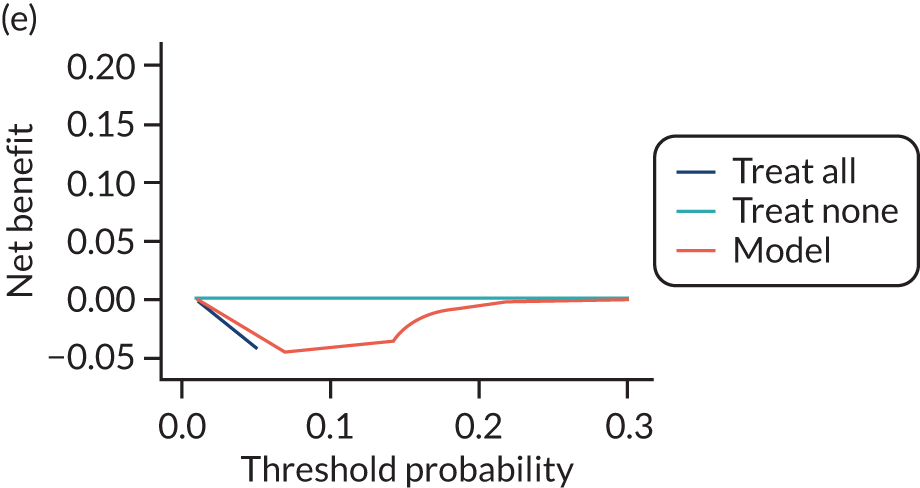
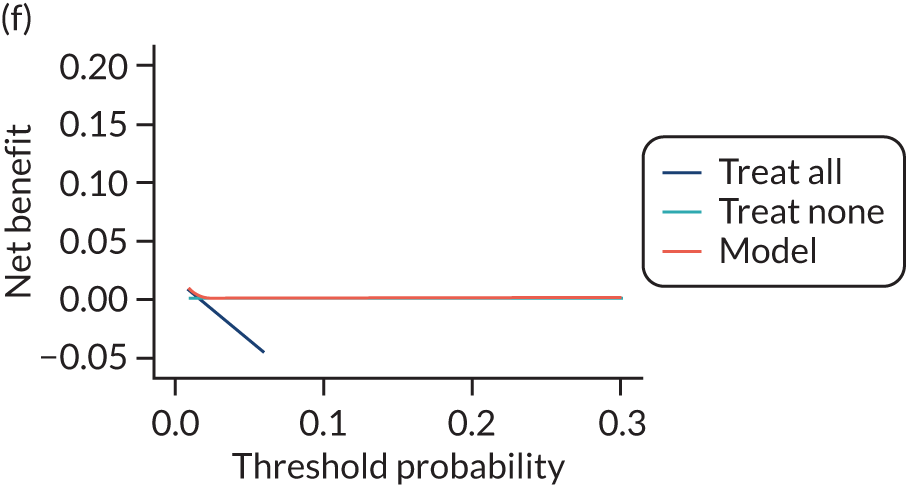
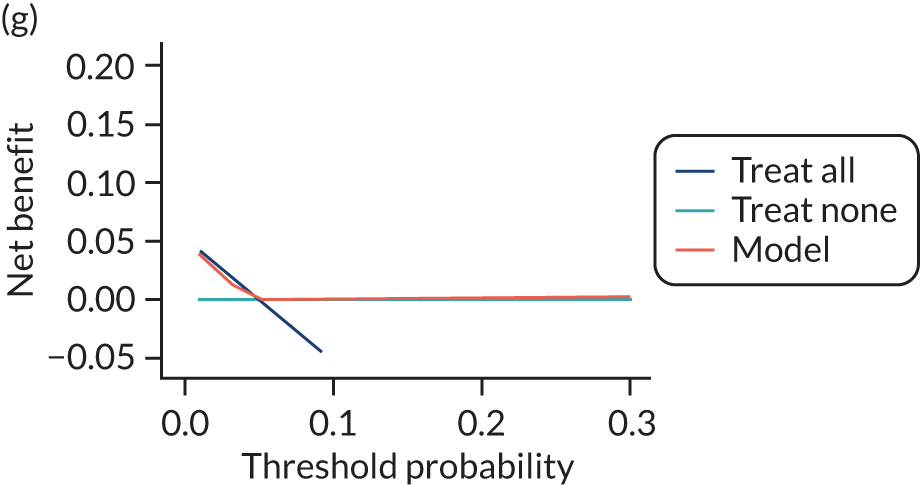
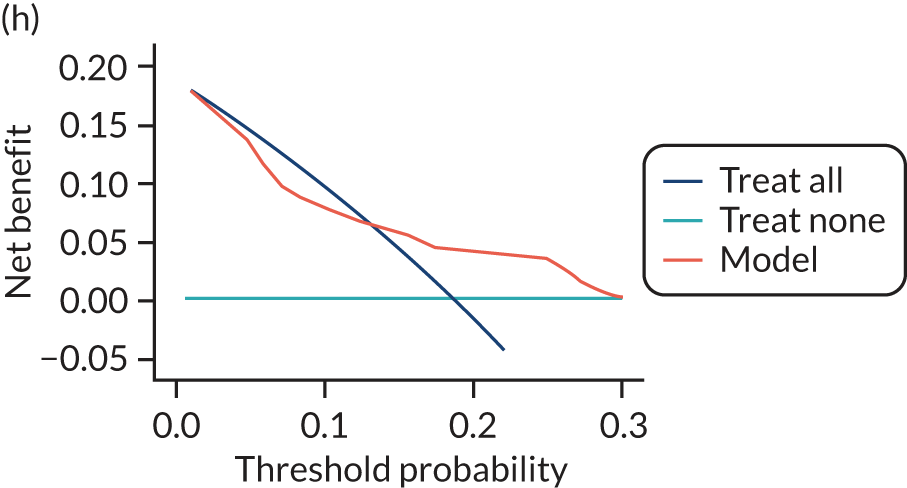
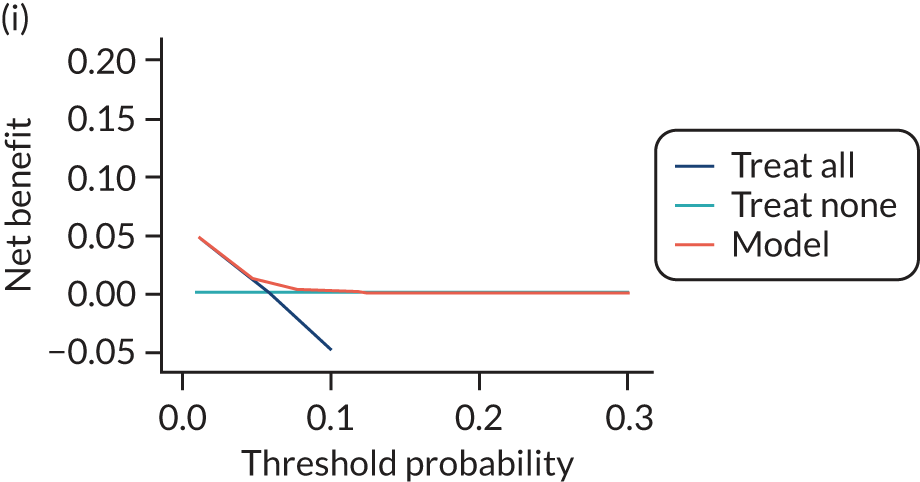
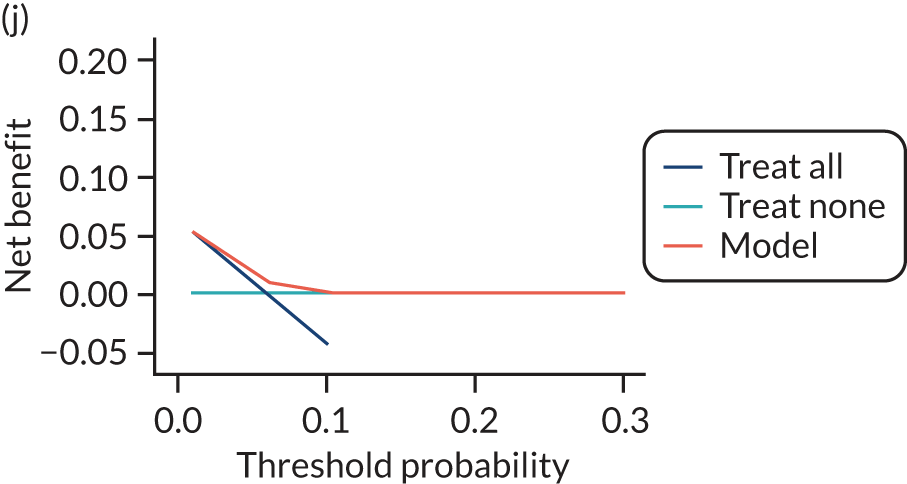
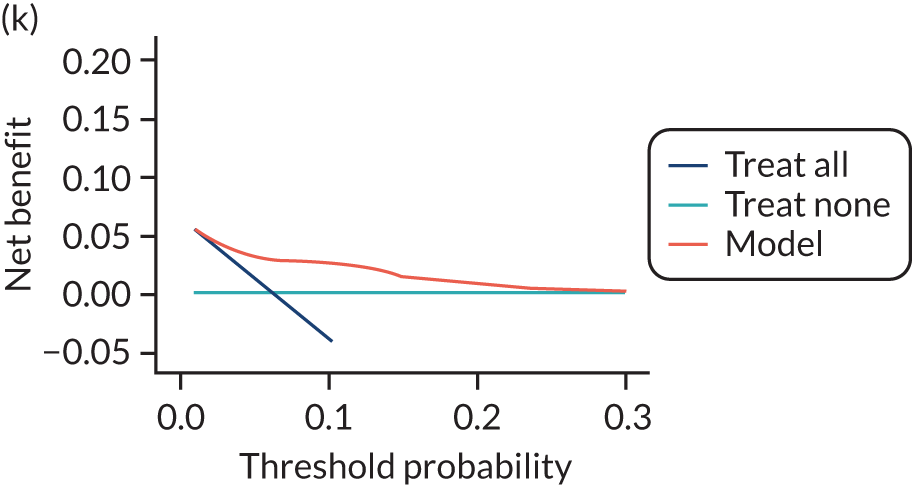
FIGURE 15.
Decision curves for the final (shrunken) model predicting any pre-eclampsia using second-trimester clinical characteristics, in data sets used in the development and validation of the model. (a) SCOPE;42 (b) Poston et al. 2015;149 (c) Antsaklis et al. ;110 (d) WHO;175 (e) NICH LR;159 (f) POUCH;131 (g) van Kuijk et al. 2014;166 (h) STORK G;135 and (i) POP. 161 Net benefit values are often best multiplied by 1000 to reveal the extra number of women who would be correctly treated per 1000 women who used the model, with none treated incorrectly.
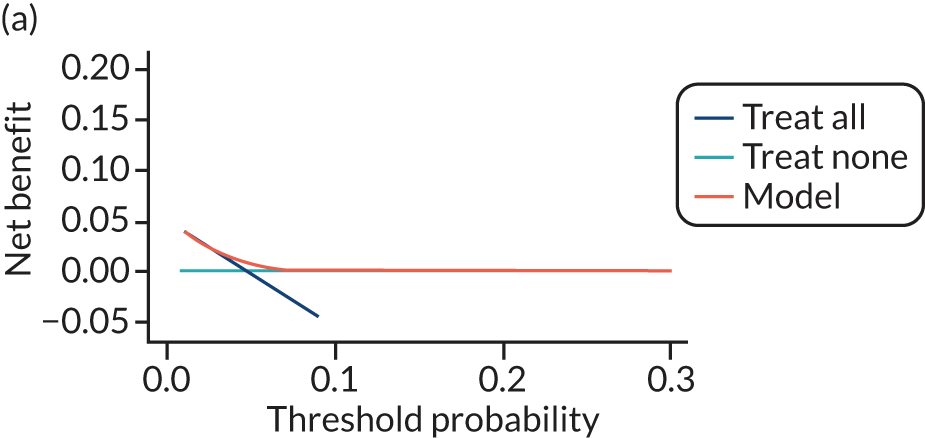
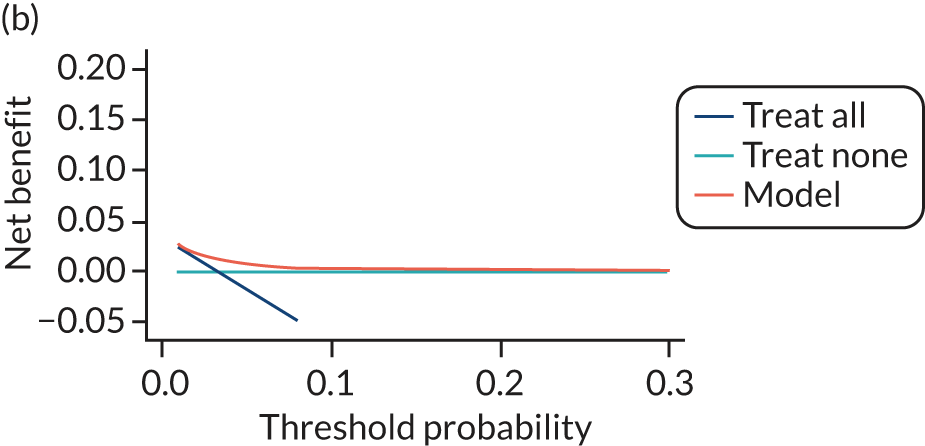
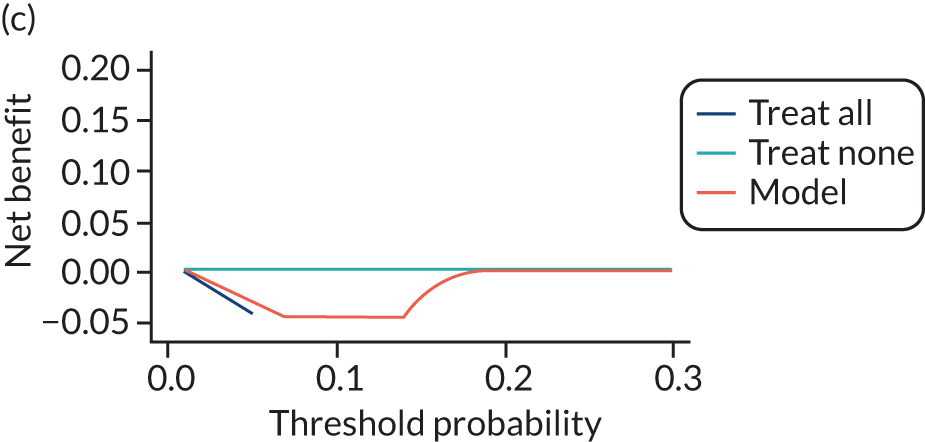
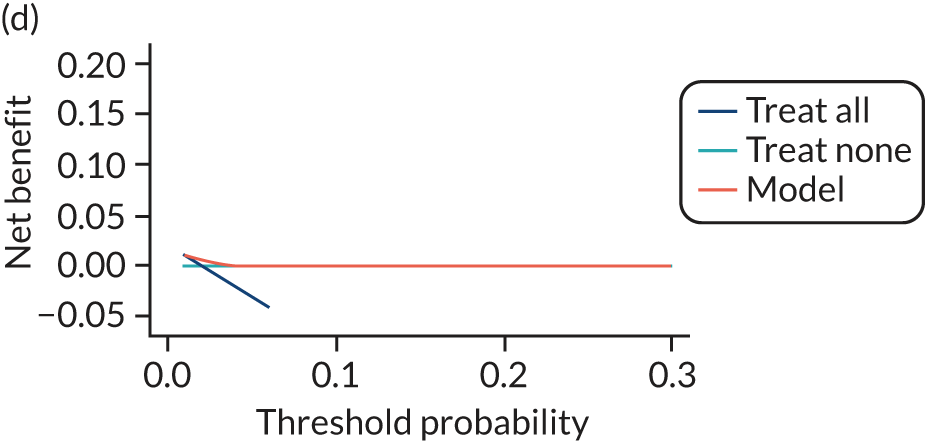
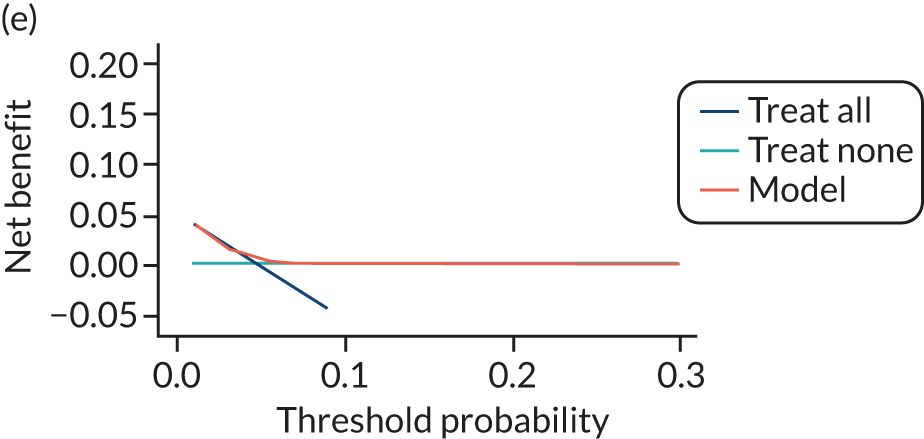
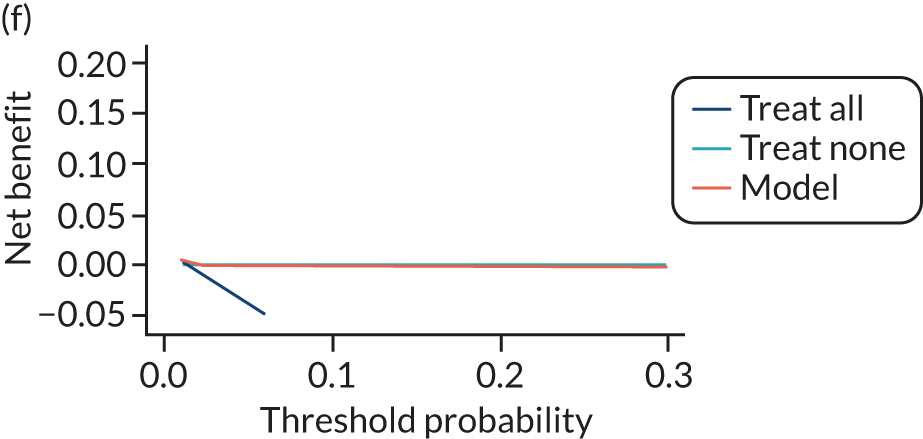
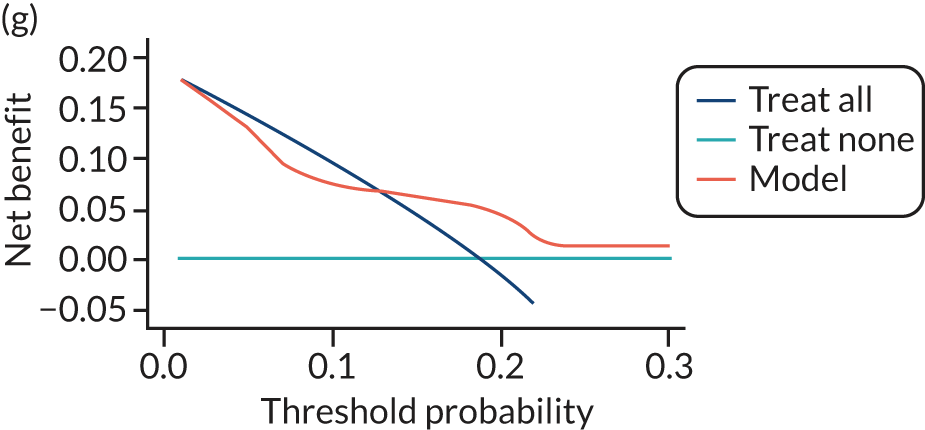
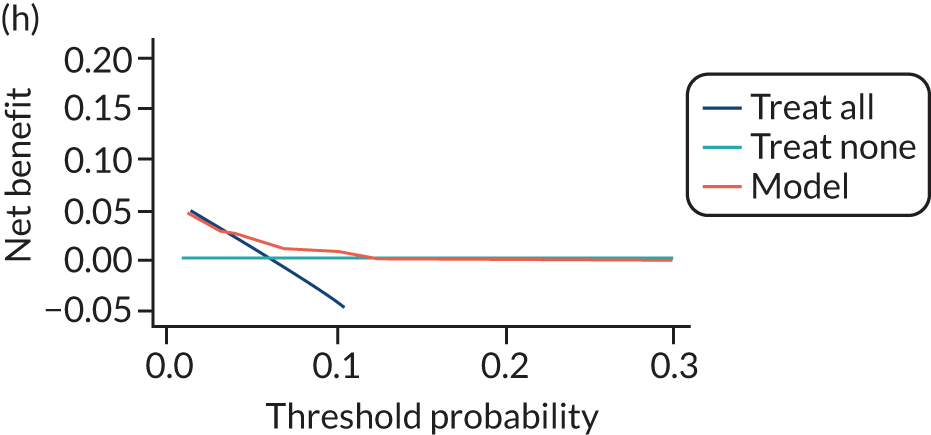
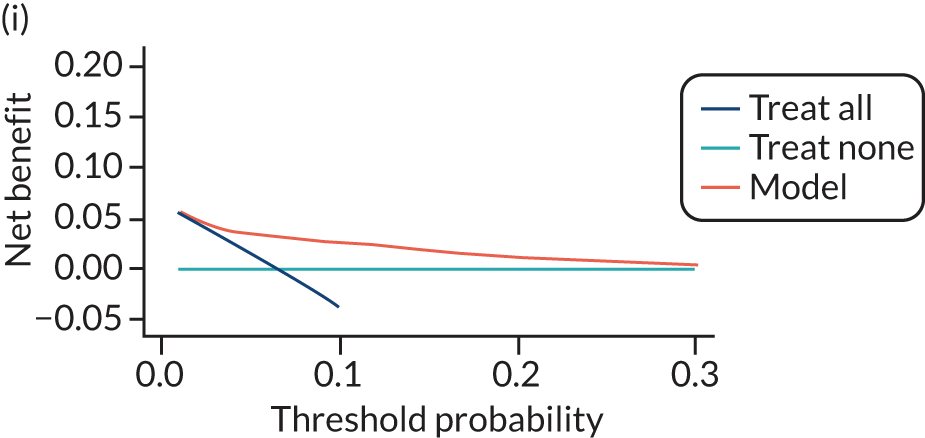
FIGURE 16.
Decision curves for the final (shrunken) model predicting any pre-eclampsia using first-trimester clinical characteristics and biochemical markers, in data sets used in the development and validation of the model. (a) SCOPE;42 (b) WHO;175 (c) POUCH;131 and (d) POP. 161 Net benefit values are often best multiplied by 1000 to reveal the extra number of women who would be correctly treated per 1000 women who used the model, with none treated incorrectly.
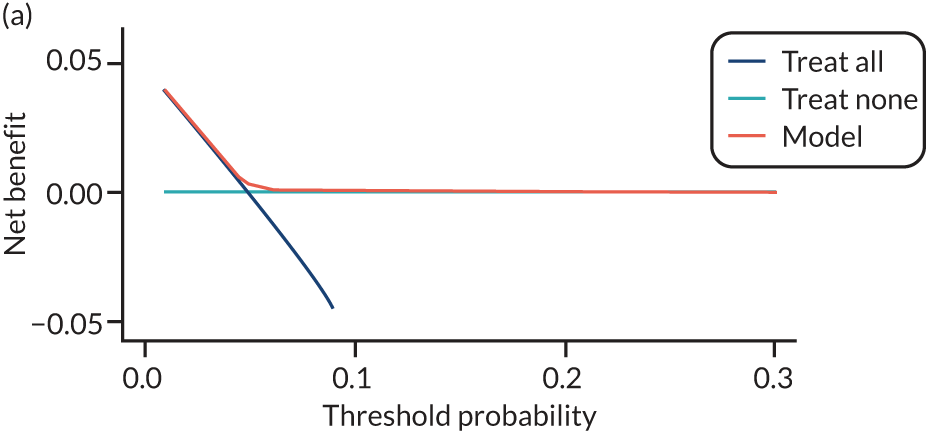
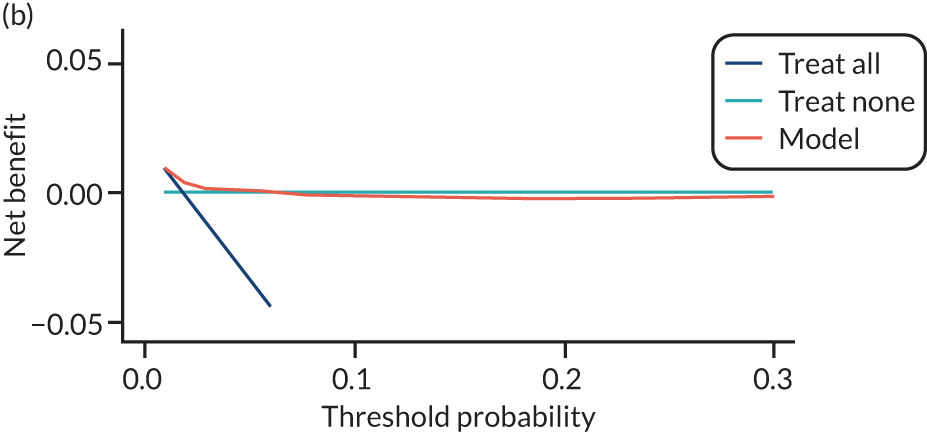
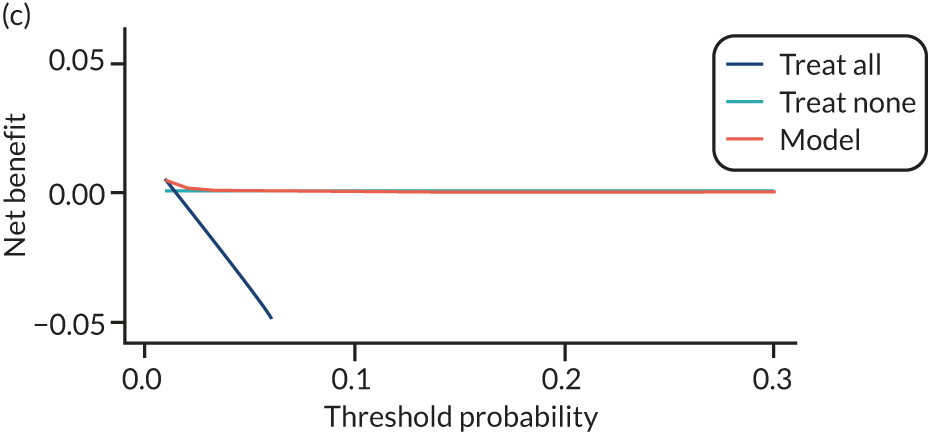
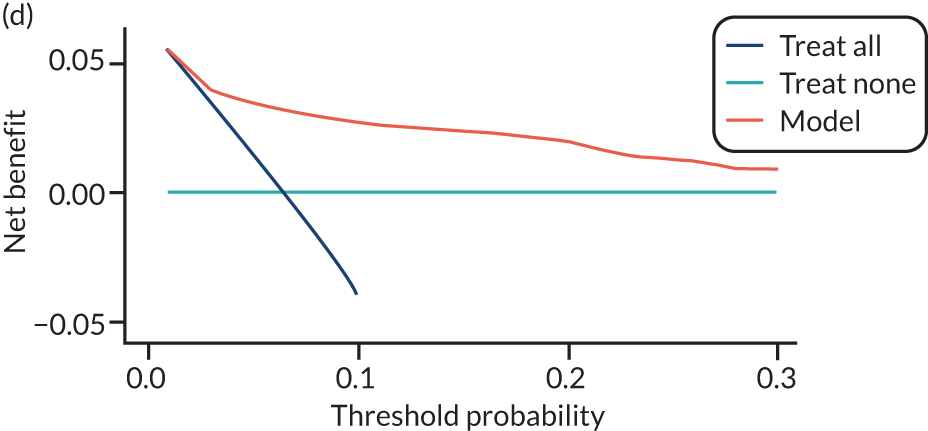
FIGURE 17.
Decision curves for the final (shrunken) model predicting any pre-eclampsia using second-trimester clinical characteristics and biochemical markers, in data sets used in the development and validation of the model. (a) SCOPE;42 (b) WHO;175 and (c) POP. 161 Net benefit values are often best multiplied by 1000 to reveal the extra number of women who would be correctly treated per 1000 women who used the model, with none treated incorrectly.
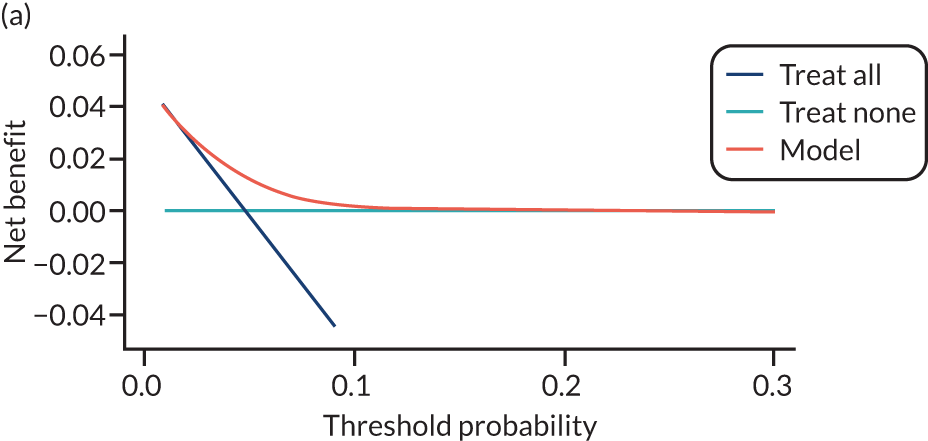
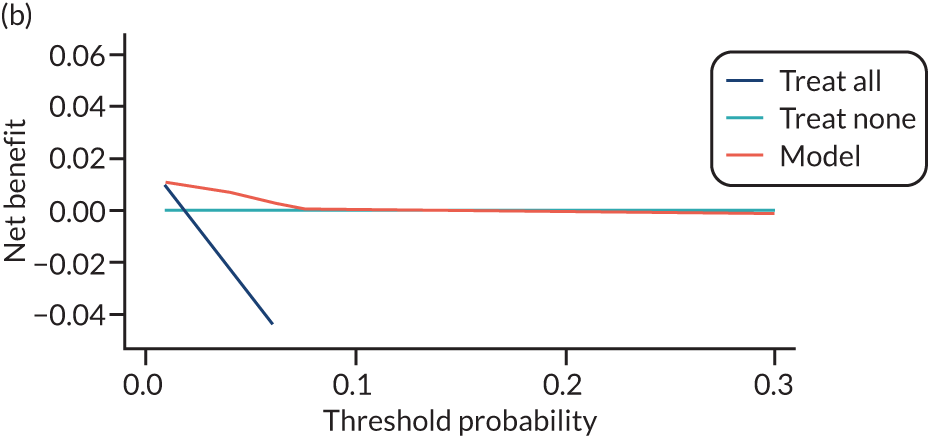
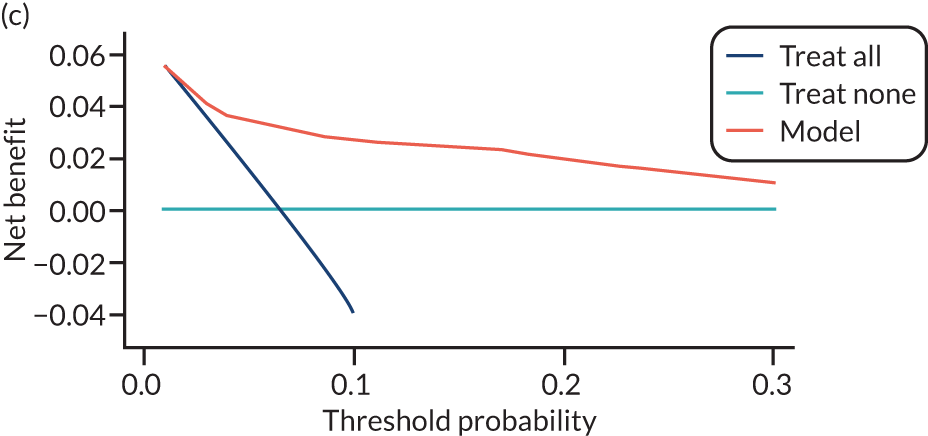
Using first-trimester clinical characteristics (model 1; see Figure 14) shows some net benefit at thresholds around 0.05 in the Poston et al. 2015,149 Baschat et al. ,115 STORK G,135 Vinter et al. 173 and POP161 cohorts. The model shows little benefit or harm in SCOPE,42 Allen et al. ,108 Antsaklis et al. ,110 WHO175 and NICH LR. 159 Using second-trimester clinical characteristics (model 4) shows some improvement in net benefit in SCOPE161 but little improvement in the other data sets (see Figure 15). In the data sets used for developing and validating models with first-trimester clinical characteristics and biochemical markers (model 7), there may be greater net benefit in POP161 but little improvement in the SCOPE42 or WHO175 cohorts (see Figure 16) compared with using clinical characteristics alone. Using second-trimester clinical characteristics and biochemical markers (model 10) shows a slight improvement in the SCOPE42 and POP161 cohorts (compared with first-trimester predictors), and the addition of second-trimester biochemical markers has slightly greater net benefit than in models including only second-trimester clinical characteristics (see Figure 17).
Decision curves for models predicting early-onset pre-eclampsia show no net benefit in most of the data sets, and decision curves for models predicting late-onset pre-eclampsia are similar to those for any-onset pre-eclampsia (see Appendix 21, Figures 38–45).
Summary
We used IPD from the IPPIC data sets to develop and validate new prediction models for early-onset, late-onset and any-onset pre-eclampsia using clinical characteristics alone or with the addition of biochemical markers. The IPPIC data sets used for model development and validation are heterogeneous, with different case mix in different data sets (e.g. owing to different inclusion and exclusion criteria). When the models were validated using an average intercept across data sets, the model performed better in some IPPIC data sets than in others even if the average performance was good. The same was observed in terms of net benefit in the individual data sets. In some data sets, there was potential for net benefit at certain thresholds, whereas there was very little or no net benefit when the models were applied in other data sets.
In summary, these prediction models have the potential to be useful in predicting pre-eclampsia in some populations; however, additional predictors may be needed, or the models may need to be tailored to improve the predictive performance across different settings and populations.
Chapter 7 Predictive performance of individual risk factors for pre-eclampsia
An unadjusted two-stage IPD meta-analysis of the prognostic effect of each candidate predictor prioritised in Chapter 4, Prioritisation of predictors of pre-eclampsia, was performed for each outcome of early-onset, late-onset and any-onset pre-eclampsia, using complete cases of singleton pregnancies in the IPPIC international data set. All analyses were carried out on raw values for ultrasound and biochemical markers.
Any-onset pre-eclampsia
We investigated the prognostic value of individual clinical, biochemical and ultrasound markers for predicting pre-eclampsia using a two-stage IPD meta-analysis. The analysis could not be performed for the candidate factors PAPP-A, PlGF, sFlt-1, PCR and umbilical artery pulsatility index because the first-stage logistic regression models could not be fitted owing to complete or quasi-complete separation. 239 This happens when the outcome separates the predictor variable completely or partially, stopping the model from fitting. The results of the two-stage IPD meta-analysis for any-onset pre-eclampsia and each predictor variable are shown in Table 20.
| Predictor | Number of studies | OR | 95% CI | 95% prediction interval | I2 (%) |
|---|---|---|---|---|---|
| Maternal clinical characteristics | |||||
| Age | 71 | 1.06 | 0.97 to 1.17 | 0.58 to 1.94 | 95.96 |
| Multiple current pregnancy | 23 | 2.56 | 1.86 to 3.51 | 0.67 to 9.75 | 98.62 |
| Previous pre-eclampsia | 35 | 3.40 | 2.55 to 4.53 | 0.87 to 13.26 | 90.62 |
| Parity | 48 | 0.88 | 0.79 to 0.99 | 0.47 to 1.67 | 96.60 |
| History of hypertension | 49 | 4.76 | 3.56 to 6.35 | 0.69 to 32.93 | 98.39 |
| History of renal disease | 21 | 3.28 | 2.15 to 5.01 | 0.70 to 15.44 | 93.16 |
| History of diabetes | 34 | 2.89 | 2.15 to 3.88 | 0.64 to 12.95 | 95.62 |
| History of autoimmune disease | 16 | 1.94 | 1.19 to 3.17 | 0.49 to 7.69 | 75.43 |
| Family history of pre-eclampsia | 15 | 1.40 | 1.00 to 1.95 | 0.55 to 3.58 | 53.87 |
| Previous SGA | 11 | 1.22 | 0.82 to 1.82 | 0.53 to 2.83 | 48.18 |
| Smoking during pregnancy | 54 | 0.84 | 0.76 to 0.93 | 0.50 to 1.42 | 86.46 |
| Spontaneous mode of conception | 22 | 0.73 | 0.64 to 0.84 | 0.49 to 1.09 | 58.67 |
| Cocaine, heroin or methamphetamine use | 8 | 1.13 | 0.70 to 1.81 | 0.47 to 2.68 | 54.03 |
| BMI | 43 | 1.07 | 1.06 to 1.08 | 1.01 to 1.13 | 96.75 |
| BMI trimester 1 | 48 | 1.08 | 1.06 to 1.09 | 1.00 to 1.16 | 93.33 |
| BMI trimester 2 | 21 | 1.07 | 1.05 to 1.09 | 1.00 to 1.15 | 86.36 |
| BMI trimester 3 | 11 | 1.09 | 1.06 to 1.14 | 0.98 to 1.22 | 87.89 |
| SBP | 19 | 1.60 | 1.42 to 1.80 | 0.99 to 2.58 | 89.98 |
| SBP trimester 1 | 39 | 1.46 | 1.39 to 1.53 | 1.15 to 1.85 | 76.57 |
| SBP trimester 2 | 28 | 1.55 | 1.46 to 1.64 | 1.24 to 1.94 | 73.11 |
| SBP trimester 3 | 19 | 2.04 | 1.76 to 2.36 | 1.13 to 3.66 | 92.86 |
| DBP | 19 | 1.78 | 1.55 to 2.05 | 0.99 to 3.20 | 88.45 |
| DBP trimester 1 | 39 | 1.70 | 1.57 to 1.85 | 1.19 to 2.45 | 79.61 |
| DBP trimester 2 | 28 | 1.80 | 1.62 to 1.99 | 1.18 to 2.75 | 84.16 |
| DBP trimester 3 | 18 | 2.59 | 2.04 to 3.29 | 1.06 to 6.30 | 94.44 |
| MAP | 25 | 2.00 | 1.75 to 2.30 | 1.08 to 3.71 | 89.06 |
| MAP trimester 1 | 41 | 1.79 | 1.65 to 1.93 | 1.24 to 2.58 | 80.57 |
| MAP trimester 2 | 29 | 1.92 | 1.74 to 2.11 | 1.27 to 2.89 | 83.52 |
| MAP trimester 3 | 19 | 2.71 | 2.19 to 3.34 | 1.17 to 6.24 | 93.89 |
| PCR | 3 | 1.11 | 0.72 to 1.73 | 0.48 to 2.60 | 99.92 |
| PCR trimester 1 | 1 | 0.75 | 0.34 to 1.67 | N/A | N/A |
| PCR trimester 2 | 1 | 0.98 | 0.93 to 1.03 | N/A | N/A |
| Urine dipstick | 7 | 2.72 | 0.83 to 8.87 | 0.10 to 75.23 | 98.34 |
| Urine dipstick trimester 1 | 6 | 2.27 | 1.52 to 3.38 | 1.20 to 4.29 | 27.67 |
| Urine dipstick trimester 2 | 10 | 1.82 | 1.30 to 2.54 | 0.73 to 4.52 | 74.77 |
| Urine dipstick trimester 3 | 6 | 3.46 | 1.83 to 6.55 | 0.74 to 16.18 | 92.92 |
| Ultrasound markers | |||||
| Uterine artery PI | 11 | 3.11 | 1.86 to 5.18 | 0.71 to 13.66 | 74.32 |
| Uterine artery PI trimester 1 | 12 | 1.67 | 1.18 to 2.38 | 0.64 to 4.37 | 70.27 |
| Uterine artery PI trimester 2 | 13 | 4.18 | 3.25 to 5.38 | 2.27 to 7.70 | 47.16 |
| Uterine artery PI trimester 3 | 3 | 3.99 | 0.02 to 688.38 | 0.00 to 37,752.95 | 75.04 |
| Umbilical artery PI | 5 | 1.08 | 0.44 to 2.65 | 0.44 to 2.65 | 0.00 |
| Umbilical artery PI trimester 2 | 8 | 1.94 | 0.89 to 4.23 | 0.52 to 7.26 | 33.52 |
| Umbilical artery PI trimester 3 | 5 | 1.60 | 0.48 to 5.38 | 0.21 to 12.31 | 37.99 |
| Biochemical markers | |||||
| PIGF trimester 1 | 17 | 0.22 | 0.09 to 0.50 | 0.01 to 4.34 | 85.44 |
| PIGF trimester 2 | 16 | 0.66 | 0.53 to 0.83 | 0.34 to 1.29 | 87.27 |
| PIGF trimester 3 | 12 | 0.59 | 0.45 to 0.77 | 0.25 to 1.36 | 96.78 |
| sFlt-1 trimester 1 | 12 | 0.98 | 0.97 to 1.00 | 0.96 to 1.01 | 51.24 |
| sFlt-1 trimester 2 | 13 | 1.02 | 0.99 to 1.04 | 0.96 to 1.08 | 89.91 |
| sFlt-1 trimester 3 | 9 | 1.03 | 1.02 to 1.04 | 1.00 to 1.06 | 85.70 |
| PAPP-A trimester 1 | 11 | 0.83 | 0.54 to 1.29 | 0.21 to 3.24 | 99.99 |
| PAPP-A trimester 2 | 4 | 1.00 | 0.99 to 1.02 | 0.98 to 1.02 | 65.03 |
The study estimates varied considerably in terms of heterogeneity, with most predictors showing heterogeneity of ≥ 70%. Variation between the 95% CI for the mean effect and the 95% prediction interval for the distribution of true effects was great for many predictors, which was expected given the varying level of heterogeneity.
The strongest association with the outcome was observed for history of hypertension (OR 4.76, 95% CI 3.56 to 6.35; I2 = 98.39%), with a fivefold increase in the odds of pre-eclampsia in women with a history of hypertension compared with women without a history of hypertension. Most predictors had evidence of an association at the 5% level. However, maternal age, family history of pre-eclampsia, PCR (for unspecified trimester estimates and trimester 1 and 2 estimates), urine dipstick (unspecified trimester estimates), previous small for gestational age fetus, cocaine, heroin or methamphetamine use, uterine artery pulsatility index (trimester 3), umbilical artery pulsatility index (for unspecified trimester estimates and trimester 2 and 3 estimates), sFlt-1 (all trimesters estimates) and PAPP-A (all trimester estimates) were not statistically significant at the 5% level (see Table 20).
Most statistically significant predictors had evidence of an increase in odds of pre-eclampsia over the course of pregnancy with increasing values, except multiparity (OR 0.88, 95% CI 0.79 to 0.99; I2 = 96.6%), smoking during pregnancy (OR 0.84, 95% CI 0.76 to 0.93; I2 = 86.46%), spontaneous mode of conception (OR 0.73, 95% CI 0.64 to 0.84; I2 = 58.67%) and increasing levels of PIGF measured in the first (OR 0.22, 95% CI 0.09 to 0.50; I2 = 85.44), second (OR 0.66, 95% CI 0.53 to 0.83; I2 = 87.27%) and third trimester (OR 0.59, 95% CI 0.45 to 0.77; I2 = 96.78%), which showed a decrease in the odds of pre-eclampsia, thereby providing a possible protective effect.
Early-onset pre-eclampsia
Table 21 shows the results of the two-stage meta-analysis for early-onset (delivery at < 34 weeks’ gestation) pre-eclampsia and each predictor variable. Third-trimester predictors were not explored because they do not predate the outcome. The two-stage IPD meta-analysis could not be performed for the following predictors: previous pre-eclampsia, history of hypertension, history of renal disease, history of diabetes, history of autoimmune disease, family history of pre-eclampsia, PCR measured in the first trimester, previous small for gestational age fetus, smoking during pregnancy, spontaneous mode of conception, cocaine, heroin or methamphetamine use, umbilical artery pulsatility index measured in the first trimester, PlGF, sFlt-1 and PAPP-A. 239
| Predictor | Number of studies | OR | 95% CI | 95% prediction interval | I2 (%) |
|---|---|---|---|---|---|
| Maternal clinical characteristics | |||||
| Age | 64 | 1.21 | 1.01 to 1.44 | 0.45 to 3.22 | 89.83 |
| Multiple current pregnancy | 18 | 3.63 | 2.35 to 5.60 | 0.66 to 19.82 | 96.96 |
| Parity | 37 | 0.99 | 0.87 to 1.12 | 0.56 to 1.74 | 86.04 |
| BMI | 40 | 1.06 | 1.04 to 1.08 | 0.99 to 1.13 | 85.89 |
| BMI trimester 1 | 41 | 1.07 | 1.05 to 1.09 | 0.99 to 1.15 | 73.36 |
| BMI trimester 2 | 20 | 1.05 | 1.02 to 1.08 | 0.95 to 1.15 | 66.81 |
| SBP | 18 | 1.65 | 1.43 to 1.92 | 0.98 to 2.80 | 75.60 |
| SBP trimester 1 | 31 | 1.50 | 1.39 to 1.63 | 1.11 to 2.05 | 57.89 |
| SBP trimester 2 | 23 | 1.73 | 1.51 to 1.97 | 1.15 to 2.60 | 68.74 |
| DBP | 18 | 1.92 | 1.53 to 2.42 | 0.86 to 4.31 | 80.98 |
| DBP trimester 1 | 30 | 1.85 | 1.61 to 2.12 | 1.10 to 3.10 | 71.92 |
| DBP trimester 2 | 23 | 2.15 | 1.79 to 2.59 | 1.23 to 3.76 | 69.28 |
| MAP | 24 | 2.18 | 1.83 to 2.60 | 1.11 to 4.30 | 74.08 |
| MAP trimester 1 | 33 | 2.00 | 1.73 to 2.31 | 1.13 to 3.55 | 74.01 |
| MAP trimester 2 | 24 | 2.25 | 1.89 to 2.69 | 1.28 to 3.97 | 70.79 |
| PCR | 3 | 1.00 | 1.00 to 1.01 | 1.00 to 1.01 | 0.00 |
| PCR trimester 2 | 1 | 1.02 | 0.90 to 1.14 | N/A | N/A |
| Urine dipstick | 5 | 3.30 | 0.31 to 35.27 | 0.01 to 922.24 | 96.69 |
| Urine dipstick trimester 1 | 3 | 3.29 | 0.28 to 38.31 | 0.04 to 300.79 | 86.11 |
| Urine dipstick trimester 2 | 8 | 3.20 | 1.79 to 5.72 | 0.93 to 10.94 | 67.54 |
| Ultrasound markers | |||||
| Uterine artery PI | 11 | 6.27 | 2.70 to 14.58 | 0.82 to 48.23 | 54.17 |
| Uterine artery PI trimester 1 | 11 | 2.14 | 1.24 to 3.67 | 0.56 to 8.24 | 50.70 |
| Uterine artery PI trimester 2 | 13 | 14.73 | 8.12 to 26.72 | 3.10 to 69.96 | 60.11 |
| Umbilical artery PI | 5 | 5.44 | 3.27 to 9.06 | 3.27 to 9.06 | 0.00 |
| Umbilical artery PI trimester 2 | 8 | 6.29 | 2.85 to 13.92 | 2.85 to 13.92 | 0.00 |
| Biochemical markers | |||||
| PIGF trimester 1 | 15 | 0.08 | 0.02 to 0.35 | 0.00 to 5.94 | 55.69 |
| PIGF trimester 2 | 13 | 0.07 | 0.01 to 0.43 | 0.00 to 13.16 | 97.18 |
| sFlt-1 trimester 1 | 10 | 0.99 | 0.97 to 1.01 | 0.97 to 1.01 | 0.21 |
| sFlt-1 trimester 2 | 12 | 1.05 | 1.01 to 1.09 | 0.96 to 1.15 | 90.28 |
| PAPP-A trimester 1 | 9 | 0.99 | 0.97 to 1.01 | 0.97 to 1.01 | 0.43 |
| PAPP-A trimester 2 | 3 | 1.00 | 1.00 to 1.00 | 1.00 to 1.00 | 0.00 |
Heterogeneity for predictors varied considerably, and ranged from 0% to 97% for PlGF. The strongest association with the outcome was observed for uterine artery pulsatility index measured in the second trimester (OR 14.73, 95% CI 8.12 to 26.72; I2 = 60.11%), which suggests an increase in the odds of early-onset pre-eclampsia of about 15 times, with a unit increase in the mean uterine artery pulsatility index. The confidence and prediction intervals for this were wide (95% prediction interval 3.10 to 69.96), demonstrating the variability within the studies. Most predictors had evidence of an association at the 5% level, but parity, PCR measured in the second trimester, urine dipstick (unspecified trimester estimates and first trimester), sFlt-1 (first-trimester estimates) and PAPP-A (all trimester estimates) were not statistically significant at the 5% level. All statistically significant predictors had evidence of an increase in the odds of early pre-eclampsia with increasing values, except PlGF measured in the first (OR 0.08, 95% CI 0.02 to 0.35; I2 = 55.69%) or the second trimester (OR 0.07, 95% CI 0.01 to 0.43; I2 = 97.18%), which showed a decrease in the odds of pre-eclampsia with increasing levels.
Late-onset pre-eclampsia
The results of the two-stage meta-analysis for late-onset (delivery at ≥ 34 weeks’ gestation) pre-eclampsia and each predictor variable are shown in Table 22. The two-stage IPD meta-analysis could not be performed for the following predictors: previous pre-eclampsia, history of hypertension, history of renal disease, history of diabetes, history of autoimmune disease, family history of pre-eclampsia, PCR measured in the first trimester, previous small for gestational age fetus, smoking during pregnancy, spontaneous mode of conception, cocaine, heroin or methamphetamine use, umbilical artery pulsatility index measured in the first trimester, PlGF, sFlt-1, PAPP-A and PAPP-A measured in the third trimester. 239
| Predictor | Number of studies | OR | 95% CI | 95% prediction interval | I2 (%) |
|---|---|---|---|---|---|
| Maternal clinical characteristics | |||||
| Age | 68 | 1.04 | 0.94 to 1.14 | 0.60 to 1.81 | 94.66 |
| Multiple current pregnancy | 22 | 2.18 | 1.59 to 2.99 | 0.62 to 7.64 | 98.07 |
| Parity | 46 | 0.87 | 0.78 to 0.97 | 0.48 to 1.58 | 95.16 |
| BMI | 42 | 1.07 | 1.06 to 1.08 | 1.02 to 1.12 | 94.96 |
| BMI trimester 1 | 45 | 1.07 | 1.06 to 1.09 | 1.00 to 1.15 | 92.31 |
| BMI trimester 2 | 19 | 1.07 | 1.06 to 1.09 | 1.01 to 1.14 | 82.19 |
| BMI trimester 3 | 10 | 1.09 | 1.06 to 1.13 | 0.99 to 1.21 | 82.31 |
| SBP | 19 | 1.44 | 1.29 to 1.61 | 0.94 to 2.22 | 87.60 |
| SBP trimester 1 | 36 | 1.39 | 1.31 to 1.47 | 1.08 to 1.80 | 77.48 |
| SBP trimester 2 | 26 | 1.43 | 1.33 to 1.54 | 1.09 to 1.88 | 76.58 |
| SBP trimester 3 | 15 | 1.83 | 1.54 to 2.18 | 0.96 to 3.48 | 93.60 |
| DBP | 19 | 1.56 | 1.38 to 1.77 | 0.93 to 2.62 | 85.49 |
| DBP trimester 1 | 36 | 1.58 | 1.44 to 1.73 | 1.09 to 2.30 | 79.70 |
| DBP trimester 2 | 26 | 1.59 | 1.41 to 1.8 | 0.97 to 2.61 | 85.68 |
| DBP trimester 3 | 15 | 2.20 | 1.68 to 2.88 | 0.84 to 5.78 | 95.00 |
| MAP | 25 | 1.74 | 1.51 to 2.01 | 0.94 to 3.22 | 88.77 |
| MAP trimester 1 | 38 | 1.65 | 1.51 to 1.80 | 1.12 to 2.42 | 81.05 |
| MAP trimester 2 | 27 | 1.70 | 1.51 to 1.92 | 1.02 to 2.85 | 86.86 |
| MAP trimester 3 | 15 | 2.26 | 1.77 to 2.89 | 0.91 to 5.60 | 94.78 |
| PCR | 3 | 1.01 | 0.98 to 1.04 | 0.95 to 1.07 | 96.67 |
| PCR trimester 1 | 1 | 0.75 | 0.34 to 1.67 | N/A | N/A |
| PCR trimester 2 | 1 | 0.97 | 0.92 to 1.03 | N/A | N/A |
| Urine dipstick | 7 | 2.45 | 0.79 to 7.65 | 0.10 to 59.76 | 98.01 |
| Urine dipstick trimester 1 | 5 | 1.85 | 1.48 to 2.32 | 1.48 to 2.32 | 0.00 |
| Urine dipstick trimester 2 | 9 | 1.44 | 1.04 to 1.99 | 0.63 to 3.27 | 59.56 |
| Urine dipstick trimester 3 | 5 | 2.73 | 1.47 to 5.04 | 0.72 to 10.34 | 81.85 |
| Ultrasound markers | |||||
| Uterine artery PI | 11 | 2.19 | 1.36 to 3.54 | 0.59 to 8.12 | 64.30 |
| Uterine artery PI trimester 1 | 12 | 1.61 | 1.09 to 2.39 | 0.54 to 4.83 | 71.53 |
| Uterine artery PI trimester 2 | 12 | 2.95 | 2.31 to 3.76 | 1.94 to 4.48 | 20.77 |
| Uterine artery PI trimester 3 | 3 | 3.98 | 0.04 to 403.56 | 0.00 to 11,096.61 | 67.65 |
| Umbilical artery PI | 5 | 0.91 | 0.05 to 15.46 | 0.00 to 375.27 | 88.83 |
| Umbilical artery PI trimester 2 | 8 | 1.25 | 0.56 to 2.78 | 0.48 to 3.25 | 8.11 |
| Umbilical artery PI trimester 3 | 5 | 0.91 | 0.26 to 3.25 | 0.11 to 7.36 | 32.31 |
| Biochemical markers | |||||
| PIGF trimester 1 | 17 | 0.33 | 0.16 to 0.68 | 0.03 to 3.93 | 82.67 |
| PIGF trimester 2 | 16 | 0.81 | 0.69 to 0.94 | 0.52 to 1.25 | 76.39 |
| PIGF trimester 3 | 12 | 0.68 | 0.57 to 0.81 | 0.38 to 1.20 | 93.60 |
| sFlt-1 trimester 1 | 11 | 0.98 | 0.97 to 0.99 | 0.95 to 1.01 | 37.07 |
| sFlt-1 trimester 2 | 13 | 1.00 | 0.99 to 1.00 | 0.99 to 1.01 | 8.29 |
| sFlt-1 trimester 3 | 9 | 1.02 | 1.00 to 1.03 | 0.99 to 1.04 | 91.03 |
| PAPP-A trimester 1 | 10 | 0.83 | 0.53 to 1.31 | 0.22 to 3.18 | 99.99 |
| PAPP-A trimester 2 | 4 | 1.00 | 0.99 to 1.02 | 0.98 to 1.02 | 57.98 |
There was considerable variation in the heterogeneity for predictors, and the strongest association with the outcome was observed for uterine artery pulsatility index measured in the second trimester (OR 2.95, 95% CI 2.31 to 3.76; I2 = 20.77%), which suggests that a 1-unit increase in the uterine artery pulsatility index will lead to an approximate increase of three times in the odds of late-onset pre-eclampsia. Most predictors assessed showed evidence of an association at the 5% level. However, maternal age, PCR (for all trimester estimates), urine dipstick (unspecified trimester), third-trimester uterine artery pulsatility index, umbilical artery pulsatility index (unspecified trimester and third trimester), second-trimester sFlt-1 and PAPP-A (all trimesters estimates) were not statistically significant at the 5% level. All statistically significant predictors had evidence of an increase in odds of late-onset pre-eclampsia except parity (OR 0.87, 95% CI 0.78 to 0.97; I2 = 95.16%), first-trimester (OR 0.33, 95% CI 0.16 to 0.68; I2 = 82.67%), second-trimester (OR 0.81, 95% CI 0.69 to 0.94; I2 = 76.39%) or third-trimester (OR 0.68, 95% CI 0.57 to 0.81; I2 = 93.60%) measurement of PlGF and sFlt-1 measured in the first trimester (OR 0.98, 95% CI 0.97 to 0.99; I2 = 37.07%), which showed a reduction in the odds of late-onset pre-eclampsia.
Forest plots for each predictor assessed and for each outcome are presented in Report Supplementary Material 1.
Summary
This unadjusted two-stage IPD meta-analysis of the prognostic effect of candidate predictors of early-, late- and any-onset pre-eclampsia was limited to complete singleton records only and included all 78 studies in the IPPIC database, with missing data assumed to be missing completely at random. 84 Most predictors had evidence of an association at the 5% level for each outcome. Increasing the values of second-trimester measurement of uterine artery pulsatility index had the strongest statistically significant association with early-onset (OR 14.73, 95% CI 8.12 to 26.72; I2 = 60.11%) or late-onset (OR 2.95, 95% CI 2.31 to 3.76; I2 = 20.77%) pre-eclampsia. The predictor with the strongest statistically significant association with any-onset pre-eclampsia was history of hypertension (OR 4.76, 95% CI 3.56 to 6.35; I2 = 98.39%). Smoking during pregnancy and spontaneous mode of conception were associated with a reduction in odds of any-onset pre-eclampsia only; however, parity was associated with reduced odds of late-onset and any-onset pre-eclampsia. Increases in PlGF measurements were associated with a reduction in the odds of early-, late- and any-onset pre-eclampsia, whereas sFlt-1 was associated with a reduction in the odds of late-onset pre-eclampsia only. Conclusions drawn about the association of predictors with outcomes should take into account the uncertainty resulting from multiple testing type I errors. Further research should consider imputing for missing data to control for the introduction of bias, and this would also allow associations to be adjusted for potential confounders, giving a better evaluation of associations.
Chapter 8 Discussion
Summary of the findings
Existing prediction models for any-, early- and late-onset pre-eclampsia IPD have poor to average predictive performance when externally validated in the combined IPPIC-UK data sets. All models that could be validated showed suboptimal predictive performance across data sets. The clinical utility of the published models was poor.
Most of the IPPIC pre-eclampsia models showed good to average discrimination across data sets; all had good to average calibration across data sets. The models varied in the predictive performance between data sets. The clinical characteristics only and the clinical and biochemical first- and second-trimester models to predict any pre-eclampsia showed consistent net benefit over a strategy of considering all women to have pre-eclampsia for a wide range of probability thresholds beyond 5% in cohort of nulliparous women with singleton pregnancies in the UK. Very few risk factors were associated with pre-eclampsia, with significance in both the CIs and the prediction intervals including BMI, SBP and DBP and MAP for any-, early- and late-onset pre-eclampsia; and urine dipstick, uterine artery pulsatility index and umbilical artery pulsatility index for any- and early-onset pre-eclampsia.
Strengths and limitations
We used the largest data set to date to validate existing prediction models for pre-eclampsia, and to further develop models when required. The IPPIC data set consists of information on predictor variables at various trimesters. We used raw data to determine the presence or absence of pre-eclampsia and the type of onset, ensuring the reliability of the findings. Our comprehensive search identified all published models. By validating them in UK data sets, we were able to assess the extent of transportability of existing models to women managed in the NHS. We assessed the quality of the data sets and studies using robust tools. Rather than develop further models, we ensured that the performances of the existing models were robustly evaluated. The models were validated not only using evidence synthesis, but also within individual data sets. The large sample size provided us with sufficient events for the rare but important outcome of early-onset pre-eclampsia. In addition to reporting the performances of the models in terms of discrimination and calibration, we determined their clinical utility using decision curve analysis.
Prior to the development of the IPPIC pre-eclampsia models, we prioritised the predictors for importance to clinical practice by consensus to ensure face validity. We used multiple imputation to deal with missing values for both predictors and outcomes to avoid the loss of useful information84,240 and explored complex associations such as the non-linearity of predictor effects. We were able to report the association between individual clinical, biochemical and ultrasound predictors, measured in the first, second or third trimester, and rates of early-, late- and any-onset pre-eclampsia with very precise estimates. We pooled data from a very large sample size using IPD meta-analysis, and explored a considerable number of risk factors thought to be predictors of pre-eclampsia. We did not dichotomise any of the continuous predictive factors and we also considered the predictive accuracy of these risk factors along with their association.
Our findings were limited by the variations in population mix, the definitions of the predictors in each study and the outcomes reported. Some studies included only nulliparous women, some strictly included low-risk pregnancies and some included all pregnancies. The prioritisation of predictors of pre-eclampsia by members of the collaborative network who contributed data to the project could also be considered a limitation in the identification of predictors to be considered for model development. It was possible that participants in the survey would rank predictors as important based on their particular research interest, and this method could potentially hamper the identification of new candidate predictors. However, we had a good representation of responses to the survey, and respondents were able to suggest possible factors not already assessed in the survey to be considered as predictors. There was also good consensus among respondents about the importance of predictors assessed, and no new candidate predictors were identified. The individual UK data sets measured different sets of variables (potential predictors) and measured them at different times (e.g. first, second or third trimester). Our validation was carried out considering only UK data sets to reduce the heterogeneity in the outcome definition and to allow existing models’ predictive performance to be assessed in the UK health-care system context. However, this limited our ability to validate many of the existing prediction models and meant that we could validate models across the studies only if all of them reported the same variables. We were, therefore, also unable to validate all existing models in our IPD because of the unavailability of predictors in the models in our UK IPD. It is possible that a significant predictor may not have been evaluated if it was not provided across varied data sets. Some studies used data from the same cohort of women to report various prediction models in multiple combinations. The sources of the data also differed across data sets. Some were collected prospectively with the explicit purpose of predicting pre-eclampsia, whereas others were routine registry data. All of the above accounted for the heterogeneity observed in the performance of the models across the data sets. We validated the performances of published models across only UK data sets, but included all data sets for model development. It is likely that the transportability performances may have differed if all available data had been included. Furthermore, some models, such as the North et al. model42 for any-onset pre-eclampsia, the Poon 2009 model234 for early-onset pre-eclampsia and the Akolekar et al. model241 for late-onset pre-eclampsia, could not be validated as the predictor variables in the models were not available in any of the IPPIC-UK data sets.
To ensure that the relevant data were included in the analysis, we dealt with missing data by imputing both within and between studies. We also made assumptions such as using early second-trimester values of BMI and MAP if the first-trimester values were missing. Our analysis of the association of risk factors and the different pre-eclampsia outcomes was limited to complete records only. Our assumption that data missing were missing completely at random is unlikely to be true. Applying multilevel multiple imputation would reduce this bias, and also allow estimates to be adjusted for potential confounders, giving a better evaluation of associations. However, the complexity of modelling the missing data mechanism would make this demanding (or impossible). We therefore present this as the most robustly available assessment of risk factors for pre-eclampsia.
Comparison with existing evidence
Current guidelines such as those by the National Institute for Health and Care Excellence242 in the UK and by the American College of Obstetricians and Gynecologists73 in the USA provide a list of risk factors rather than a prediction model to determine an individual’s risk of pre-eclampsia. The predictive performance of both approaches has been shown to be inferior to that of multivariable prediction models. 236,243,244 However, these models had not been externally validated in multiple data sets until now, with resulting suboptimal predictive performance.
Until now, only a small fraction of the 131 pre-eclampsia prediction models identified have been externally validated (11%, 15/131), and an even smaller proportion (4%, 5/131) have been evaluated for their clinical utility. 44,108,160,245,246 Studies reporting on the external validation of these models often have not reported performance measures in terms of calibration, which has more value in assessing the predictive performance of the model than discrimination estimates or detection rates. 247 Some existing models also used multiples of the mean to standardise biochemical and ultrasound markers, but this is of limited use in real-world settings as the estimates of adjustment factors used to standardise these measurements to multiple of the mean values in the models are not always known in the population in which the model is to be used. For some of the models that we could validate, the summary performance measures have a lot of uncertainty (wide CIs), reflecting the small numbers of events and/or the heterogeneity. Even with larger numbers of events, CIs can be wide, especially for calibration, so it is possible that, for some of the models, miscalibration is due to chance. However, we must also look at the broader picture emerging across all of the validations. For most models, the majority of summary results for calibration, and the study-specific results for calibration, are suggestive of overfitting (slopes < 1). This is something that the field needs to address as whole. In our IPPIC models, we have examined overfitting in our model development, and adjusted for it.
Recently, the ASPRE trial34 showed that women at high risk of preterm pre-eclampsia who were started on 150 mg of aspirin early in pregnancy had their risk lowered, and their high-risk status was determined using a prediction model. However, a few questions need to be answered before this approach is implemented. First, the extent to which the model over- or underpredicted women’s pre-eclampsia risk is not reported in the study, because women assessed as being at low risk of pre-eclampsia using the model were not followed up further. This is also shown by the significant difference in the incidence of preterm pre-eclampsia between the placebo group and the population used to develop the model. Second, it is likely that some women in the group categorised as ‘low risk’ using the model may have benefited from the intervention. In the absence of follow-up of this group for pre-eclampsia outcomes, we cannot robustly confirm that they would not have benefited from the intervention. Third, the clinical utility of the prediction model has not been assessed, limiting our ability to recommend its routine use in clinical practice. We were unable to validate the exact model used in the ASPRE trial because the multiple of the mean predictors were unavailable in our IPD data set. The authors who developed this model recently validated it in three data sets with ‘appropriately trained staff and quality control of measurement’. 248 This showed that the model discriminated well, with a large C-statistic in all three validation data sets. However, further independent validation of this model is needed to evaluate its performance in ‘real-world’ settings.
Although some of the published models showed a promising ability to discriminate between women who had a pre-eclampsia outcome and those who did not, calibration performance was generally poor across the data sets and there was large heterogeneity in the calibration performance across different IPPIC data sets. Although the CIs (e.g. for calibration slope) were sometimes very wide, a general picture is that most models demonstrated overfitting at model development with predictions that were too extreme compared with the observed risk in the data sets (calibration slope < 1). Model predictions were also systematically too low or too high depending on the data set used to validate the model (calibration-in-the-large ≠ 0). The models were validated in a rather heterogeneous group of data sets with different eligibility criteria. These findings suggest that the differences between women in the data sets were not adequately captured by the set of predictors included in the models. There was also little difference in predictive performance when biochemical markers or ultrasound markers were combined with maternal and clinical characteristics, compared with models with only maternal and clinical characteristics. Some of the heterogeneity in predictive performance of the models is likely to be due to different methods and timing of measurement, for example in blood pressure and biochemical marker values. Going forward, standardisation of measurement methods, for example across laboratories and hospitals, might reduce heterogeneity in calibration performance. A related point is that prediction models in this field need to be clearer with regard to how and exactly when included predictors should be measured.
For IPPIC models, summary predictive performance was promising, and net benefit was demonstrated in some data sets across clinically relevant thresholds of predicted risk. However, large heterogeneity remained in all performance statistics across data sets. Heterogeneity in calibration performance could be reduced if, when applying the models in practice, model parameters (e.g. intercept) could be recalibrated to each population and setting. This would require local data for recalibration and model updating.
Relevance to clinical practice
Existing models that were externally validated had poor calibration performance and their utility was limited, with no model identified that can be recommended for clinical use. The IPPIC models showed promising performance for predicting pre-eclampsia, in particular in both low- and middle-income countries where only clinical characteristics may be available, and in high-income countries where there is access to additional biochemical markers. However, on application, the predictive performance of the models needs to be improved by recalibration to particular settings and populations; this would require local data. Ultrasound markers did not add any additional information or improve the performance of the prediction models beyond the clinical characteristic only models. This suggests a lack of need for additional time or resources in carrying out these assessments for screening of women at risk of pre-eclampsia. The thresholds of risk on which decision-making is based are likely to vary with the planned intervention (aspirin or calcium), as well as following shared decision-making through discussions between the clinician and the woman.
Relevance to research
Validation, including examination of calibration heterogeneity, is still required for the models we could not validate. For these and the IPPIC models, we need validation in multiple large data sets across different settings and populations to properly assess their transportability. 249 The impact of using the models in clinical practice needs to be evaluated beyond predicting pre-eclampsia, but also in the identification of women with pre-eclampsia who are also most likely to have severe complications such as HELLP syndrome, eclampsia, abruption or renal failure. The acceptability of the models to both women and health-care professionals needs to be assessed, including elucidation of their preferred threshold probability for treatment decisions. A decision-analytic model of resource implications, including the cost utilities of consequences of decisions for various false-positive and false-negative cases, is also needed. Updated models may be needed in local populations, for example using recalibration of the IPPIC models in local data sets, to improve calibration performance. Furthermore, additional strong predictors need to be identified to improve model performance and consistency. New cohorts need to standardise the predictors and outcomes measured, including their timing and measurement methods, to enable more homogenous data sets to be combined in IPD meta-analyses.
Conclusion
Among the 24 existing prediction models that could be validated in the IPD meta-analysis, generally their predictive performance was poor across data sets. To address this, IPPIC models were developed with adjustment for overfitting, which show good predictive performance on average across data sets and may have net benefit in singleton nulliparous populations in the UK. However, heterogeneity across settings is likely in calibration performance, and thus the models need to be recalibrated in local settings and populations of application. Ultrasound markers did not improve the predictive performance of the developed IPPIC clinical characteristic-only models. We did not identify any new predictors for our model development that were not considered previously in existing models. Further work is therefore needed to validate other models, identify new predictors and improve calibration performance in all settings of intended use.
Acknowledgements
We would like to acknowledge all researchers who contributed data to this IPD meta-analysis, including the original teams involved in the collection of the data, and the participants who took part in the research studies.
We are extremely grateful to all of the families who took part in this study, the midwives for their help in recruiting them, and the whole ALSPAC team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists and nurses.
We acknowledge the National Institute of Child and Human Development Data and Specimen Hub for providing the MFMU HRA and MFMU LRA studies data that were used for this research, the contribution of the principal investigator(s) who conducted the original study from which the data were generated, and NICHD and other funding organisations, if applicable, that supported the original study.
We are thankful to members of the Independent Steering Committee, which included Professor Arri Coomarasamy (Chairperson, University of Birmingham), Dr Aris Papageorghiou (St George’s University Hospital), Mrs Ngawai Moss (Katie’s Team), Professor Sarosh Rana (University of Chicago) and Dr Thomas Debray (University Medical Center Utrecht), for their guidance and support throughout the project.
Members of the IPPIC Collaborative Network
Alex Kwong, University of Bristol; Ary I Savitri, University Medical Center Utrecht; Kjell Åsmund Salvesen, Norwegian University of Science and Technology; Sohinee Bhattacharya, University of Aberdeen; Cuno SPM Uiterwaal, University Medical Center Utrecht; Annetine C Staff, University of Oslo; Louise Bjoerkholt Andersen, University of Southern Denmark; Elisa Llurba Olive, Hospital Universitari Vall d’Hebron; Christopher Redman, University of Oxford; George Daskalakis, University of Athens; Maureen Macleod, University of Dundee; Baskaran Thilaganathan, St George’s University of London; Javier Arenas Ramírez, University Hospital de Cabueñes; Jacques Massé, Laval University; Asma Khalil, St George’s University of London; Francois Audibert, Université de Montréal; Per Minor Magnus, Norwegian Institute of Public Health; Anne Karen Jenum, University of Oslo; Ahmet Baschat, Johns Hopkins University School of Medicine; Akihide Ohkuchi, University School of Medicine, Shimotsuke-shi; Fionnuala M. McAuliffe, University College Dublin; Jane West, University of Bristol; Lisa M Askie, University of Sydney; Fionnuala Mone, University College Dublin; Diane Farrar, Bradford Teaching Hospitals; Peter A Zimmerman, Päijät-Häme Central Hospital; Luc JM Smits, Maastricht University Medical Centre; Catherine Riddell, Better Outcomes Registry & Network (BORN); John C Kingdom, University of Toronto; Joris van de Post, Academisch Medisch Centrum; Sebastián E Illanes, University of the Andes; Claudia Holzman, Michigan State University; Sander MJ van Kuijk, Maastricht University Medical Centre; Lionel Carbillon, Assistance Publique-Hôpitaux de Paris Université; Pia M Villa, University of Helsinki and Helsinki University Hospital; Anne Eskild, University of Oslo; Lucy Chappell, King’s College London; Federico Prefumo, University of Brescia; Luxmi Velauthar, Queen Mary University of London; Paul Seed, King’s College London; Miriam van Oostwaard, IJsselland Hospital; Stefan Verlohren, Charité University Medicine; Lucilla Poston, King’s College London; Enrico Ferrazzi, University of Milan; Christina A Vinter, University of Southern Denmark; Chie Nagata, National Center for Child Health and Development; Mark Brown, University of New South Wales; Karlijn C Vollebregt, Academisch Medisch Centrum; Satoru Takeda, Juntendo University; Josje Langenveld, Atrium Medisch Centrum Parkstad; Mariana Widmer, WHO; Shigeru Saito, University of Toyama; Camilla Haavaldsen, Akershus University Hospital; Guillermo Carroli, Centro Rosarino De Estudios Perinatales; Jørn Olsen, Aarhus University; Hans Wolf, Academisch Medisch Centrum; Nelly Zavaleta, Instituto Nacional De Salud; Inge Eisensee, Aarhus University; Patrizia Vergani, University of Milano-Bicocca; Pisake Lumbiganon, Khon Kaen University; Maria Makrides, South Australian Health and Medical Research Institute; Fabio Facchinetti, Università degli Studi di Modena e Reggio Emilia; Evan Sequeira, Aga Khan University; Robert Gibson, University of Adelaide; Sergio Ferrazzani, Università Cattolica del Sacro Cuore; Tiziana Frusca, Università degli Studi di Parma; Jane E Norman, University of Edinburgh; Ernesto A Figueiró-Filho, Mount Sinai Hospital; Olav Lapaire, Universitätsspital Basel; Hannele Laivuori, University of Helsinki and Helsinki University Hospital; Jacob A Lykke, Rigshospitalet; Agustin Conde-Agudelo, Eunice Kennedy Shriver National Institute of Child Health and Human Development; Alberto Galindo, Universidad Complutense de Madrid; Alfred Mbah, University of South Florida; Ana Pilar Betran, WHO; Ignacio Herraiz, Universidad Complutense de Madrid; Lill Trogstad, Norwegian Institute of Public Health; Gordon GS Smith, University of Cambridge; Eric AP Steegers, University Hospital Nijmegen; Read Salim, HaEmek Medical Center; Tianhua Huang, North York General Hospital; Annemarijne Adank, Erasmus Medical Centre; Jun Zhang, National Institute of Child Health and Human Development; Wendy S Meschino, North York General Hospital; Joyce L Browne, University Medical Centre Utrecht; Rebecca E Allen, Queen Mary University of London; Fabricio Da Silva Costa, University of São Paulo; Kerstin Klipstein-Grobusch Browne, University Medical Center Utrecht; Caroline A Crowther, University of Adelaide; Jan Stener Jørgensen, Syddansk Universitet; Jean-Claude Forest, Centre hospitalier universitaire de Québec; Alice R Rumbold, University of Adelaide; Ben W Mol, Monash University; Yves Giguère, Laval University; Louise C Kenny, University of Liverpool; Wessel Ganzevoort, Academisch Medisch Centrum; Anthony O Odibo, University of South Florida; Jenny Myers, University of Manchester; SeonAe Yeo, University of North Carolina at Chapel Hill; Helena J Teede, Monash University and Monash Health; Francois Goffinet, Assistance Publique, Hôpitaux de Paris; Lesley McCowan, University of Auckland; Eva Pajkrt, Academisch Medisch Centrum; Bassam G Haddad, Portland State University; Gustaaf Dekker, University of Adelaide; Emily C Kleinrouweler, Academisch Medisch Centrum; Édouard LeCarpentier, Centre Hospitalier Intercommunal de Créteil; Claire T Roberts, University of Adelaide; Henk Groen, University Medical Center Groningen; Ragnhild Bergene Skråstad, St Olav’s Hospital; Seppo Heinonen, University of Helsinki and Helsinki University Hospital; and Kajantie Eero, University of Helsinki and Helsinki University Hospital.
Contributions of authors
Shakila Thangaratinam (https://orcid.org/0000-0002-4254-460X) (Professor, Maternal and Perinatal Health), Richard D Riley (https://orcid.org/0000-0001-8699-0735) (Professor, Biostatistics), Khalid S Khan (https://orcid.org/0000-0001-5084-7312) (Professor, Women’s Health & Clinical Epidemiology), Karel GM Moons (https://orcid.org/0000-0003-2118-004X) (Professor, Clinical Epidemiology), Richard Hooper (https://orcid.org/0000-0002-1063-0917) (Professor, Medical Statistics), Basky Thilaganathan (https://orcid.org/0000-0002-5531-4301) (Professor, Fetal Medicine) and Asma Khalil (https://orcid.org/0000-0003-2802-7670) (Professor, Obstetrics and Maternal–Fetal Medicine) developed the protocol. John Allotey (https://orcid.org/0000-0003-4134-6246) (Senior Research Fellow, Women’s Health), Shakila Thangaratinam, Melanie Smuk (https://orcid.org/0000-0002-1594-1458) (Lecturer, Medical Statistics) and Kym IE Snell (https://orcid.org/0000-0001-9373-6591) (Lecturer, Biostatistics) undertook the literature searches, study selection, drafted the manuscript, acquired IPD and led the project.
Asif Ahmed (https://orcid.org/0000-0002-8755-8546) (Pro-Vice Chancellor for Health, Medicine), Lucy C Chappell (https://orcid.org/0000-0001-6219-3379) (Professor, Obstetrics), Peter von Dadelszen (https://orcid.org/0000-0003-4136-3070) (Professor, Global Women’s Health), Lucilla Poston (https://orcid.org/0000-0003-1100-2821) (Professor, Maternal & Fetal Health), Marcus Green (https://orcid.org/0000-0002-4561-8256) (CEO APEC, PPI), Louise Kenny (https://orcid.org/0000-0002-9011-759X) (Executive Pro-Vice Chancellor, Health and Life Sciences), Jenny Myers (https://orcid.org/0000-0003-0913-2096) (Clinical Senior Lecturer, Maternal & Fetal Health), Anne C Staff (https://orcid.org/0000-0001-9247-5721) (Professor, Obstetrics and Gynaecology), Ben WMol (https://orcid.org/0000-0001-8337-550X) (Professor, Obstetrics & Gynaecology), Gordon CS Smith (https://orcid.org/0000-0003-2124-0997) (Professor and Head of the Department, Obstetrics and Gynaecology), Wessel Ganzevoort (https://orcid.org/0000-0002-7243-2115) (Specialist, Obstetrician and Gynaecologist), Hannele Laivuori (https://orcid.org/0000-0003-3212-7826) (Professor, Clinical Genetics), Anthony O Odibo (https://orcid.org/0000-0003-4340-450X) (Professor, Obstetrics and Gynaecology), Ahmet A Baschat (https://orcid.org/0000-0003-1927-2084) (Professor, Gynaecology and Obstetrics), Paul T Seed (https://orcid.org/0000-0001-7904-7933) (Senior Lecturer, Medical Statistician), Federico Prefumo (https://orcid.org/0000-0001-7793-714X) (Specialist, Gynaecology and Obstetrics), Fabricio da Silva Costa (https://orcid.org/0000-0002-0765-7780) (Adjunct Clinical Assistant Professor, Obstetrics and Gynaecology), Henk Groen (https://orcid.org/0000-0002-6629-318X) (Assistant Professor, Epidemiology), Francois Audibert (https://orcid.org/0000-0002-2697-3826) (Professor, Obstetrics & Gynaecology), Camilla Haavaldsen (https://orcid.org/0000-0002-4708-3267) (Scientist, Obstetrics & Gynaecology), Chie Nagata (https://orcid.org/0000-0003-4897-2119) (Chief of Clinical Research, Maternal and Child Health), Alice R Rumbold (https://orcid.org/0000-0002-4453-9425) (Associate Professor, Reproductive Epidemiology), Seppo Heinonen (https://orcid.org/0000-0001-5949-0874) (Professor, Obstetrics and Gynaecology), Lisa M Askie (https://orcid.org/0000-0002-8934-5544) (Professorial Research Fellow, Medicine), Luc JM Smits (https://orcid.org/0000-0003-0785-1345) (Professor, Epidemiology), Christina A Vinter (https://orcid.org/0000-0001-5084-6053) (Clinical Associate Professor, Gynaecology and Obstetrics), Per M Magnus (https://orcid.org/0000-0002-6427-4735) (Professor, Community Medicine and Global Health), Pia M Villa (https://orcid.org/0000-0002-0317-0713) (Researcher, Obstetrics and Gynaecology), Anne K Jenum (https://orcid.org/0000-0003-0304-7800) (Professor, General Practice), Julie Dodds (https://orcid.org/0000-0002-6041-1456) (Senior Research Manager, Women’s Health), Louise B Andersen (https://orcid.org/0000-0002-3331-7149) (Researcher, Obstetrics and Gynaecology), Jane E Norman (https://orcid.org/0000-0001-6031-6953) (Professor, Health Sciences), Akihide Ohkuchi (https://orcid.org/0000-0002-8861-1572) (Professor, Obstetrics and Gynaecology), Anne Eskild (https://orcid.org/0000-0002-2756-1583) (Professor, Clinic Medicine), Sohinee Bhattacharya (https://orcid.org/0000-0002-2358-5860) (Senior Lecturer, Medicine), Fionnuala M McAuliffe (https://orcid.org/0000-0002-3477-6494) (Professor, Obstetrics and Gynaecology), Alberto Galindo (https://orcid.org/0000-0002-1308-1474) (Researcher, Obstetrics and Gynaecology), Ignacio Herraiz (https://orcid.org/0000-0001-6807-4944) (Researcher, Obstetrics and Gynaecology), Lionel Carbillon (https://orcid.org/0000-0001-6367-4828) (Researcher, Obstetrics and Gynaecology), Kerstin Klipstein-Grobusch (https://orcid.org/0000-0002-5462-9889) (Associate Professor, Global Health) and SeonAe Yeo (https://orcid.org/0000-0002-0721-0997) (Professor, Nursing) provided input into the protocol development and the drafting of the initial manuscript.
Basky Thilaganathan, Asma Khalil, Louise Kenny, Lucy C Chappell, Jacques Massé (https://orcid.org/0000-0002-8257-5478), Anne C Staff, Gordon CS Smith, Wessel Ganzevoort, Hannelle Laivuori, Anthony O Odibo, Ahmet A Baschat, Paul T Seed, Federico Profumo, Fabricio da Silva Costa, Henk Groen, Francois Audibert, Chie Nagata, Alice R Rumbold, Seppo Heinonen, Lisa M Askie, Luc JM Smits, Christina A Vinter, Ben W Mol, Lucilla Poston, Javier A Ramírez (https://orcid.org/0000-0003-2291-720X) (Researcher, Obstetrics and Gynaecology), John Kingdom (https://orcid.org/0000-0002-7411-5535) (Professor, Maternal-Fetal Medicine), George Daskalakis (https://orcid.org/0000-0001-7108-211X) (Researcher, Obstetrics and Gynaecology), Dianne Farrar (https://orcid.org/0000-0002-5625-761X) (NIHR Post-doctoral Research Fellow, Maternal Health), Helena J Teede (https://orcid.org/0000-0001-7609-577X) (Director, Monash Centre for Health Research, Women’s and Children’s Health), Kjell Å Salvesen (https://orcid.org/0000-0002-1788-4063) (Professor, Clinical and Molecular Medicine), Joyce L Browne (https://orcid.org/0000-0001-7048-3245) (Assistant Professor, Maternal and Global Health), Kajantie Eero (https://orcid.org/0000-0001-7081-8391) (Professor, Clinical Research), Ragnhild B Skråstad (https://orcid.org/0000-0003-3810-0413) (Associate Professor, Clinical and Molecular Medicine) and Camilla Haavaldsen contributed data to the project and provided input at all stages of the project.
John Allotey and Melanie Smuk mapped the variables in the available data sets, and cleaned and quality checked the data.
Melanie Smuk and Claire Chan (https://orcid.org/0000-0002-0821-4068) (Statistician, Statistics) harmonised the data.
Kym IE Snell, Melanie Smuk, Richard D Riley and Richard Hooper conducted the data analysis.
All authors critically appraised and provided feedback and input into the drafts and final version of the report.
Publications
Allotey J, Snell KIE, Chan C, Hooper R, Dodds J, Rogozinska E, et al. External validation, update and development of prediction models for pre-eclampsia using an individual participant data (IPD) meta-analysis: the International Prediction of Pregnancy Complication Network (IPPIC pre-eclampsia) protocol. Diagn Progn Res 2017;1:16.
Snell KIE, Allotey J, Smuk M, Hooper R, Chan C, Ahmed A, et al. External validation of prognostic models predicting pre-eclampsia: individual participant data meta-analysis. BMC Med 2020;18:302.
Data-sharing statement
All data requests should be submitted to the corresponding author for consideration. Access to available anonymised data may be granted following review and appropriate agreements being in place.
Disclaimers
This report presents independent research funded by the National Institute for Health Research (NIHR). The views and opinions expressed by authors in this publication are those of the authors and do not necessarily reflect those of the NHS, the NIHR, NETSCC, the HTA programme or the Department of Health and Social Care. If there are verbatim quotations included in this publication the views and opinions expressed by the interviewees are those of the interviewees and do not necessarily reflect those of the authors, those of the NHS, the NIHR, NETSCC, the HTA programme or the Department of Health and Social Care.
References
- Brown MA, Hague WM, Higgins J, Lowe S, McCowan L, Oats J, et al. The detection, investigation and management of hypertension in pregnancy: executive summary. Aust N Z J Obstet Gynaecol 2000;40:133-8. https://doi.org/10.1111/j.1479-828x.2000.tb01136.x.
- Sibai B, Dekker G, Kupferminc M. Pre-eclampsia. Lancet 2005;365:785-99. https://doi.org/10.1016/S0140-6736(05)17987-2.
- Sibai BM. Diagnosis and management of gestational hypertension and preeclampsia. Obstet Gynecol 2003;102:181-92. https://doi.org/10.1097/00006250-200307000-00033.
- Tranquilli AL, Dekker G, Magee L, Roberts J, Sibai BM, Steyn W, et al. The classification, diagnosis and management of the hypertensive disorders of pregnancy: a revised statement from the ISSHP. Pregnancy Hypertens 2014;4:97-104. https://doi.org/10.1016/j.preghy.2014.02.001.
- Staff AC, Benton SJ, von Dadelszen P, Roberts JM, Taylor RN, Powers RW, et al. Redefining preeclampsia using placenta-derived biomarkers. Hypertension 2013;61:932-42. https://doi.org/10.1161/HYPERTENSIONAHA.111.00250.
- Duley L. The global impact of pre-eclampsia and eclampsia. Semin Perinatol 2009;33:130-7. https://doi.org/10.1053/j.semperi.2009.02.010.
- Gardosi J, Kady SM, McGeown P, Francis A, Tonks A. Classification of stillbirth by relevant condition at death (ReCoDe): population based cohort study. BMJ 2005;331:1113-17. https://doi.org/10.1136/bmj.38629.587639.7C.
- Iams JD, Goldenberg RL, Mercer BM, Moawad A, Thom E, Meis PJ, et al. The Preterm Prediction Study: recurrence risk of spontaneous preterm birth. National Institute of Child Health and Human Development Maternal-Fetal Medicine Units Network. Am J Obstet Gynecol 1998;178:1035-40. https://doi.org/10.1016/S0002-9378(98)70544-7.
- Davies EL, Bell JS, Bhattacharya S. Preeclampsia and preterm delivery: a population-based case–control study. Hypertens Pregnancy 2016;35:510-19. https://doi.org/10.1080/10641955.2016.1190846.
- Tang LC, Kwok AC, Wong AY, Lee YY, Sun KO, So AP. Critical care in obstetrical patients: an eight-year review. Chin Med J 1997;110:936-41.
- Khan KS, Wojdyla D, Say L, Gülmezoglu AM, Van Look PF. WHO analysis of causes of maternal death: a systematic review. Lancet 2006;367:1066-74. https://doi.org/10.1016/S0140-6736(06)68397-9.
- World Health Organization . The World Health Report: 2005 – Make Every Mother and Child Count 2005. www.who.int/whr/2005/whr2005_en.pdf (accessed 6 December 2018).
- Carty DM, Delles C, Dominiczak AF. Preeclampsia and future maternal health. J Hypertens 2010;28:1349-55. https://doi.org/10.1097/HJH.0b013e32833a39d0.
- Bellamy L, Casas JP, Hingorani AD, Williams DJ. Pre-eclampsia and risk of cardiovascular disease and cancer in later life: systematic review and meta-analysis. BMJ 2007;335. https://doi.org/10.1136/bmj.39335.385301.BE.
- Thoulass JC, Robertson L, Denadai L, Black C, Crilly M, Iversen L, et al. Hypertensive disorders of pregnancy and adult offspring cardiometabolic outcomes: a systematic review of the literature and meta-analysis. J Epidemiol Community Health 2016;70:414-22. https://doi.org/10.1136/jech-2015-205483.
- Davis EF, Lazdam M, Lewandowski AJ, Worton SA, Kelly B, Kenworthy Y, et al. Cardiovascular risk factors in children and young adults born to preeclamptic pregnancies: a systematic review. Pediatrics 2012;129:e1552-61. https://doi.org/10.1542/peds.2011-3093.
- Tuovinen S, Eriksson JG, Kajantie E, Lahti J, Pesonen AK, Heinonen K, et al. Maternal hypertensive disorders in pregnancy and self-reported cognitive impairment of the offspring 70 years later: the Helsinki Birth Cohort Study. Am J Obstet Gynecol 2013;208:200-9. https://doi.org/10.1016/j.ajog.2012.12.017.
- Tuovinen S, Räikkönen K, Kajantie E, Pesonen AK, Heinonen K, Osmond C, et al. Depressive symptoms in adulthood and intrauterine exposure to pre-eclampsia: the Helsinki Birth Cohort Study. BJOG 2010;117:1236-42. https://doi.org/10.1111/j.1471-0528.2010.02634.x.
- Crispi F, Domínguez C, Llurba E, Martín-Gallán P, Cabero L, Gratacós E. Placental angiogenic growth factors and uterine artery Doppler findings for characterization of different subsets in preeclampsia and in isolated intrauterine growth restriction. Am J Obstet Gynecol 2006;195:201-7. https://doi.org/10.1016/j.ajog.2006.01.014.
- Crispi F, Llurba E, Domínguez C, Martín-Gallán P, Cabero L, Gratacós E. Predictive value of angiogenic factors and uterine artery Doppler for early- versus late-onset pre-eclampsia and intrauterine growth restriction. Ultrasound Obstet Gynecol 2008;31:303-9. https://doi.org/10.1002/uog.5184.
- Valensise H, Vasapollo B, Gagliardi G, Novelli GP. Early and late preeclampsia: two different maternal hemodynamic states in the latent phase of the disease. Hypertension 2008;52:873-80. https://doi.org/10.1161/HYPERTENSIONAHA.108.117358.
- Redman CW, Sargent IL, Staff AC. IFPA Senior Award Lecture: making sense of pre-eclampsia – two placental causes of preeclampsia?. Placenta 2014:S20-5. https://doi.org/10.1016/j.placenta.2013.12.008.
- MacKay AP, Berg CJ, Atrash HK. Pregnancy-related mortality from preeclampsia and eclampsia. Obstet Gynecol 2001;97:533-8. https://doi.org/10.1097/00006250-200104000-00011.
- von Dadelszen P, Payne B, Li J, Ansermino JM, Broughton Pipkin F, Côté AM, et al. Prediction of adverse maternal outcomes in pre-eclampsia: development and validation of the fullPIERS model. Lancet 2011;377:219-27. https://doi.org/10.1016/S0140-6736(10)61351-7.
- von Dadelszen P, Magee LA, Roberts JM. Subclassification of preeclampsia. Hypertens Pregnancy 2003;22:143-8. https://doi.org/10.1081/PRG-120021060.
- Lisonkova S, Joseph KS. Incidence of preeclampsia: risk factors and outcomes associated with early- versus late-onset disease. Am J Obstet Gynecol 2013;209:544-544.e12. https://doi.org/10.1016/j.ajog.2013.08.019.
- Murphy DJ, Stirrat GM. Mortality and morbidity associated with early-onset preeclampsia. Hypertens Pregnancy 2000;19:221-31. https://doi.org/10.1081/PRG-100100138.
- Sibai BM, Ramadan MK, Usta I, Salama M, Mercer BM, Friedman SA. Maternal morbidity and mortality in 442 pregnancies with hemolysis, elevated liver enzymes, and low platelets (HELLP syndrome). Am J Obstet Gynecol 1993;169:1000-6. https://doi.org/10.1016/0002-9378(93)90043-I.
- Churchill D, Duley L. Interventionist versus expectant care for severe pre-eclampsia before term. Cochrane Database Syst Rev 2002;3. https://doi.org/10.1002/14651858.CD003106.
- Mangham LJ, Petrou S, Doyle LW, Draper ES, Marlow N. The cost of preterm birth throughout childhood in England and Wales. Pediatrics 2009;123:e312-27. https://doi.org/10.1542/peds.2008-1827.
- Douglas KA, Redman CW. Eclampsia in the United Kingdom. BMJ 1994;309:1395-400. https://doi.org/10.1136/bmj.309.6966.1395.
- Sibai BM. Management of late preterm and early-term pregnancies complicated by mild gestational hypertension/pre-eclampsia. Semin Perinatol 2011;35:292-6. https://doi.org/10.1053/j.semperi.2011.05.010.
- Bujold E, Roberge S, Lacasse Y, Bureau M, Audibert F, Marcoux S, et al. Prevention of preeclampsia and intrauterine growth restriction with aspirin started in early pregnancy: a meta-analysis. Obstet Gynecol 2010;116:402-14. https://doi.org/10.1097/AOG.0b013e3181e9322a.
- Rolnik DL, Wright D, Poon LC, O’Gorman N, Syngelaki A, de Paco Matallana C, et al. Aspirin versus placebo in pregnancies at high risk for preterm preeclampsia. N Engl J Med 2017;377:613-22. https://doi.org/10.1056/NEJMoa1704559.
- National Institute for Health and Care Excellence . Hypertension in Pregnancy: The Management of Hypertensive Disorders During Pregnancy 2010. www.nice.org.uk/guidance/cg107 (accessed 31 March 2016).
- National Institute for Health and Care Excellence . Hypertension in Pregnancy: Diagnosis and Management 2019. www.nice.org.uk/guidance/cg107.
- Townsend R, Khalil A, Premakumar Y, Allotey J, Snell KIE, Chan C, et al. Prediction of pre-eclampsia: review of reviews. Ultrasound Obstet Gynecol 2019;54:16-27. https://doi.org/10.1002/uog.20117.
- Allen RE, Rogozinska E, Cleverly K, Aquilina J, Thangaratinam S. Abnormal blood biomarkers in early pregnancy are associated with preeclampsia: a meta-analysis. Eur J Obstet Gynecol Reprod Biol 2014;182:194-201. https://doi.org/10.1016/j.ejogrb.2014.09.027.
- Velauthar L, Plana MN, Kalidindi M, Zamora J, Thilaganathan B, Illanes SE, et al. First-trimester uterine artery Doppler and adverse pregnancy outcome: a meta-analysis involving 55,974 women. Ultrasound Obstet Gynecol 2014;43:500-7. https://doi.org/10.1002/uog.13275.
- Conde-Agudelo A, Villar J, Lindheimer M. World Health Organization systematic review of screening tests for preeclampsia. Obstet Gynecol 2004;104:1367-91. https://doi.org/10.1097/01.AOG.0000147599.47713.5d.
- Giguère Y, Charland M, Bujold E, Bernard N, Grenier S, Rousseau F, et al. Combining biochemical and ultrasonographic markers in predicting preeclampsia: a systematic review. Clin Chem 2010;56:361-75. https://doi.org/10.1373/clinchem.2009.134080.
- North RA, McCowan LM, Dekker GA, Poston L, Chan EH, Stewart AW, et al. Clinical risk prediction for pre-eclampsia in nulliparous women: development of model in international prospective cohort. BMJ 2011;342. https://doi.org/10.1136/bmj.d1875.
- Kleinrouweler CE, Cheong-See FM, Collins GS, Kwee A, Thangaratinam S, Khan KS, et al. Prognostic models in obstetrics: available, but far from applicable. Am J Obstet Gynecol 2016;214:79-90. https://doi.org/10.1016/j.ajog.2015.06.013.
- Meertens LJE, Scheepers HCJ, van Kuijk SMJ, Aardenburg R, van Dooren IMA, Langenveld J, et al. External validation and clinical usefulness of first trimester prediction models for the risk of preeclampsia: a prospective cohort study. Fetal Diagn Ther 2019;45:381-93. https://doi.org/10.1159/000490385.
- Farina A, Rapacchia G, Freni Sterrantino A, Pula G, Morano D, Rizzo N. Prospective evaluation of ultrasound and biochemical-based multivariable models for the prediction of late pre-eclampsia. Prenat Diagn 2011;31:1147-52. https://doi.org/10.1002/pd.2849.
- Herraiz I, Arbués J, Camaño I, Gómez-Montes E, Grañeras A, Galindo A. Application of a first-trimester prediction model for pre-eclampsia based on uterine arteries and maternal history in high-risk pregnancies. Prenat Diagn 2009;29:1123-9. https://doi.org/10.1002/pd.2383.
- Debray TP, Moons KG, Ahmed I, Koffijberg H, Riley RD. A framework for developing, implementing, and evaluating clinical prediction models in an individual participant data meta-analysis. Stat Med 2013;32:3158-80. https://doi.org/10.1002/sim.5732.
- Riley RD, Ensor J, Snell KI, Debray TP, Altman DG, Moons KG, et al. External validation of clinical prediction models using big datasets from e-health records or IPD meta-analysis: opportunities and challenges. BMJ 2016;353. https://doi.org/10.1136/bmj.i3140.
- Debray TP, Riley RD, Rovers MM, Reitsma JB, Moons KG, Cochrane IPD. Meta-analysis Methods group. Individual participant data (IPD) meta-analyses of diagnostic and prognostic modelling studies: guidance on their use. PLOS Med 2015;12. https://doi.org/10.1371/journal.pmed.1001886.
- Riley RD, Lambert PC, Abo-Zaid G. Meta-analysis of individual participant data: rationale, conduct, and reporting. BMJ 2010;340. https://doi.org/10.1136/bmj.c221.
- Allotey J, Snell KIE, Chan C, Hooper R, Dodds J, Rogozinska E, et al. External validation, update and development of prediction models for pre-eclampsia using an Individual Participant Data (IPD) meta-analysis: the International Prediction of Pregnancy Complication Network (IPPIC pre-eclampsia) protocol. Diagn Progn Res 2017;1. https://doi.org/10.1186/s41512-017-0016-z.
- Snell KIE, Allotey J, Smuk M, Hooper R, Chan C, Ahmed A, et al. External validation of prognostic models predicting pre-eclampsia: individual participant data meta-analysis. BMC Med 2020;18. https://doi.org/10.1186/s12916-020-01766-9.
- Altman DG, Vergouwe Y, Royston P, Moons KG. Prognosis and prognostic research: validating a prognostic model. BMJ 2009;338. https://doi.org/10.1136/bmj.b605.
- Moons KG, Royston P, Vergouwe Y, Grobbee DE, Altman DG. Prognosis and prognostic research: what, why, and how?. BMJ 2009;338. https://doi.org/10.1136/bmj.b375.
- Royston P, Moons KG, Altman DG, Vergouwe Y. Prognosis and prognostic research: developing a prognostic model. BMJ 2009;338. https://doi.org/10.1136/bmj.b604.
- Moons KG, Altman DG, Reitsma JB, Ioannidis JP, Macaskill P, Steyerberg EW, et al. Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD): explanation and elaboration. Ann Intern Med 2015;162:W1-73. https://doi.org/10.7326/M14-0698.
- Stewart LA, Clarke M, Rovers M, Riley RD, Simmonds M, Stewart G, et al. PRISMA-IPD Development Group . Preferred Reporting Items for Systematic Review and Meta-Analyses of individual participant data: the PRISMA-IPD statement. JAMA 2015;313:1657-65. https://doi.org/10.1001/jama.2015.3656.
- Accuracy of Clinical Characteristics, Biochemical and Ultrasound Markers in the Prediction of Pre-Eclampsia: An Individual Participant Data (IPD) Meta-Analysis 2015. www.crd.york.ac.uk/PROSPERO/display_record.asp?ID=CRD42015029349 (accessed 27 March 2017).
- Gardosi J, Clausson B, Francis A. The value of customised centiles in assessing perinatal mortality risk associated with parity and maternal size. BJOG 2009;116:1356-63. https://doi.org/10.1111/j.1471-0528.2009.02245.x.
- Ahmed A. New insights into the etiology of preeclampsia: identification of key elusive factors for the vascular complications. Thromb Res 2011;127:72-5. https://doi.org/10.1016/S0049-3848(11)70020-2.
- Bloomenthal D, von Dadelszen P, Liston R, Magee L, Tsang P. The effect of factor V Leiden carriage on maternal and fetal health. CMAJ 2002;167:48-54.
- Bramham K, Parnell B, Nelson-Piercy C, Seed PT, Poston L, Chappell LC. Chronic hypertension and pregnancy outcomes: systematic review and meta-analysis. BMJ 2014;348. https://doi.org/10.1136/bmj.g2301.
- Cnossen JS, Morris RK, ter Riet G, Mol BW, van der Post JA, Coomarasamy A, et al. Use of uterine artery Doppler ultrasonography to predict pre-eclampsia and intrauterine growth restriction: a systematic review and bivariable meta-analysis. CMAJ 2008;178:701-11. https://doi.org/10.1503/cmaj.070430.
- Gallos ID, Sivakumar K, Kilby MD, Coomarasamy A, Thangaratinam S, Vatish M. Pre-eclampsia is associated with, and preceded by, hypertriglyceridaemia: a meta-analysis. BJOG 2013;120:1321-32. https://doi.org/10.1111/1471-0528.12375.
- Kleinrouweler CE, Wiegerinck MM, Ris-Stalpers C, Bossuyt PM, van der Post JA, von Dadelszen P, et al. Accuracy of circulating placental growth factor, vascular endothelial growth factor, soluble fms-like tyrosine kinase 1 and soluble endoglin in the prediction of pre-eclampsia: a systematic review and meta-analysis. BJOG 2012;119:778-87. https://doi.org/10.1111/j.1471-0528.2012.03311.x.
- Leeflang MM, Cnossen JS, van der Post JA, Mol BW, Khan KS, ter Riet G. Accuracy of fibronectin tests for the prediction of pre-eclampsia: a systematic review. Eur J Obstet Gynecol Reprod Biol 2007;133:12-9. https://doi.org/10.1016/j.ejogrb.2007.01.003.
- Meads CA, Cnossen JS, Meher S, Juarez-Garcia A, ter Riet G, Duley L, et al. Methods of prediction and prevention of pre-eclampsia: systematic reviews of accuracy and effectiveness literature with economic modelling. Health Technol Assess 2008;12. https://doi.org/10.3310/hta12060.
- Morris RK, Cnossen JS, Langejans M, Robson SC, Kleijnen J, Ter Riet G, et al. Serum screening with Down’s syndrome markers to predict pre-eclampsia and small for gestational age: systematic review and meta-analysis. BMC Pregnancy Childbirth 2008;8. https://doi.org/10.1186/1471-2393-8-33.
- Steegers EA, von Dadelszen P, Duvekot JJ, Pijnenborg R. Pre-eclampsia. Lancet 2010;376:631-44. https://doi.org/10.1016/S0140-6736(10)60279-6.
- Thangaratinam S, Langenveld J, Mol BW, Khan KS. Prediction and primary prevention of pre-eclampsia. Best Pract Res Clin Obstet Gynaecol 2011;25:419-33. https://doi.org/10.1016/j.bpobgyn.2011.02.008.
- van der Tuuk K, Koopmans CM, Groen H, Aarnoudse JG, van den Berg PP, van Beek JJ, et al. Prediction of progression to a high risk situation in women with gestational hypertension or mild pre-eclampsia at term. Aust N Z J Obstet Gynaecol 2011;51:339-46. https://doi.org/10.1111/j.1479-828X.2011.01311.x.
- von Dadelszen P, Firoz T, Donnay F, Gordon R, Justus Hofmeyr G, Lalani S, et al. Preeclampsia in low and middle income countries-health services lessons learned from the PRE-EMPT (PRE-Eclampsia-Eclampsia Monitoring, Prevention and Treatment) project. J Obstet Gynaecol Can 2012;34:917-26. https://doi.org/10.1016/S1701-2163(16)35405-6.
- American College of Obstetricians and Gynecologists; Task Force on Hypertension in Pregnancy . Hypertension in pregnancy. Report of the American College of Obstetricians and Gynecologists’ Task Force on Hypertension in Pregnancy. Obstet Gynecol 2013;122:1122-31.
- von Dadelszen P, Sawchuck D, Justus Hofmeyr G, Magee LA, Bracken H, Mathai M, et al. PRE-EMPT (PRE-eclampsia-Eclampsia Monitoring, Prevention and Treatment): a low and middle income country initiative to reduce the global burden of maternal, fetal and infant death and disease related to pre-eclampsia. Pregnancy Hypertens 2013;3:199-202. https://doi.org/10.1016/j.preghy.2013.06.002.
- GONet: The Global Obstetrics Network n.d. www.globalobstetricsnetwork.org/ (accessed 14 March 2019).
- Myatt L, Redman CW, Staff AC, Hansson S, Wilson ML, Laivuori H, et al. Strategy for standardization of preeclampsia research study design. Hypertension 2014;63:1293-301. https://doi.org/10.1161/HYPERTENSIONAHA.113.02664.
- Gardosi J, Francis A. Customised Centile Calculator. GROW version 8.0.4. Gestation Network; 2018.
- Wolff RF, Moons KGM, Riley RD, Whiting PF, Westwood M, Collins GS, et al. PROBAST: a tool to assess the risk of bias and applicability of prediction model studies. Ann Intern Med 2019;170:51-8. https://doi.org/10.7326/M18-1376.
- Resche-Rigon M, White IR. Multiple imputation by chained equations for systematically and sporadically missing multilevel data. Stat Methods Med Res 2018;27:1634-49. https://doi.org/10.1177/0962280216666564.
- Jolani S, Debray TP, Koffijberg H, van Buuren S, Moons KG. Imputation of systematically missing predictors in an individual participant data meta-analysis: a generalized approach using MICE. Stat Med 2015;34:1841-63. https://doi.org/10.1002/sim.6451.
- Meng XL. Multiple-imputation inferences with uncongenial sources of input. Statist Sci 1994;9:538-58. https://doi.org/10.1214/ss/1177010269.
- Little RJA, Rubin DB. Statistical Analysis with Missing Data. New York, NY: John Wiley; 2002.
- Hosmer DW, Lemeshow S. Applied Logistic Regression. Hoboken, NJ: John Wiley & Sons; 2000.
- Rubin DB. Multiple Imputation for Nonresponse in Surveys. New York, NY: Wiley; 1987.
- Snell KI, Ensor J, Debray TP, Moons KG, Riley RD. Meta-analysis of prediction model performance across multiple studies: which scale helps ensure between-study normality for the C-statistic and calibration measures?. Stat Methods Med Res 2018;27:3505-22. https://doi.org/10.1177/0962280217705678.
- Debray TP, Damen JA, Snell KI, Ensor J, Hooft L, Reitsma JB, et al. A guide to systematic review and meta-analysis of prediction model performance. BMJ 2017;356. https://doi.org/10.1136/bmj.i6460.
- Snell KI, Hua H, Debray TP, Ensor J, Look MP, Moons KG, et al. Multivariate meta-analysis of individual participant data helped externally validate the performance and implementation of a prediction model. J Clin Epidemiol 2016;69:40-5. https://doi.org/10.1016/j.jclinepi.2015.05.009.
- Hartung J, Knapp G. A refined method for the meta-analysis of controlled clinical trials with binary outcome. Stat Med 2001;20:3875-89. https://doi.org/10.1002/sim.1009.
- Vickers AJ, Elkin EB. Decision curve analysis: a novel method for evaluating prediction models. Med Decis Making 2006;26:565-74. https://doi.org/10.1177/0272989X06295361.
- Vickers AJ, Van Calster B, Steyerberg EW. Net benefit approaches to the evaluation of prediction models, molecular markers, and diagnostic tests. BMJ 2016;352. https://doi.org/10.1136/bmj.i6.
- Quartagno M, Carpenter J. Multilevel Joint Modelling Multiple Imputation: Package ‘jomo’, Version 2.6–6, License GPL-2 n.d. https://cran.r-project.org/web/packages/jomo/jomo.pdf (accessed 3 May 2018).
- Quartagno M, Carpenter JR. Multiple imputation for IPD meta-analysis: allowing for heterogeneity and studies with missing covariates. Stat Med 2016;35:2938-54. https://doi.org/10.1002/sim.6837.
- Sauerbrei W. The use of resampling methods to simplify regression models in medical statistics. J Royal Stat Soc 1999;48:313-29. https://doi.org/10.1111/1467-9876.00155.
- Heinze G, Wallisch C, Dunkler D. Variable selection – a review and recommendations for the practicing statistician. Biom J 2018;60:431-49. https://doi.org/10.1002/bimj.201700067.
- Wood AM, Royston P, White IR. The estimation and use of predictions for the assessment of model performance using large samples with multiply imputed data. Biom J 2015;57:614-32. https://doi.org/10.1002/bimj.201400004.
- Royston P, Parmar MK, Sylvester R. Construction and validation of a prognostic model across several studies, with an application in superficial bladder cancer. Stat Med 2004;23:907-26. https://doi.org/10.1002/sim.1691.
- Harrell FE. Regression Modeling Strategies, with Applications to Linear Models, Logistic Regression, and Survival Analysis. New York, NY: Springer; 2015.
- Steyerberg EW. Clinical Prediction Models. A Practical Approach to Development, Validation, and Updating. New York, NY: Springer; 2009.
- Riley RD, van der Windt D, Croft P, Moons KGM. Prognosis Research in Healthcare: Concepts, Methods and Impact. Oxford: Oxford University Press; 2019.
- IntHout J, Ioannidis JP, Rovers MM, Goeman JJ. Plea for routinely presenting prediction intervals in meta-analysis. BMJ Open 2016;6. https://doi.org/10.1136/bmjopen-2015-010247.
- IntHout J, Ioannidis JP, Borm GF. The Hartung-Knapp-Sidik-Jonkman method for random effects meta-analysis is straightforward and considerably outperforms the standard DerSimonian-Laird method. BMC Med Res Methodol 2014;14. https://doi.org/10.1186/1471-2288-14-25.
- Knapp G, Hartung J. Improved tests for a random effects meta-regression with a single covariate. Stat Med 2003;22:2693-710. https://doi.org/10.1002/sim.1482.
- Fraser A, Macdonald-Wallis C, Tilling K, Boyd A, Golding J, Davey Smith G, et al. Cohort profile: the Avon Longitudinal Study of Parents and Children: ALSPAC mothers cohort. Int J Epidemiol 2013;42:97-110. https://doi.org/10.1093/ije/dys066.
- Better Outcomes Registry &Amp; Network (BORN) Ontario n.d. www.bornontario.ca/en/about-born/ (accessed 14 March 2019).
- Japan Society of Obstetrics and Gynecology n.d. www.jsog.or.jp/modules/en/index.php?content_id=1 (accessed 14 March 2019).
- Carter J, Seed PT, Tribe RM, David A, Lachelin G, Shennan AH, et al. Saliva progesterone for prediction of spontaneous preterm birth: the POPPY study. Pregnancy Outcome Poster Abstracts. BJOG 2017;124:122-54. https://doi.org/10.1111/1471-0528.14589.
- Al-Amin A, Rolnik DL, Black C, White A, Stolarek C, Brennecke S, et al. Accuracy of second trimester prediction of preterm preeclampsia by three different screening algorithms. Aust N Z J Obstet Gynaecol 2018;58:192-6. https://doi.org/10.1111/ajo.12689.
- Allen RE, Zamora J, Arroyo-Manzano D, Velauthar L, Allotey J, Thangaratinam S, et al. External validation of preexisting first trimester preeclampsia prediction models. Eur J Obstet Gynecol Reprod Biol 2017;217:119-25. https://doi.org/10.1016/j.ejogrb.2017.08.031.
- Andersen LB, Dechend R, Jørgensen JS, Luef BM, Nielsen J, Barington T, et al. Prediction of preeclampsia with angiogenic biomarkers. Results from the prospective Odense Child Cohort. Hypertens Pregnancy 2016;35:405-19. https://doi.org/10.3109/10641955.2016.1167219.
- Antsaklis A, Daskalakis G, Tzortzis E, Michalas S. The effect of gestational age and placental location on the prediction of pre-eclampsia by uterine artery Doppler velocimetry in low-risk nulliparous women. Ultrasound Obstet Gynecol 2000;16:635-9. https://doi.org/10.1046/j.1469-0705.2000.00288.x.
- Arenas J, Fernández-iñarea J, Rodríguez-mon C, Duplá B, Díez E, González-garcía A. Cribado con doppler de las arterias uterinas para la predicción de complicaciones de la gestación. Clínicae Investigación En Ginecología Y Obstetricia 2003;30:178-84. https://doi.org/10.1016/S0210-573X(03)77255-4.
- Askie LM, Duley L, Henderson-Smart DJ, Stewart LA. PARIS Collaborative Group . Antiplatelet agents for prevention of pre-eclampsia: a meta-analysis of individual patient data. Lancet 2007;369:1791-8. https://doi.org/10.1016/S0140-6736(07)60712-0.
- Audibert F, Boucoiran I, An N, Aleksandrov N, Delvin E, Bujold E, et al. Screening for preeclampsia using first-trimester serum markers and uterine artery Doppler in nulliparous women. Am J Obstet Gynecol 2010;203:383-8. https://doi.org/10.1016/j.ajog.2010.06.014.
- Ayorinde AA, Wilde K, Lemon J, Campbell D, Bhattacharya S. Data resource profile: the Aberdeen Maternity and Neonatal Databank (AMND). Int J Epidemiol 2016;45:389-94. https://doi.org/10.1093/ije/dyv356.
- Baschat AA, Magder LS, Doyle LE, Atlas RO, Jenkins CB, Blitzer MG. Prediction of preeclampsia utilizing the first trimester screening examination. Am J Obstet Gynecol 2014;211:514-7. https://doi.org/10.1016/j.ajog.2014.04.018.
- Brown MA, Mackenzie C, Dunsmuir W, Roberts L, Ikin K, Matthews J, et al. Can we predict recurrence of pre-eclampsia or gestational hypertension?. BJOG 2007;114:984-93. https://doi.org/10.1111/j.1471-0528.2007.01376.x.
- Cameroni I, Roncaglia N, Crippa I, Orsenigo F, Locatelli A, Vergani P, et al. P32.05: Uterine artery Doppler in a risk population: what’s its role in the prediction of severe pregnancy complications?. Ultrasound Obstet Gynecol 2008;32:421-2. https://doi.org/10.1002/uog.5992.
- Caradeux J, Serra R, Nien JK, Pérez-Sepulveda A, Schepeler M, Guerra F, et al. First trimester prediction of early onset preeclampsia using demographic, clinical, and sonographic data: a cohort study. Prenat Diagn 2013;33:732-6. https://doi.org/10.1002/pd.4113.
- Carbillon L. The imbalance of circulating angiogenic/antiangiogenic factors is mild or absent in obese women destined to develop preeclampsia. Hypertens Pregnancy 2014;33. https://doi.org/10.3109/10641955.2013.872252.
- Caritis S, Sibai B, Hauth J, Lindheimer MD, Klebanoff M, Thom E, et al. Low-dose aspirin to prevent preeclampsia in women at high risk. National Institute of Child Health and Human Development Network of Maternal-Fetal Medicine Units. N Engl J Med 1998;338:701-5. https://doi.org/10.1056/NEJM199803123381101.
- Chappell LC, Seed PT, Briley AL, Kelly FJ, Lee R, Hunt BJ, et al. Effect of antioxidants on the occurrence of pre-eclampsia in women at increased risk: a randomised trial. Lancet 1999;354:810-16. https://doi.org/10.1016/S0140-6736(99)80010-5.
- Chiswick C, Reynolds RM, Denison F, Drake AJ, Forbes S, Newby DE, et al. Effect of metformin on maternal and fetal outcomes in obese pregnant women (EMPOWaR): a randomised, double-blind, placebo-controlled trial. Lancet Diabetes Endocrinol 2015;3:778-86. https://doi.org/10.1016/S2213-8587(15)00219-3.
- Conserva V, Muggiasca M, Arrigoni L, Mantegazza V, Rossi E, Ferrazzi E. Recurrence and severity of abnormal pregnancy outcome in patients treated by low-molecular-weight heparin: a prospective pilot study. J Matern Fetal Neonatal Med 2012;25:1467-73. https://doi.org/10.3109/14767058.2011.643326.
- Facchinetti F, Marozio L, Frusca T, Grandone E, Venturini P, Tiscia GL, et al. Maternal thrombophilia and the risk of recurrence of preeclampsia. Am J Obstet Gynecol 2009;200:46-5. https://doi.org/10.1016/j.ajog.2008.07.032.
- Figueiró-Filho EA, Oliveira VMd, Coelho LR, Breda I. Marcadores séricos de trombofilias hereditárias e anticorpos antifosfolípides em gestantes com antecedentes de pré-eclâmpsia grave. Revista Brasileira De Ginecologia E Obstetrícia 2012;34:40-6. https://doi.org/10.1590/S0100-72032012000100008.
- Giguère Y, Massé J, Thériault S, Bujold E, Lafond J, Rousseau F, et al. Screening for pre-eclampsia early in pregnancy: performance of a multivariable model combining clinical characteristics and biochemical markers. BJOG 2015;122:402-10. https://doi.org/10.1111/1471-0528.13050.
- Girchenko P, Lahti M, Tuovinen S, Savolainen K, Lahti J, Binder EB, et al. Cohort profile: prediction and prevention of preeclampsia and intrauterine growth restriction (PREDO) study. Int J Epidemiol 2017;46:1380-1381g. https://doi.org/10.1093/ije/dyw154.
- Goetzinger KR, Singla A, Gerkowicz S, Dicke JM, Gray DL, Odibo AO. Predicting the risk of pre-eclampsia between 11 and 13 weeks’ gestation by combining maternal characteristics and serum analytes, PAPP-A and free β-hCG. Prenat Diagn 2010;30:1138-42. https://doi.org/10.1002/pd.2627.
- Goffinet F, Aboulker D, Paris-Llado J, Bucourt M, Uzan M, Papiernik E, et al. Screening with a uterine Doppler in low risk pregnant women followed by low dose aspirin in women with abnormal results: a multicenter randomised controlled trial. BJOG 2001;108:510-18. https://doi.org/10.1111/j.1471-0528.2001.00116.x.
- Gurgel Alves JA, Praciano de Sousa PC, Bezerra Maia e, Holanda Moura S, Kane SC, da Silva Costa F. First-trimester maternal ophthalmic artery Doppler analysis for prediction of pre-eclampsia. Ultrasound Obstet Gynecol 2014;44:411-18. https://doi.org/10.1002/uog.13338.
- Holzman C, Bullen B, Fisher R, Paneth N, Reuss L. Prematurity Study Group . Pregnancy outcomes and community health: the POUCH study of preterm delivery. Paediatr Perinat Epidemiol 2001;15:136-58. https://doi.org/10.1046/j.1365-3016.2001.00014.x.
- Huang T, Hoffman B, Meschino W, Kingdom J, Okun N. Prediction of adverse pregnancy outcomes by combinations of first and second trimester biochemistry markers used in the routine prenatal screening of Down syndrome. Prenat Diagn 2010;30:471-7. https://doi.org/10.1002/pd.2505.
- Jääskeläinen T, Heinonen S, Hämäläinen E, Pulkki K, Romppanen J, Laivuori H. FINNPEC . Angiogenic profile in the Finnish Genetics of Pre-Eclampsia Consortium (FINNPEC) cohort. Pregnancy Hypertens 2018;14:252-9. https://doi.org/10.1016/j.preghy.2018.03.004.
- Jaddoe VW, van Duijn CM, Franco OH, van der Heijden AJ, van Iizendoorn MH, de Jongste JC, et al. The Generation R Study: design and cohort update 2012. Eur J Epidemiol 2012;27:739-56. https://doi.org/10.1007/s10654-012-9735-1.
- Jenum AK, Sletner L, Voldner N, Vangen S, Mørkrid K, Andersen LF, et al. The STORK Groruddalen research programme: a population-based cohort study of gestational diabetes, physical activity, and obesity in pregnancy in a multiethnic population. Rationale, methods, study population, and participation rates. Scand J Public Health 2010;38:60-7. https://doi.org/10.1177/1403494810378921.
- Olsen J, Melbye M, Olsen SF, Sørensen TI, Aaby P, Andersen AM, et al. The Danish National Birth Cohort – its background, structure and aim. Scand J Public Health 2001;29:300-7. https://doi.org/10.1177/14034948010290040201.
- Khan F, Belch JJ, MacLeod M, Mires G. Changes in endothelial function precede the clinical disease in women in whom preeclampsia develops. Hypertension 2005;46:1123-8. https://doi.org/10.1161/01.HYP.0000186328.90667.95.
- Langenveld J, Buttinger A, van der Post J, Wolf H, Mol BW, Ganzevoort W. Recurrence risk and prediction of a delivery under 34 weeks of gestation after a history of a severe hypertensive disorder. BJOG 2011;118:589-95. https://doi.org/10.1111/j.1471-0528.2010.02842.x.
- Lecarpentier E, Tsatsaris V, Goffinet F, Cabrol D, Sibai B, Haddad B. Risk factors of superimposed preeclampsia in women with essential chronic hypertension treated before pregnancy. PLOS ONE 2013;8. https://doi.org/10.1371/journal.pone.0062140.
- Llurba E, Carreras E, Gratacós E, Juan M, Astor J, Vives A, et al. Maternal history and uterine artery Doppler in the assessment of risk for development of early- and late-onset preeclampsia and intrauterine growth restriction. Obstet Gynecol Int 2009;2009. https://doi.org/10.1155/2009/275613.
- Lykke JA, Paidas MJ, Langhoff-Roos J. Recurring complications in second pregnancy. Obstet Gynecol 2009;113:1217-24. https://doi.org/10.1097/AOG.0b013e3181a66f2d.
- Magnus P, Irgens LM, Haug K, Nystad W, Skjaerven R, Stoltenberg C. MoBa Study Group . Cohort profile: the Norwegian Mother and Child cohort study (MoBa). Int J Epidemiol 2006;35:1146-50. https://doi.org/10.1093/ije/dyl170.
- Makrides M, Gibson RA, McPhee AJ, Yelland L, Quinlivan J, Ryan P. DOMInO Investigative Team . Effect of DHA supplementation during pregnancy on maternal depression and neurodevelopment of young children: a randomized controlled trial. JAMA 2010;304:1675-83. https://doi.org/10.1001/jama.2010.1507.
- Massé J, Forest JC, Moutquin JM, Marcoux S, Brideau NA, Bélanger M. A prospective study of several potential biologic markers for early prediction of the development of preeclampsia. Am J Obstet Gynecol 1993;169:501-8. https://doi.org/10.1016/0002-9378(93)90608-L.
- Mbah AK, Sharma PP, Alio AP, Fombo DW, Bruder K, Salihu HM. Previous cesarean section, gestational age at first delivery and subsequent risk of pre-eclampsia in obese mothers. Arch Gynecol Obstet 2012;285:1375-81. https://doi.org/10.1007/s00404-011-2161-x.
- Mone F, Mulcahy C, McParland P, Stanton A, Culliton M, Downey P, et al. An open-label randomized-controlled trial of low dose aspirin with an early screening test for pre-eclampsia and growth restriction (TEST): trial protocol. Contemp Clin Trials 2016;49:143-8. https://doi.org/10.1016/j.cct.2016.07.003.
- Odibo AO, Zhong Y, Goetzinger KR, Odibo L, Bick JL, Bower CR, et al. First-trimester placental protein 13, PAPP-A, uterine artery Doppler and maternal characteristics in the prediction of pre-eclampsia. Placenta 2011;32:598-602. https://doi.org/10.1016/j.placenta.2011.05.006.
- Ohkuchi A, Minakami H, Sato I, Mori H, Nakano T, Tateno M. Predicting the risk of pre-eclampsia and a small-for-gestational-age infant by quantitative assessment of the diastolic notch in uterine artery flow velocity waveforms in unselected women. Ultrasound Obstet Gynecol 2000;16:171-8. https://doi.org/10.1046/j.1469-0705.2000.00192.x.
- Poston L, Bell R, Croker H, Flynn AC, Godfrey KM, Goff L, et al. Effect of a behavioural intervention in obese pregnant women (the UPBEAT study): a multicentre, randomised controlled trial. Lancet Diabetes Endocrinol 2015;3:767-77. https://doi.org/10.1016/S2213-8587(15)00227-2.
- Poston L, Briley AL, Seed PT, Kelly FJ, Shennan AH. Vitamins in Pre-eclampsia (VIP) Trial Consortium. Vitamin C and vitamin E in pregnant women at risk for pre-eclampsia (VIP trial): randomised placebo-controlled trial. Lancet 2006;367:1145-54. https://doi.org/10.1016/S0140-6736(06)68433-X.
- Prefumo F, Fratelli N, Ganapathy R, Bhide A, Frusca T, Thilaganathan B. First trimester uterine artery Doppler in women with previous pre-eclampsia. Acta Obstet Gynecol Scand 2008;87:1271-5. https://doi.org/10.1080/00016340802460347.
- Rang S, van Montfrans GA, Wolf H. Serial hemodynamic measurement in normal pregnancy, preeclampsia, and intrauterine growth restriction. Am J Obstet Gynecol 2008;198:519-9. https://doi.org/10.1016/j.ajog.2007.11.014.
- Rocha RS, Gurgel Alves JA, Bezerra Maia e, Holanda Moura S, Araujo Júnior E, Peixoto AB, et al. Simple approach based on maternal characteristics and mean arterial pressure for the prediction of preeclampsia in the first trimester of pregnancy. J Perinat Med 2017;45:843-9. https://doi.org/10.1515/jpm-2016-0418.
- Rocha RS, Gurgel Alves JA, Bezerra Maia e, Holanda Moura S, Araujo Júnior E, Martins WP, et al. Comparison of three algorithms for prediction preeclampsia in the first trimester of pregnancy. Pregnancy Hypertens 2017;10:113-17. https://doi.org/10.1016/j.preghy.2017.07.146.
- Rumbold AR, Crowther CA, Haslam RR, Dekker GA, Robinson JS. ACTS Study Group . Vitamins C and E and the risks of preeclampsia and perinatal complications. N Engl J Med 2006;354:1796-806. https://doi.org/10.1056/NEJMoa054186.
- Salim R, Czarnowicki T, Nachum Z, Shalev E. The impact of close surveillance on pregnancy outcome among women with a prior history of antepartum complications attributed to thrombosis: a cohort study. Reprod Biol Endocrinol 2008;6. https://doi.org/10.1186/1477-7827-6-55.
- Savitri AI, Zuithoff P, Browne JL, Amelia D, Baharuddin M, Grobbee DE, et al. Does pre-pregnancy BMI determine blood pressure during pregnancy? A prospective cohort study. BMJ Open 2016;6. https://doi.org/10.1136/bmjopen-2016-011626.
- Ferrazzani S, D’Alessio MC, Fatigante G, Soreca G, De Carolis S, Paradisi G, et al. Prophylaxis of recurrent preeclampsia: low-molecular-weight heparin plus low-dose aspirin versus low-dose aspirin alone. Hypertens Pregnancy 2006;25:115-27. https://doi.org/10.1080/10641950600745517.
- Sibai BM, Caritis SN, Thom E, Klebanoff M, McNellis D, Rocco L, et al. Prevention of preeclampsia with low-dose aspirin in healthy, nulliparous pregnant women. The National Institute of Child Health and Human Development Network of Maternal-Fetal Medicine Units. N Engl J Med 1993;329:1213-8. https://doi.org/10.1056/NEJM199310213291701.
- Skråstad RB, Hov GG, Blaas HG, Romundstad PR, Salvesen KÅ. Risk assessment for preeclampsia in nulliparous women at 11–13 weeks gestational age: prospective evaluation of two algorithms. BJOG 2015;122:1781-8. https://doi.org/10.1111/1471-0528.13194.
- Sovio U, White IR, Dacey A, Pasupathy D, Smith GCS. Screening for fetal growth restriction with universal third trimester ultrasonography in nulliparous women in the Pregnancy Outcome Prediction (POP) study: a prospective cohort study. Lancet 2015;386:2089-97. https://doi.org/10.1016/S0140-6736(15)00131-2.
- Staff AC, Braekke K, Harsem NK, Lyberg T, Holthe MR. Circulating concentrations of sFlt1 (soluble fms-like tyrosine kinase 1) in fetal and maternal serum during pre-eclampsia. Eur J Obstet Gynecol Reprod Biol 2005;122:33-9. https://doi.org/10.1016/j.ejogrb.2004.11.015.
- Stirrup OT, Khalil A, D’Antonio F, Thilaganathan B. Southwest Thames Obstetric Research Collaborative (STORK) . Fetal growth reference ranges in twin pregnancy: analysis of the Southwest Thames Obstetric Research Collaborative (STORK) multiple pregnancy cohort. Ultrasound Obstet Gynecol 2015;45:301-7. https://doi.org/10.1002/uog.14640.
- Trogstad L, Skrondal A, Stoltenberg C, Magnus P, Nesheim BI, Eskild A. Recurrence risk of preeclampsia in twin and singleton pregnancies. Am J Med Genet A 2004;126A:41-5. https://doi.org/10.1002/ajmg.a.20512.
- Van Der Linden EL, Browne JL, Vissers KM, Antwi E, Agyepong IA, Grobbee DE, et al. Maternal body mass index and adverse pregnancy outcomes: a Ghanaian cohort study. Obesity 2016;24:215-22. https://doi.org/10.1002/oby.21210.
- van Kuijk SM, Delahaije DH, Dirksen CD, Scheepers HC, Spaanderman ME, Ganzevoort W, et al. External validation of a model for periconceptional prediction of recurrent early-onset preeclampsia. Hypertens Pregnancy 2014;33:265-76. https://doi.org/10.3109/10641955.2013.872253.
- van Kuijk SM, Nijdam ME, Janssen KJ, Sep SJ, Peeters LL, Delahaije DH, et al. A model for preconceptional prediction of recurrent early-onset preeclampsia: derivation and internal validation. Reprod Sci 2011;18:1154-9. https://doi.org/10.1177/1933719111410708.
- van Oostwaard MF, Langenveld J, Bijloo R, Wong KM, Scholten I, Loix S, et al. Prediction of recurrence of hypertensive disorders of pregnancy between 34 and 37 weeks of gestation: a retrospective cohort study. BJOG 2012;119:840-7. https://doi.org/10.1111/j.1471-0528.2012.03312.x.
- van Oostwaard MF, Langenveld J, Schuit E, Wigny K, Van Susante H, Beune I, et al. Prediction of recurrence of hypertensive disorders of pregnancy in the term period, a retrospective cohort study. Pregnancy Hypertens 2014;4:194-202. https://doi.org/10.1016/j.preghy.2014.04.001.
- Vatten LJ, Eskild A, Nilsen TI, Jeansson S, Jenum PA, Staff AC. Changes in circulating level of angiogenic factors from the first to second trimester as predictors of preeclampsia. Am J Obstet Gynecol 2007;196:239-6. https://doi.org/10.1016/j.ajog.2006.10.909.
- Verlohren S, Galindo A, Schlembach D, Zeisler H, Herraiz I, Moertl MG, et al. An automated method for the determination of the sFlt-1/PIGF ratio in the assessment of preeclampsia. Am J Obstet Gynecol 2010;202:161-161.e11. https://doi.org/10.1016/j.ajog.2009.09.016.
- Verlohren S, Herraiz I, Lapaire O, Schlembach D, Moertl M, Zeisler H, et al. The sFlt-1/PlGF ratio in different types of hypertensive pregnancy disorders and its prognostic potential in preeclamptic patients. Am J Obstet Gynecol 2012;206:58-8. https://doi.org/10.1016/j.ajog.2011.07.037.
- Vinter CA, Jensen DM, Ovesen P, Beck-Nielsen H, Jørgensen JS. The LiP (Lifestyle in Pregnancy) study: a randomized controlled trial of lifestyle intervention in 360 obese pregnant women. Diabetes Care 2011;34:2502-7. https://doi.org/10.2337/dc11-1150.
- Vollebregt KC, Gisolf J, Guelen I, Boer K, van Montfrans G, Wolf H. Limited accuracy of the hyperbaric index, ambulatory blood pressure and sphygmomanometry measurements in predicting gestational hypertension and preeclampsia. J Hypertens 2010;28:127-34. https://doi.org/10.1097/HJH.0b013e32833266fc.
- Widmer M, Cuesta C, Khan KS, Conde-Agudelo A, Carroli G, Fusey S, et al. Accuracy of angiogenic biomarkers at ≤ 20 weeks’ gestation in predicting the risk of pre-eclampsia: a WHO multicentre study. Pregnancy Hypertens 2015;5:330-8. https://doi.org/10.1016/j.preghy.2015.09.004.
- Wright E, Audette MC, Ye XY, Keating S, Hoffman B, Lye SJ, et al. Maternal vascular malperfusion and adverse perinatal outcomes in low-risk nulliparous women. Obstet Gynecol 2017;130:1112-20. https://doi.org/10.1097/AOG.0000000000002264.
- Wright J, Small N, Raynor P, Tuffnell D, Bhopal R, Cameron N, et al. Cohort Profile: the Born in Bradford multi-ethnic family cohort study. Int J Epidemiol 2013;42:978-91. https://doi.org/10.1093/ije/dys112.
- Zhang J, Troendle JF, Levine RJ. Risks of hypertensive disorders in the second pregnancy. Paediatr Perinat Epidemiol 2001;15:226-31. https://doi.org/10.1046/j.1365-3016.2001.00347.x.
- Cnossen JS, Leeflang MM, de Haan EE, Mol BW, van der Post JA, Khan KS, et al. Accuracy of body mass index in predicting pre-eclampsia: bivariate meta-analysis. BJOG 2007;114:1477-85. https://doi.org/10.1111/j.1471-0528.2007.01483.x.
- O’Brien TE, Ray JG, Chan WS. Maternal body mass index and the risk of preeclampsia: a systematic overview. Epidemiology 2003;14:368-74. https://doi.org/10.1097/00001648-200305000-00020.
- Wang Z, Wang P, Liu H, He X, Zhang J, Yan H, et al. Maternal adiposity as an independent risk factor for pre-eclampsia: a meta-analysis of prospective cohort studies. Obes Rev 2013;14:508-21. https://doi.org/10.1111/obr.12025.
- Duckitt K, Harrington D. Risk factors for pre-eclampsia at antenatal booking: systematic review of controlled studies. BMJ 2005;330. https://doi.org/10.1136/bmj.38380.674340.E0.
- Alpoim PN, de Barros Pinheiro M, Junqueira DR, Freitas LG, das Graças Carvalho M, Fernandes AP, et al. Preeclampsia and ABO blood groups: a systematic review and meta-analysis. Mol Biol Rep 2013;40:2253-61. https://doi.org/10.1007/s11033-012-2288-2.
- England L, Zhang J. Smoking and risk of preeclampsia: a systematic review. Front Biosci 2007;12:2471-83. https://doi.org/10.2741/2248.
- Rebelo F, Schlüssel MM, Vaz JS, Franco-Sena AB, Pinto TJ, Bastos FI, et al. C-reactive protein and later preeclampsia: systematic review and meta-analysis taking into account the weight status. J Hypertens 2013;31:16-2. https://doi.org/10.1097/HJH.0b013e32835b0556.
- Luo ZC, An N, Xu HR, Larante A, Audibert F, Fraser WD. The effects and mechanisms of primiparity on the risk of pre-eclampsia: a systematic review. Paediatr Perinat Epidemiol 2007;21:36-45. https://doi.org/10.1111/j.1365-3016.2007.00836.x.
- Cnossen JS, Vollebregt KC, de Vrieze N, ter Riet G, Mol BW, Franx A, et al. Accuracy of mean arterial pressure and blood pressure measurements in predicting pre-eclampsia: systematic review and meta-analysis. BMJ 2008;336:1117-20. https://doi.org/10.1136/bmj.39540.522049.BE.
- Sgolastra F, Petrucci A, Severino M, Gatto R, Monaco A. Relationship between periodontitis and pre-eclampsia: a meta-analysis. PLOS ONE 2013;8. https://doi.org/10.1371/journal.pone.0071387.
- Kunnen A, van Doormaal JJ, Abbas F, Aarnoudse JG, van Pampus MG, Faas MM. Periodontal disease and pre-eclampsia: a systematic review. J Clin Periodontol 2010;37:1075-87. https://doi.org/10.1111/j.1600-051X.2010.01636.x.
- Morris RK, Riley RD, Doug M, Deeks JJ, Kilby MD. Diagnostic accuracy of spot urinary protein and albumin to creatinine ratios for detection of significant proteinuria or adverse pregnancy outcome in patients with suspected pre-eclampsia: systematic review and meta-analysis. BMJ 2012;345. https://doi.org/10.1136/bmj.e4342.
- Sanchez-Ramos L, Gillen G, Zamora J, Stenyakina A, Kaunitz AM. The protein-to-creatinine ratio for the prediction of significant proteinuria in patients at risk for preeclampsia: a meta-analysis. Ann Clin Lab Sci 2013;43:211-20.
- Wolf HT, Owe KM, Juhl M, Hegaard HK. Leisure time physical activity and the risk of pre-eclampsia: a systematic review. Matern Child Health J 2014;18:899-910. https://doi.org/10.1007/s10995-013-1316-8.
- Palmer KT, Bonzini M, Harris EC, Linaker C, Bonde JP. Work activities and risk of prematurity, low birth weight and pre-eclampsia: an updated review with meta-analysis. Occup Environ Med 2013;70:213-22. https://doi.org/10.1136/oemed-2012-101032.
- Bonzini M, Coggon D, Palmer KT. Risk of prematurity, low birthweight and pre-eclampsia in relation to working hours and physical activities: a systematic review. Occup Environ Med 2007;64:228-43. https://doi.org/10.1136/oem.2006.026872.
- Cnossen JS, de Ruyter-Hanhijärvi H, van der Post JA, Mol BW, Khan KS, ter Riet G. Accuracy of serum uric acid determination in predicting pre-eclampsia: a systematic review. Acta Obstet Gynecol Scand 2006;85:519-25. https://doi.org/10.1080/00016340500342037.
- Chien PF, Arnott N, Gordon A, Owen P, Khan KS. How useful is uterine artery Doppler flow velocimetry in the prediction of pre-eclampsia, intrauterine growth retardation and perinatal death? An overview. BJOG 2000;107:196-208. https://doi.org/10.1111/j.1471-0528.2000.tb11690.x.
- Kleinrouweler CE, Bossuyt PM, Thilaganathan B, Vollebregt KC, Arenas Ramírez J, Ohkuchi A, et al. Value of adding second-trimester uterine artery Doppler to patient characteristics in identification of nulliparous women at increased risk for pre-eclampsia: an individual patient data meta-analysis. Ultrasound Obstet Gynecol 2013;42:257-67. https://doi.org/10.1002/uog.12435.
- Pedrosa AC, Matias A. Screening for pre-eclampsia: a systematic review of tests combining uterine artery Doppler with other markers. J Perinat Med 2011;39:619-35. https://doi.org/10.1515/JPM.2011.077.
- Kosmas IP, Tatsioni A, Ioannidis JP. Association of Leiden mutation in factor V gene with hypertension in pregnancy and pre-eclampsia: a meta-analysis. J Hypertens 2003;21:1221-8. https://doi.org/10.1097/00004872-200307000-00002.
- Dudding T, Heron J, Thakkinstian A, Nurk E, Golding J, Pembrey M, et al. Factor V Leiden is associated with pre-eclampsia but not with fetal growth restriction: a genetic association study and meta-analysis. J Thromb Haemost 2008;6:1869-75. https://doi.org/10.1111/j.1538-7836.2008.03134.x.
- Rodger MA, Betancourt MT, Clark P, Lindqvist PG, Dizon-Townson D, Said J, et al. The association of factor V leiden and prothrombin gene mutation and placenta-mediated pregnancy complications: a systematic review and meta-analysis of prospective cohort studies. PLOS Med 2010;7. https://doi.org/10.1371/journal.pmed.1000292.
- Xia XP, Chang WW, Cao YX. Meta-analysis of the methylenetetrahydrofolate reductase C677T polymorphism and susceptibility to pre-eclampsia. Hypertens Res 2012;35:1129-34. https://doi.org/10.1038/hr.2012.117.
- Kosmas IP, Tatsioni A, Ioannidis JP. Association of C677T polymorphism in the methylenetetrahydrofolate reductase gene with hypertension in pregnancy and pre-eclampsia: a meta-analysis. J Hypertens 2004;22:1655-62. https://doi.org/10.1097/00004872-200409000-00004.
- Zusterzeel PL, Visser W, Blom HJ, Peters WH, Heil SG, Steegers EA. Methylenetetrahydrofolate reductase polymorphisms in preeclampsia and the HELLP syndrome. Hypertens Pregnancy 2000;19:299-307. https://doi.org/10.1081/PRG-100101991.
- Li X, Shen L, Tan H. Polymorphisms and plasma level of transforming growth factor-Beta 1 and risk for preeclampsia: a systematic review. PLOS ONE 2014;9. https://doi.org/10.1371/journal.pone.0097230.
- Wang XM, Wu HY, Qiu XJ. Methylenetetrahydrofolate reductase (MTHFR) gene C677T polymorphism and risk of preeclampsia: an updated meta-analysis based on 51 studies. Arch Med Res 2013;44:159-68. https://doi.org/10.1016/j.arcmed.2013.01.011.
- Widmer M, Villar J, Benigni A, Conde-Agudelo A, Karumanchi SA, Lindheimer M. Mapping the theories of preeclampsia and the role of angiogenic factors: a systematic review. Obstet Gynecol 2007;109:168-80. https://doi.org/10.1097/01.AOG.0000249609.04831.7c.
- Jacobs M, Nassar N, Roberts CL, Hadfield R, Morris JM, Ashton AW. Levels of soluble fms-like tyrosine kinase one in first trimester and outcomes of pregnancy: a systematic review. Reprod Biol Endocrinol 2011;9. https://doi.org/10.1186/1477-7827-9-77.
- Huppertz B, Meiri H, Gizurarson S, Osol G, Sammar M. Placental protein 13 (PP13): a new biological target shifting individualized risk assessment to personalized drug design combating pre-eclampsia. Hum Reprod Update 2013;19:391-405. https://doi.org/10.1093/humupd/dmt003.
- Schneuer FJ, Nassar N, Khambalia AZ, Tasevski V, Guilbert C, Ashton AW, et al. First trimester screening of maternal placental protein 13 for predicting preeclampsia and small for gestational age: in-house study and systematic review. Placenta 2012;33:735-40. https://doi.org/10.1016/j.placenta.2012.05.012.
- Lau SY, Guild SJ, Barrett CJ, Chen Q, McCowan L, Jordan V, et al. Tumour necrosis factor-alpha, interleukin-6, and interleukin-10 levels are altered in preeclampsia: a systematic review and meta-analysis. Am J Reprod Immunol 2013;70:412-27. https://doi.org/10.1111/aji.12138.
- Tabesh M, Salehi-Abargouei A, Tabesh M, Esmaillzadeh A. Maternal vitamin D status and risk of pre-eclampsia: a systematic review and meta-analysis. J Clin Endocrinol Metab 2013;98:3165-73. https://doi.org/10.1210/jc.2013-1257.
- Morgan JA, Bombell S, McGuire W. Association of plasminogen activator inhibitor-type 1 (–675 4G/5G) polymorphism with pre-eclampsia: systematic review. PLOS ONE 2013;8. https://doi.org/10.1371/journal.pone.0056907.
- Dai B, Liu T, Zhang B, Zhang X, Wang Z. The polymorphism for endothelial nitric oxide synthase gene, the level of nitric oxide and the risk for pre-eclampsia: a meta-analysis. Gene 2013;519:187-93. https://doi.org/10.1016/j.gene.2013.01.004.
- Chen Z, Xu F, Wei Y, Liu F, Qi H. Angiotensin-converting enzyme insertion/deletion polymorphism and risk of pregnancy hypertensive disorders: a meta-analysis. J Renin Angiotensin Aldosterone Syst 2012;13:184-95. https://doi.org/10.1177/1470320311427755.
- Qi HP, Fraser WD, Luo ZC, Julien P, Audibert F, Wei SQ. Endothelial nitric oxide synthase gene polymorphisms and risk of preeclampsia. Am J Perinatol 2013;30:795-804. https://doi.org/10.1055/s-0032-1333406.
- Zhao L, Bracken MB, Dewan AT, Chen S. Association between the SERPINE1 (PAI-1) 4G/5G insertion/deletion promoter polymorphism (rs1799889) and pre-eclampsia: a systematic review and meta-analysis. Mol Hum Reprod 2013;19:136-43. https://doi.org/10.1093/molehr/gas056.
- Zhao L, Dewan AT, Bracken MB. Association of maternal AGTR1 polymorphisms and preeclampsia: a systematic review and meta-analysis. J Matern Fetal Neonatal Med 2012;25:2676-80. https://doi.org/10.3109/14767058.2012.708370.
- Zhong WG, Wang Y, Zhu H, Zhao X. Meta analysis of angiotensin-converting enzyme I/D polymorphism as a risk factor for preeclampsia in Chinese women. Genet Mol Res 2012;11:2268-76. https://doi.org/10.4238/2012.May.21.1.
- Ni S, Zhang Y, Deng Y, Gong Y, Huang J, Bai Y, et al. AGT M235T polymorphism contributes to risk of preeclampsia: evidence from a meta-analysis. J Renin Angiotensin Aldosterone Syst 2012;13:379-86. https://doi.org/10.1177/1470320312440903.
- Hui D, Okun N, Murphy K, Kingdom J, Uleryk E, Shah PS. Combinations of maternal serum markers to predict preeclampsia, small for gestational age, and stillbirth: a systematic review. J Obstet Gynaecol Can 2012;34:142-53. https://doi.org/10.1016/S1701-2163(16)35157-X.
- Giguère Y, Charland M, Bujold E, Bernard N, Grenier S, Rousseau F, et al. Combining biochemical and ultrasonographic markers in predicting preeclampsia: a systematic review. Ann Biol Clin 2011;69:257-71. https://doi.org/10.1684/abc.2011.0572.
- Abou-Nassar K, Carrier M, Ramsay T, Rodger MA. The association between antiphospholipid antibodies and placenta mediated complications: a systematic review and meta-analysis. Thromb Res 2011;128:77-85. https://doi.org/10.1016/j.thromres.2011.02.006.
- do Prado AD, Piovesan DM, Staub HL, Horta BL. Association of anticardiolipin antibodies with preeclampsia: a systematic review and meta-analysis. Obstet Gynecol 2010;116:1433-43. https://doi.org/10.1097/AOG.0b013e3181fe02ec.
- Gupta S, Aziz N, Sekhon L, Agarwal R, Mansour G, Li J, et al. Lipid peroxidation and antioxidant status in preeclampsia: a systematic review. Obstet Gynecol Surv 2009;64:750-9. https://doi.org/10.1097/OGX.0b013e3181bea0ac.
- Bombell S, McGuire W. Tumour necrosis factor (–308A) polymorphism in pre-eclampsia: meta-analysis of 16 case–control studies. Aust N Z J Obstet Gynaecol 2008;48:547-51. https://doi.org/10.1111/j.1479-828X.2008.00924.x.
- Zafarmand MH, Nijdam ME, Franx A, Grobbee DE, Bots ML. The angiotensinogen gene M235T polymorphism and development of preeclampsia/eclampsia: a meta-analysis and meta-regression of observational studies. J Hypertens 2008;26:1726-34. https://doi.org/10.1097/HJH.0b013e3283009ca5.
- Wiwanitkit V. Correlation between plasminogen activator inhibitor-1 4G/5G polymorphism and pre-eclampsia: an appraisal. Arch Gynecol Obstet 2006;273:322-4. https://doi.org/10.1007/s00404-005-0117-8.
- Crovetto F, Figueras F, Triunfo S, Crispi F, Rodriguez-Sureda V, Dominguez C, et al. First trimester screening for early and late preeclampsia based on maternal characteristics, biophysical parameters, and angiogenic factors. Prenat Diagn 2015;35:183-91. https://doi.org/10.1002/pd.4519.
- Kuc S, Koster MP, Franx A, Schielen PC, Visser GH. Maternal characteristics, mean arterial pressure and serum markers in early prediction of preeclampsia. PLOS ONE 2013;8. https://doi.org/10.1371/journal.pone.0063546.
- Plasencia W, Maiz N, Bonino S, Kaihura C, Nicolaides KH. Uterine artery Doppler at 11 + 0 to 13 + 6 weeks in the prediction of pre-eclampsia. Ultrasound Obstet Gynecol 2007;30:742-9. https://doi.org/10.1002/uog.5157.
- Poon LC, Kametas NA, Chelemen T, Leal A, Nicolaides KH. Maternal risk factors for hypertensive disorders in pregnancy: a multivariate approach. J Hum Hypertens 2010;24:104-10. https://doi.org/10.1038/jhh.2009.45.
- Poon LC, Kametas NA, Pandeva I, Valencia C, Nicolaides KH. Mean arterial pressure at 11(+0) to 13(+6) weeks in the prediction of preeclampsia. Hypertension 2008;51:1027-33. https://doi.org/10.1161/HYPERTENSIONAHA.107.104646.
- Poon LC, Maiz N, Valencia C, Plasencia W, Nicolaides KH. First-trimester maternal serum pregnancy-associated plasma protein-A and pre-eclampsia. Ultrasound Obstet Gynecol 2009;33:23-3. https://doi.org/10.1002/uog.6280.
- Scazzocchio E, Figueras F, Crispi F, Meler E, Masoller N, Mula R, et al. Performance of a first-trimester screening of preeclampsia in a routine care low-risk setting. Am J Obstet Gynecol 2013;208:203-203.e10. https://doi.org/10.1016/j.ajog.2012.12.016.
- Wright D, Syngelaki A, Akolekar R, Poon LC, Nicolaides KH. Competing risks model in screening for preeclampsia by maternal characteristics and medical history. Am J Obstet Gynecol 2015;213:62-62.e10. https://doi.org/10.1016/j.ajog.2015.02.018.
- Yu CK, Smith GC, Papageorghiou AT, Cacho AM, Nicolaides KH. Fetal Medicine Foundation Second Trimester Screening Group . An integrated model for the prediction of pre-eclampsia using maternal factors and uterine artery Doppler velocimetry in unselected low-risk women. Am J Obstet Gynecol 2006;195. https://doi.org/10.1016/j.ajog.2006.06.010.
- Yu CK, Smith GC, Papageorghiou AT, Cacho AM, Nicolaides KH. Fetal Medicine Foundation Second Trimester Screening Group. Am J Obstet Gynecol 2005;193:429-36. https://doi.org/10.1016/j.ajog.2004.12.014.
- Albert A, Anderson JA. On the existence of maximum likelihood estimates in logistic regression models. Biometrika 1984;71. https://doi.org/10.1093/biomet/71.1.1.
- White IR, Royston P, Wood AM. Multiple imputation using chained equations: issues and guidance for practice. Stat Med 2011;30:377-99. https://doi.org/10.1002/sim.4067.
- Akolekar R, Zaragoza E, Poon LC, Pepes S, Nicolaides KH. Maternal serum placental growth factor at 11 + 0 to 13 + 6 weeks of gestation in the prediction of pre-eclampsia. Ultrasound Obstet Gynecol 2008;32:732-9. https://doi.org/10.1002/uog.6244.
- National Institute for Health and Care Excellence . Hypertension in Pregnancy: The Management of Hypertensive Disorders During Pregnancy 2010. www.nice.org.uk/guidance/cg107 (accessed 12 February 2015).
- O’Gorman N, Wright D, Syngelaki A, Akolekar R, Wright A, Poon LC, et al. Competing risks model in screening for preeclampsia by maternal factors and biomarkers at 11–13 weeks gestation. Am J Obstet Gynecol 2016;214:103-103.e12. https://doi.org/10.1016/j.ajog.2015.08.034.
- O’Gorman N, Wright D, Poon LC, Rolnik DL, Syngelaki A, de Alvarado M, et al. Multicenter screening for pre-eclampsia by maternal factors and biomarkers at 11–13 weeks’ gestation: comparison with NICE guidelines and ACOG recommendations. Ultrasound Obstet Gynecol 2017;49:756-60. https://doi.org/10.1002/uog.17455.
- Oliveira N, Magder LS, Blitzer MG, Baschat AA. First-trimester prediction of pre-eclampsia: external validity of algorithms in a prospectively enrolled cohort. Ultrasound Obstet Gynecol 2014;44:279-85. https://doi.org/10.1002/uog.13435.
- Park FJ, Leung CH, Poon LC, Williams PF, Rothwell SJ, Hyett JA. Clinical evaluation of a first trimester algorithm predicting the risk of hypertensive disease of pregnancy. Aust N Z J Obstet Gynaecol 2013;53:532-9. https://doi.org/10.1111/ajo.12126.
- Steyerberg EW, Vickers AJ, Cook NR, Gerds T, Gonen M, Obuchowski N, et al. Assessing the performance of prediction models: a framework for traditional and novel measures. Epidemiology 2010;21:128-38. https://doi.org/10.1097/EDE.0b013e3181c30fb2.
- Wright D, Tan MY, O’Gorman N, Poon LC, Syngelaki A, Wright A, et al. Predictive performance of the competing risk model in screening for preeclampsia. Am J Obstet Gynecol 2019;220:199-199.e13. https://doi.org/10.1016/j.ajog.2018.11.1087.
- Debray TP, Vergouwe Y, Koffijberg H, Nieboer D, Steyerberg EW, Moons KG. A new framework to enhance the interpretation of external validation studies of clinical prediction models. J Clin Epidemiol 2015;68:279-89. https://doi.org/10.1016/j.jclinepi.2014.06.018.
Appendix 1 Full list of authors
John Allotey,1,2† Kym IE Snell,3*† Melanie Smuk,2 Richard Hooper,2 Claire L Chan,2 Asif Ahmed,4 Lucy C Chappell,5 Peter von Dadelszen,5 Julie Dodds,1,2 Marcus Green,6 Louise Kenny,7 Asma Khalil,8 Khalid S Khan,1,2 Ben W Mol,9 Jenny Myers,10 Lucilla Poston,5 Basky Thilaganathan,8 Anne C Staff,11,12 Gordon CS Smith,13 Wessel Ganzevoort,14 Hannele Laivuori,15,16,17,18 Anthony O Odibo,19 Javier A Ramírez,20 John Kingdom,21 George Daskalakis,22 Diane Farrar,23 Ahmet A Baschat,24 Paul T Seed,5 Federico Prefumo,25 Fabricio da Silva Costa,26 Henk Groen,27 Francois Audibert,28 Jacques Massé,29 Ragnhild B Skråstad,30,31 Kjell Å Salvesen,32,33 Camilla Haavaldsen,34 Chie Nagata,35 Alice R Rumbold,36 Seppo Heinonen,37 Lisa M Askie,38 Luc JM Smits,39 Christina A Vinter,40 Per M Magnus,41 Kajantie Eero,42,43 Pia M Villa,37 Anne K Jenum,44 Louise B Andersen,45,46 Jane E Norman,47 Akihide Ohkuchi,48 Anne Eskild,34,49 Sohinee Bhattacharya,50 Fionnuala M McAuliffe,51 Alberto Galindo,52,53 Ignacio Herraiz,54 Lionel Carbillon,55 Kerstin Klipstein-Grobusch,56 SeonAe Yeo,57 Helena J Teede,58 Joyce L Browne,56 Karel GM Moons,56,59 Richard D Riley3 and Shakila Thangaratinam1,2 on behalf of the IPPIC Collaborative Network§
Appendix 2 Search strategies
| Set# | Searched for |
|---|---|
| S1 | MESH.EXACT(“Pre-Eclampsia”) OR MESH.EXACT(“Hypertension, Pregnancy-Induced”) |
| S2 | (MESH.EXACT.EXPLODE(“Pregnancy”) OR MESH.EXACT.EXPLODE(“Pregnancy Trimesters”) OR MESH.EXACT(“Pregnancy Complications”) OR MESH.EXACT(“Pregnancy Complications, Cardiovascular”) OR MESH.EXACT(“Pregnant Women”)) and MESH.EXACT(“Hypertension”) |
| S3 | (MESH.EXACT.EXPLODE(“Pregnancy”) OR MESH.EXACT.EXPLODE(“Pregnancy Trimesters”) OR MESH.EXACT(“Pregnancy Complications”) OR MESH.EXACT(“Pregnancy Complications, Cardiovascular”) OR MESH.EXACT(“Pregnant Women”)) and ti,ab(hypertens[*4]) |
| S4 | ti,ab(pregnan*) and MESH.EXACT(“Hypertension”) |
| S5 | EMB.EXACT(“eclampsia and preeclampsia”) OR EMB.EXACT(“preeclampsia”) OR EMB.EXACT(“pregnancy toxemia”) OR EMB.EXACT(“maternal hypertension”) |
| S6 | (EMB.EXACT.EXPLODE(“pregnancy”) OR EMB.EXACT(“pregnancy complication”) OR EMB.EXACT(“pregnancy disorder”) OR EMB.EXACT(“pregnant woman”)) and (EMB.EXACT(“essential hypertension”) OR EMB.EXACT(“hypertension”)) |
| S7 | (EMB.EXACT.EXPLODE(“pregnancy”) OR EMB.EXACT(“pregnancy complication”) OR EMB.EXACT(“pregnancy disorder”) OR EMB.EXACT(“pregnant woman”)) and ti,ab(hypertens[*4]) |
| S8 | ti,ab(pregnan*) and (EMB.EXACT(“essential hypertension”) OR EMB.EXACT(“hypertension”)) |
| S9 | ti,ab(preeclamp* or preclamp* or “pre eclamp*” or “pre clamp*”) |
| S10 | ti,ab((pregnan* or eclamp*) near/3 (toxemi[*2] or toxaemi[*2] or toxicosis)) |
| S11 | ti,ab((edema or oedema) near/3 proteinuria near/3 hypertens[*4]) |
| S12 | ti,ab(“eph gestos[*2]” or “eph toxemi[*2]” or “eph toxaemi[*2]” or “eph complex” or “eph syndrome”) |
| S13 | ti,ab(gestation* near/3 (hypertens[*4] or toxemi[*2] or toxaemi[*2] or toxicosis)) |
| S14 | ti,ab(maternal near/3 hypertens[*4]) |
| S15 | ti,ab(pregnan* near/5 hypertens[*4]) |
| S16 | rtype.exact(“Meta-Analysis”) or MESH.EXACT(“Meta-Analysis”) or EMB.EXACT(“meta analysis”) or EMB.EXACT(“systematic review”) |
| S17 | MESH.EXACT(“Meta-Analysis as Topic”) or EMB.EXACT(“meta analysis (topic)”) or EMB.EXACT(“systematic review (topic)”) |
| S18 | ti,ab(“meta analy[*3]” or metaanaly[*3] or “systematic review[*1]”) |
| S19 | pub.exact(“Cochrane Database of Systematic Reviews” OR “Cochrane Database of Systemic Reviews” OR “Cochrane Library” OR “Cochrane database of systematic reviews (Online)” OR “The Cochrane database of systematic reviews” OR “The Cochrane library”) |
| S20 | (s1 or s2 or s3 or s4 or s5 or s6 or s7 or s8 or s9 or s10 or s11 or s12 or s13 or s14 or s15) and (s16 or s17 or s18 or s19) |
| S21 | (s1 or s2 or s3 or s4 or s5 or s6 or s7 or s8 or s9 or s10 or s11 or s12 or s13 or s14 or s15) and (s16 or s17 or s18 or s19) and human(yes) |
| S22 | ((s1 or s2 or s3 or s4 or s5 or s6 or s7 or s8 or s9 or s10 or s11 or s12 or s13 or s14 or s15) and (s16 or s17 or s18 or s19)) not (human(yes) or animal(yes) or EMB.EXACT(“nonhuman”)) |
| S23 | s21 or s22 |
| Set# | Searched for |
|---|---|
| #1 | Validat*[tiab] OR Predict*[ti] OR Rule*[tiab] |
| #2 | Predict*[tiab] AND (Outcome*[tiab] OR Risk*[tiab] OR Model*[tiab]) |
| #3 | (History[tiab] OR Variable*[tiab] OR Criteria[tiab] OR Scor*[tiab] OR Characteristic*[tiab] OR Finding*[tiab] OR Factor*[tiab]) AND (Predict*[tiab] OR Model*[tiab] OR Decision*[tiab] OR Identif*[tiab] OR Prognos*[tiab]) |
| #4 | Decision*[tiab] AND (Model*[tiab] OR Clinical*[tiab] OR Logistic Model*[tiab]) |
| #5 | Prognostic[tiab] AND (History[tiab] OR Variable*[tiab] OR Criteria[tiab] OR Scor*[tiab] OR Characteristic*[tiab] OR Finding*[tiab] OR Factor*[tiab] OR Model*[tiab]) |
| #6 | “risk score”[All fields] OR “prediction model”[All fields] OR “prediction rule”[All fields] OR “risk assessment”[All fields] OR “algorithm”[All fields] |
| #7 | # 1 OR #2 OR #3 OR #4 OR #5 OR #6 |
| #8 | (pregnan*[tiab] OR obstetric*[tiab] OR woman[tiab] OR women[tiab]) AND (preeclampsia[tiab] OR pre-eclampsia [tiab]) |
| #9 | #7 AND #8 |
| #10 | #9 NOT (Animals[MeSH] NOT Humans[MeSH]) |
Appendix 3 Individual participant data extracted on the IPPIC project
| Candidate predictors | Brief description | Categories |
|---|---|---|
| Maternal clinical characteristics | ||
| Age | ‘Age in years (continuous)’ | |
| Parity | ‘Number of times giving birth before this pregnancy (continuous)’ | 0 ‘No’ 1 ‘Yes’ |
| ‘Nulliparous (binary)’ | ||
| Previous miscarriage | ‘Number of previous miscarriages (continuous)’ | |
| ‘Previous miscarriage (binary)’ | 0 ‘No’ 1 ‘Yes’ | |
| ‘Previous miscarriage trimester 1 (binary)’ | ||
| ‘Previous miscarriage trimester 2 (binary)’ | ||
| Previous stillbirth | ‘Previous stillbirth (binary)’ | 0 ‘No’ 1 ‘Yes’ |
| History of SGA birth | ‘SGA in a previous pregnancy (binary)’ | 0 ‘No’ 1 ‘Yes’ |
| Previous pre-eclampsia | ‘Previous history of any pre-eclampsia (binary)’ | 0 ‘No’ 1 ‘Yes’ |
| Previous early pre-eclampsia | ‘Previous early pre-eclampsia (binary)’ | 0 ‘No’ 1 ‘Yes’ |
| Gestational age at previous pre-eclampsia | ‘Gestational age at previous pre-eclampsia (continuous)’ | |
| Previous any preterm delivery | ‘Previous any preterm delivery (binary)’ | 0 ‘No’ 1 ‘Yes’ |
| Previous early preterm delivery | ‘Previous early preterm delivery < 34 weeks (binary)’ | 0 ‘No’ 1 ‘Yes’ |
| Previous late preterm delivery | ‘Previous late preterm delivery ≥ 34 weeks (binary)’ | 0 ‘No’ 1 ‘Yes’ |
| Gestational age at previous preterm delivery | ‘Gestational age at previous preterm delivery (continuous)’ | |
| Previous heritable thrombophilia | ‘History of heritable thrombophilia (binary)’ | 0 ‘No’ 1 ‘Yes’ |
| Previous autoimmune disease | ‘History of systemic lupus, rheumatoid arthritis or antiphospholipid syndrome (binary)’ | 0 ‘No’ 1 ‘Yes’ |
| History of pre-eclampsia in mother | ‘Mother has a history of pre-eclampsia (binary)’ | 0 ‘No’ 1 ‘Yes’ |
| Family history of pre-eclampsia | ‘Mother or sister has a history of pre-eclampsia (binary)’ | 0 ‘No’ 1 ‘Yes’ |
| Family history of cardiovascular disease | ‘Family history of cardiovascular disease (binary)’ | 0 ‘No’ 1 ‘Yes’ |
| History of renal disease | ‘History of renal disease (binary)’ | 0 ‘No’ 1 ‘Yes’ |
| History of pre-existing diabetes | ‘History of type 1, type 2 or gestational diabetes (binary)’ | 0 ‘No’ 1 ‘Yes’ |
| History of gestational diabetes mellitus | ‘Previous gestational diabetes mellitus (binary)’ | 0 ‘No’ 1 ‘Yes’ |
| Gestational diabetes in current pregnancy | ‘Gestational diabetes in current pregnancy trimester 1 (binary)’ | 0 ‘No’ 1 ‘Yes’ |
| ‘Gestational diabetes in current pregnancy trimester 1 (binary)’ | 0 ‘No’ 1 ‘Yes’ | |
| ‘Gestational diabetes in current pregnancy trimester 2 (binary)’ | 0 ‘No’ 1 ‘Yes’ | |
| ‘Gestational diabetes in current pregnancy trimester 3 (binary)’ | 0 ‘No’ 1 ‘Yes’ | |
| Early pregnancy bleeding | ‘Early pregnancy bleeding in current pregnancy (binary)’ | 0 ‘No’ 1 ‘Yes’ |
| Chronic or pre-existing hypertension | ‘History of chronic, essential or pre-existing hypertension (binary)’ | 0 ‘No’ 1 ‘Yes’ |
| Smoking | ‘Smoked during current pregnancy (binary)’ | 0 ‘No’ 1 ‘Yes’ |
| Number of cigarettes smoked per day | ‘Number of cigarettes smoked per day (continuous)’ | |
| Alcohol use | ‘Drank Alcohol (binary)’ | 0 ‘No’ 1 ‘Yes’ |
| Substance misuse in current pregnancy | ‘Cocaine, heroin or methamphetamine use in current pregnancy (binary)’ | 0 ‘No’ 1 ‘Yes’ |
| Mode of conception | ‘Spontaneous mode of conception (binary)’ | 0 ‘No’ 1 ‘Yes’ |
| Multiple pregnancy | ‘Multiple pregnancy in current pregnancy (binary)’ | 0 ‘No’ 1 ‘Yes’ |
| Number of years between pregnancies | ‘Number of years between pregnancies (continuous)’ | |
| Maternal education | ‘Highest level of maternal education attained (categorical)’ |
1 ‘Primary’ 2 ‘Secondary’ 3 ‘Tertiary’ |
| ‘Maternal years in education primary school (continuous)’ | ||
| New partner | ‘New partner (binary)’ | 0 ‘No’ 1 ‘Yes’ |
| Healthy diet in pregnancy | ‘On healthy diet in pregnancy (binary)’ | 0 ‘No’ 1 ‘Yes’ |
| Planned physical activity | ‘Planned physical activity (categorical)’ |
1 ‘Sedentary’ 2 ‘Moderate’ 3 ‘Severe’ |
| Ethnicity | ‘Ethnicity (categorical)’ |
1 ‘White’ 2 ‘Black’ 3 ‘Asian’ 4 ‘Hispanic’ 5 ‘Mixed’ 6 ‘Other’ |
| BMI | ‘BMI in kg/m2 (continuous)’ | |
| ‘BMI in kg/m2 trimester 1 (continuous)’ | ||
| ‘BMI in kg/m2 trimester 2 (continuous)‘ | ||
| ‘BMI in kg/m2 trimester 3 (continuous)’ | ||
| Height | ‘Height in cm (continuous)’ | |
| Weight | ‘Weight in kg (continuous)’ | |
| ‘Weight in kg trimester 1 (continuous)’ | ||
| ‘Weight in kg trimester 2 (continuous)’ | ||
| ‘Weight in kg trimester 3 (continuous)’ | ||
| SBP | ‘SBP in mmHg (continuous)’ | |
| ‘SBP in mmHg trimester 1 (continuous)’ | ||
| ‘SBP in mmHg trimester 2 (continuous)’ | ||
| ‘SBP in mmHg trimester 3 (continuous)’ | ||
| DBP | ‘DBP in mmHg (continuous)’ | |
| ‘DBP in mmHg trimester 1 (continuous)’ | ||
| ‘DBP in mmHg trimester 2 (continuous)’ | ||
| ‘DBP in mmHg trimester 3 (continuous)’ | ||
| MAP | ‘MAP in mmHg (continuous)’ | |
| ‘MAP in mmHg trimester 1 (continuous)’ | ||
| ‘MAP in mmHg trimester 2 (continuous)’ | ||
| ‘MAP in mmHg trimester 3 (continuous)’ | ||
| Urine dipstick | ‘Urine dipstick (categorical)’ | 0 ‘0 or trace’ 1 ‘1+’ 2 ‘≥ 2+’ |
| ‘Urine dipstick trimester 1 (categorical)’ | ||
| ‘Urine dipstick trimester 2 (categorical)’ | ||
| ‘Urine dipstick trimester 3 (categorical)’ | ||
| 24-hour protein | ‘24-hour protein in g/24hr (continuous)’ | |
| ‘24-hour protein in g/24hr trimester 1 (continuous)’ | ||
| ‘24-hour protein in g/24hr trimester 2 (continuous)’ | ||
| ‘24-hour protein in g/24hr trimester 3 (continuous)’ | ||
| PCR | ‘PCR in mg/mmol of creatinine (continuous)’ | |
| ‘PCR in mg/mmol of creatinine trimester 1 (continuous)’ | ||
| ‘PCR in mg/mmol of creatinine trimester 2 (continuous)’ | ||
| ‘PCR in mg/mmol of creatinine trimester 3 (continuous)’ | ||
| Ultrasound markers | ||
| Unilateral notching | ‘Unilateral notching (binary)’ | 0 ‘No’ 1 ‘Yes’ |
| ‘Unilateral notching trimester 1 (binary)’ | ||
| ‘Unilateral notching trimester 2 (binary)’ | ||
| ‘Unilateral notching trimester 3 (binary)’ | ||
| Bilateral notching | ‘Bilateral notching (binary)’ | 0 ‘No’ 1 ‘Yes’ |
| ‘Bilateral notching trimester 1 (binary)’ | ||
| ‘Bilateral notching trimester 2 (binary)’ | ||
| ‘Bilateral notching trimester 3 (binary)’ | ||
| Uterine artery PI | ‘Uterine artery PI (continuous)’ | |
| ‘Uterine artery PI trimester 1 (continuous)’ | ||
| ‘Uterine artery PI trimester 2 (continuous)’ | ||
| ‘Uterine artery PI trimester 3 (continuous)’ | ||
| Umbilical artery PI | ‘Umbilical artery PI (continuous)’ | |
| ‘Umbilical artery PI trimester 1 (continuous)’ | ||
| ‘Umbilical artery PI trimester 2 (continuous)’ | ||
| ‘Umbilical artery PI trimester 3 (continuous)’ | ||
| Uterine artery PI MoM | ‘Uterine artery PI MoM (continuous)’ | |
| ‘Uterine artery PI MoM trimester 1 (continuous)’ | ||
| ‘Uterine artery PI MoM trimester 2 (continuous)’ | ||
| ‘Uterine artery PI MoM trimester 3 (continuous)’ | ||
| Umbilical artery PI MoM | ‘Umbilical artery PI MoM (continuous)’ | |
| ‘Umbilical artery PI MoM trimester 1 (continuous)’ | ||
| ‘Umbilical artery PI MoM trimester 2 (continuous)’ | ||
| ‘Umbilical artery PI MoM trimester 3 (continuous)’ | ||
| Uterine artery RI | ‘Uterine artery RI (continuous)’ | |
| ‘Uterine artery RI trimester 1 (continuous)’ | ||
| ‘Uterine artery RI trimester 2 (continuous)’ | ||
| ‘Uterine artery RI trimester 3 (continuous)’ | ||
| Umbilical artery RI | ‘Umbilical artery RI (continuous)’ | |
| ‘Umbilical artery RI trimester 1 (continuous)’ | ||
| ‘Umbilical artery RI trimester 2 (continuous)’ | ||
| ‘Umbilical artery RI trimester 3 (continuous)’ | ||
| Uterine artery RI MoM | ‘Uterine artery RI MoM (continuous)’ | |
| ‘Uterine artery RI MoM trimester 1 (continuous)’ | ||
| ‘Uterine artery RI MoM trimester 2 (continuous)’ | ||
| ‘Uterine artery RI MoM trimester 3 (continuous)’ | ||
| Umbilical artery RI MoM | ‘Umbilical artery RI MoM (continuous)’ | |
| ‘Umbilical artery RI MoM trimester 1 (continuous)’ | ||
| ‘Umbilical artery RI MoM trimester 2 (continuous)’ | ||
| ‘Umbilical artery RI MoM trimester 3 (continuous)’ | ||
| Abdominal circumference | ‘Abdominal circumference (continuous)’ | |
| ‘Abdominal circumference trimester 2 (continuous)’ | ||
| ‘Abdominal circumference trimester 3 (continuous)’ | ||
| Abdominal circumference centile | ‘Abdominal circumference centile (continuous)’ | |
| ‘Abdominal circumference centile trimester 2 (continuous)’ | ||
| ‘Abdominal circumference centile trimester 3 (continuous)’ | ||
| CRL | ‘Crown–rump length in mm (continuous)’ | |
| ‘Crown–rump length in mm trimester 1 (continuous)’ | ||
| ‘Crown–rump length in mm trimester 2 (continuous)’ | ||
| Expected fetal weight | ‘Expected fetal weight in grams (continuous)’ | |
| ‘Expected fetal weight in grams trimester 1 (continuous)’ | ||
| ‘Expected fetal weight in grams trimester 2 (continuous)’ | ||
| ‘Expected fetal weight in grams trimester 3 (continuous)’ | ||
| Expected fetal weight centile | ‘Expected fetal weight centile (continuous)’ | |
| ‘Expected fetal weight centile trimester 1 (continuous)’ | ||
| ‘Expected fetal weight centile trimester 2 (continuous)’ | ||
| ‘Expected fetal weight centile trimester 3 (continuous)’ | ||
| Biochemical markers | ||
| PAPP-A | ‘PAPP-A (continuous)’ | |
| ‘PAPP-A trimester 1 (continuous)’ | ||
| ‘PAPP-A trimester 2 (continuous)’ | ||
| ‘PAPP-A trimester 3 (continuous)’ | ||
| PlGF | ‘PlGF (continuous)’ | |
| ‘PlGF trimester 1 (continuous)’ | ||
| ‘PlGF trimester 2 (continuous)’ | ||
| ‘PlGF trimester 3 (continuous)’ | ||
| sFlt-1 | ‘sFlt-1 (continuous)’ | |
| ‘sFlt-1 trimester 1 (continuous)’ | ||
| ‘sFlt-1 trimester 2 (continuous)’ | ||
| ‘sFlt-1 trimester 3 (continuous)’ | ||
| sENG | ‘sENG (continuous)’ | |
| ‘sENG trimester 1 (continuous)’ | ||
| ‘sENG trimester 2 (continuous)’ | ||
| ‘sENG trimester 3 (continuous)’ | ||
| CRP | ‘CRP (continuous)’ | |
| ‘CRP trimester 1 (continuous)’ | ||
| ‘CRP trimester 2 (continuous)’ | ||
| ‘CRP trimester 3 (continuous)’ | ||
| Hypertriglyceridaemia | ‘Hypertriglyceridaemia (continuous)’ | |
| ‘Hypertriglyceridaemia trimester 1 (continuous)’ | ||
| ‘Hypertriglyceridaemia trimester 2 (continuous)’ | ||
| ‘Hypertriglyceridaemia trimester 3 (continuous)’ | ||
| PAI-1 polymorphism | ‘PAI-1 polymorphism (continuous)’ | |
| ‘PAI-1 polymorphism trimester 1 (continuous)’ | ||
| ‘PAI-1 polymorphism trimester 2 (continuous)’ | ||
| ‘PAI-1 polymorphism trimester 3 (continuous)’ | ||
| Human chorionic gonadotropin | ‘hCG (continuous)’ | |
| ‘hCG trimester 1 (continuous)’ | ||
| ‘hCG trimester 2 (continuous)’ | ||
| ‘hCG trimester 3 (continuous)’ | ||
| AFP | ‘AFP (continuous)’ | |
| ‘AFP trimester 1 (continuous)’ | ||
| ‘AFP trimester 2 (continuous)’ | ||
| ‘AFP trimester 3 (continuous)’ | ||
| PAPP-A MoM | ‘PAPP-A MoM (continuous)’ | |
| ‘PAPP-A MoM trimester 1 (continuous)’ | ||
| ‘PAPP-A MoM trimester 2 (continuous)’ | ||
| ‘PAPP-A MoM trimester 3 (continuous)’ | ||
| PlGF MoM | ‘PlGF MoM (continuous)’ | |
| ‘PlGF MoM trimester 1 (continuous)’ | ||
| ‘PlGF MoM trimester 2 (continuous)’ | ||
| ‘PlGF MoM trimester 3 (continuous)’ | ||
| sFlt-1 MoM | ‘sFlt-1 MoM (continuous)’ | |
| ‘sFlt-1 MoM trimester 1 (continuous)’ | ||
| ‘sFlt-1 MoM trimester 2 (continuous)’ | ||
| ‘sFlt-1 MoM trimester 3 (continuous)’ | ||
| sENG MoM | ‘sENG MoM (continuous)’ | |
| ‘sENG MoM trimester 1 (continuous)’ | ||
| ‘sENG MoM trimester 2 (continuous)’ | ||
| ‘sENG MoM trimester 3 (continuous)’ | ||
| CRP MoM | ‘CRP MoM (continuous)’ | |
| ‘CRP MoM trimester 1 (continuous)’ | ||
| ‘CRP MoM trimester 2 (continuous)’ | ||
| ‘CRP MoM trimester 3 (continuous)’ | ||
| Hypertriglyceridaemia MoM | ‘Hypertriglyceridaemia MoM (continuous)’ | |
| ‘Hypertriglyceridaemia MoM trimester 1 (continuous)’ | ||
| ‘Hypertriglyceridaemia MoM trimester 2 (continuous)’ | ||
| ‘Hypertriglyceridaemia MoM trimester 3 (continuous)’ | ||
| PAI-1 polymorphism MoM | ‘PAI-1 polymorphism MoM (continuous)’ | |
| ‘PAI-1 polymorphism MoM trimester 1 (continuous)’ | ||
| ‘PAI-1 polymorphism MoM trimester 2 (continuous)’ | ||
| ‘PAI-1 polymorphism MoM trimester 3 (continuous)’ | ||
| Human chorionic gonadotropin MoM | ‘hCG MoM (continuous)’ | |
| ‘hCG MoM trimester 1 (continuous)’ | ||
| ‘hCG MoM trimester 2 (continuous)’ | ||
| ‘hCG MoM trimester 3 (continuous)’ | ||
| AFP MoM | ‘AFP MoM (continuous)’ | |
| ‘AFP MoM trimester 1 (continuous)’ | ||
| ‘AFP MoM trimester 2 (continuous)’ | ||
| ‘AFP MoM trimester 3 (continuous)’ | ||
| Primary outcome | ||
| Any-onset pre-eclampsia | ‘Any-onset pre-eclampsia (binary)’ | 0 ‘No’ 1 ‘Yes’ |
| Early-onset pre-eclampsia | ‘Early-onset pre-eclampsia < 34 weeks (binary)’ | 0 ‘No’ 1 ‘Yes’ |
| Late-onset pre-eclampsia | ‘Late-onset pre-eclampsia ≥ 34 weeks (binary)’ | 0 ‘No’ 1 ‘Yes’ |
| Superimposed pre-eclampsia | ‘Superimposed pre-eclampsia (binary)’ | 0 ‘No’ 1 ‘Yes’ |
| Gestational age at diagnosis | ‘Gestational age in weeks of diagnosis of pre-eclampsia (continuous)’ | |
| SBP | ‘SBP in mmHg (continuous)’ | |
| ‘SBP in mmHg trimester 2 (continuous)’ | ||
| ‘SBP in mmHg trimester 3 (continuous)’ | ||
| Gestational age of measurement | ‘Gestational age in weeks of measurement of SBP (continuous)’ | |
| DBP | ‘DBP in mmHg (continuous)’ | |
| ‘DBP in mmHg trimester 2 (continuous)’ | ||
| ‘DBP in mmHg trimester 3 (continuous)’ | ||
| Gestational age at measurement | ‘Gestational age in weeks of measurement of DBP (continuous)’ | |
| PCR | ‘PCR in mg/mmol of creatinine (continuous)’ | |
| ‘PCR in mg/mmol of creatinine trimester 2 (continuous)’ | ||
| ‘PCR in mg/mmol of creatinine trimester 3 (continuous)’ | ||
| 24-hour protein | ‘24-hour protein in g/24hr (continuous)’ | |
| ‘24-hour protein in g/24hr trimester 2 (continuous)’ | ||
| ‘24-hour protein in g/24hr trimester 3 (continuous)’ | ||
| Urine dipstick | ‘Urine dipstick (categorical)’ | 0 ‘0 or trace’ 1 ‘1+’ 2 ‘≥ 2+’ |
| ‘Urine dipstick trimester 2 (categorical)’ | ||
| ‘Urine dipstick trimester 3 (categorical)’ | ||
| Gestational age of measurement | ‘Gestational age in weeks of measurement of urine dipstick (continuous)’ | |
| Birthweight | ‘Birth weight in grams (continuous)’ | |
| ‘Birth weight in grams trimester 2 (continuous)’ | ||
| ‘Birth weight in grams trimester 3 (continuous)’ | ||
| Birthweight centile | ‘Birth weight centile (continuous)’ | |
| ‘Birth weight centile trimester 2 (continuous)’ | ||
| ‘Birth weight centile trimester 3 (continuous)’ | ||
| Gestational age at delivery | ‘Gestational age at delivery in weeks (continuous)’ | |
| ‘Gestational age at delivery in weeks (categorical)’ |
0 ‘≥ 37’ 1 ‘34–36 + 6’ 2 ‘30–33 + 6’ 3 ‘26–29 + 6’ 4 ‘23–25 + 6’ 5 ‘< 23’ |
|
| Secondary outcome | ||
| Eclampsia | ‘Eclampsia (binary)’ | 0 ‘No’ 1 ‘Yes’ |
| HELLP syndrome | ‘HELLP syndrome (binary)’ | 0 ‘No’ 1 ‘Yes’ |
| Placental abruption | ‘Placental abruption (binary)’ | 0 ‘No’ 1 ‘Yes’ |
| Hepatic failure | ‘Hepatic failure (binary)’ | 0 ‘No’ 1 ‘Yes’ |
| Renal failure | ‘Renal failure (binary)’ | 0 ‘No’ 1 ‘Yes’ |
| Cortical blindness | ‘Cortical blindness (binary)’ | 0 ‘No’ 1 ‘Yes’ |
| Pulmonary oedema | ‘Pulmonary oedema (binary)’ | 0 ‘No’ 1 ‘Yes’ |
| Postpartum haemorrhage | ‘Postpartum haemorrhage (binary)’ | 0 ‘No’ 1 ‘Yes’ |
| Disseminated intravascular coagulation | ‘Disseminated intravascular coagulation (binary)’ | 0 ‘No’ 1 ‘Yes’ |
| Preterm delivery | ‘Preterm delivery (binary)’ | 0 ‘No’ 1 ‘Yes’ |
| ‘Preterm delivery < 34 weeks (binary)’ | ||
| ‘Preterm delivery ≥ 34 weeks (binary)’ | ||
| Admission to high-dependency unit | ‘Admission to high-dependency unit (binary)’ | 0 ‘No’ 1 ‘Yes’ |
| Maternal death | ‘Maternal death (binary)’ | 0 ‘No’ 1 ‘Yes’ |
| Caesarean section | ‘Caesarean section (binary)’ | 0 ‘No’ 1 ‘Yes’ |
| Stillbirth | ‘Stillbirth (binary)’ | 0 ‘No’ 1 ‘Yes’ |
| ‘Stillbirth trimester 2 (binary)’ | ||
| ‘Stillbirth trimester 3 (binary)’ | ||
| Neonatal death | ‘Neonatal death (binary)’ | 0 ‘No’ 1 ‘Yes’ |
| Hypoxic-ischaemic encephalopathy | ‘Hypoxic ischaemic encephalopathy (binary)’ | 0 ‘No’ 1 ‘Yes’ |
| Respiratory distress syndrome | ‘Respiratory distress syndrome (binary)’ | 0 ‘No’ 1 ‘Yes’ |
| Septicaemia | ‘Septicaemia (binary)’ | 0 ‘No’ 1 ‘Yes’ |
| Admission to neonatal unit | ‘Admission to neonatal unit (binary)’ | 0 ‘No’ 1 ‘Yes’ |
| Treatment | ||
| Aspirin | ‘Aspirin use (binary)’ | 0 ‘No’ 1 ‘Yes’ |
| ‘Aspirin dose in mg (continuous)’ | ||
| ‘Aspirin use in trimester 1 (binary)’ | 0 ‘No’ 1 ‘Yes’ | |
| ‘Aspirin use in mg trimester 1 (continuous)’ | ||
| ‘Aspirin use in trimester 2 (binary)’ | 0 ‘No’ 1 ‘Yes’ | |
| ‘Aspirin use in mg trimester 2 (continuous)’ | ||
| ‘Aspirin use in trimester 3 (binary)’ | 0 ‘No’ 1 ‘Yes’ | |
| ‘Aspirin use in mg trimester 3 (continuous)’ | ||
| Calcium supplement | ‘Calcium supplement (binary)’ | 0 ‘No’ 1 ‘Yes’ |
| Vitamin supplement | ‘Vitamin supplement (binary)’ | 0 ‘No’ 1 ‘Yes’ |
Appendix 4 Participant summary from data sets contributing to the IPPIC project
| Study/data set | Maternal characteristics and outcomes | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total number of pregnancies | Number of singleton pregnancies | Age (years), mean (SD), range | First BMI measured, median [IQR], range | Ethnicity, n (%) | Nulliparous, n (%) | Previous PE, n (%) | Outcome, n (%) | ||||||||
| White | Black | Asian | Hispanic | Mixed | Other | Any-onset PE | Early-onset PE | Late-onset PE | |||||||
| SCOPE42 | 5628 | 5628 | 28.7 (5.5), 14–45 | 24.2 [5.6], 15.4–58.5 | 5061 (89.9) | 65 (1.2) | 304 (5.4) | 24 (0.4) | 0 (0) | 174 (3.1) | 5628 (100) | 0 (0) | 278 (4.9) | 44 (0.8) | 234 (4.2) |
| Allen108 | 1045 | 1045 | 29.9 (5.1), 15–48 | 23.6 [5.7], 14.8–51.1 | 398 (38.2) | 108 (10.4) | 495 (47.5) | 0 (0) | 12 (1.2) | 30 (2.9) | 584 (55.9) | 17 (1.6) | 14 (1.3) | 1 (0.1) | 13 (1.2) |
| ALSPAC103 | 15,242 | 15,038 | 27.7 (4.9), 13–46 | 21.5 [4], 11.7–61.3 | 11,769 (97.4) | 127 (1.1) | 113 (0.9) | 0 (0) | 0 (0) | 76 (0.6) | 5704 (45.3) | 0 (0) | 297 (2.2) | 37 (0.3) | 260 (1.9) |
| Chappell121 | 316 | 316 | 29.6 (5.9), 16.3–43 | 24.2 [5.3], 16.4–56.5 | 215 (68) | 91 (28.8) | 6 (1.9) | 3 (0.9) | 0 (0) | 1 (0.3) | 202 (63.9) | 56 (17.7) | 35 (12.4) | 6 (2.1) | 29 (10.2) |
| EMPOWaR122 | 449 | 449 | 28.7 (5.4), 17–43 | 38 [6.8], 30.6–59.7 | 449 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 291 (65) | 0 (0) | 10 (4.9) | 0 (0) | 10 (4.8) |
| POPPY106 | 1216 | 1216 | 32.1 (5.4), 17–51 | 24.6 [6.8], 14.5–57.9 | 881 (72.5) | 193 (15.9) | 85 (7) | 0 (0) | 0 (0) | 57 (4.7) | 206 (16.9) | 0 (0) | 22 (1.8) | 2 (0.2) | 20 (1.7) |
| Poston 2006150 | 2422 | 2040 | 30.8 (5.9), 16–45 | 31.2 [9.8], 16.6–65.4 | – | – | – | – | – | – | – | 657 (27.1) | 371 (15.3) | 144 (5.9) | 227 (9.4) |
| Poston 2015149 | 1554 | 1554 | 30.5 (5.5), 16–45 | 35.1 [5.8], 30–66 | 954 (61.4) | 395 (25.4) | 97 (6.2) | 0 (0) | 0 (0) | 108 (6.9) | 674 (43.4) | 69 (4.5) | 54 (3.6) | 5 (0.3) | 49 (3.3) |
| Khan137 | 222 | 222 | – | 25.3 [6.6], 18–46.1 | – | – | – | – | – | – | 146 (65.8) | 0 (0) | 27 (12.2) | 11 (5) | 16 (7.2) |
| St George’s163 | 54,656 | 54,635 | 30.5 (5.6), 13–54 | 23.5 [5.5], 11.9–79.8 | 33,257 (62.1) | 7820 (14.6) | 10388 (19.4) | 5 (0) | 1528 (2.9) | 555 (1) | 29318 (53.8) | 0 (0) | 1492 (2.7) | 152 (0.3) | 1340 (2.5) |
| PARIS112 | 35,955 | 34,609 | 23.6 (6.1), 11–48 | – | – | – | – | – | – | – | 19793 (57.5) | 6086 (18.5) | 2928 (8.8) | 498 (1.5) | 2403 (7.2) |
| AMND114 | 138,967 | 136,635 | 28.4 (5.6), 13–56 | 23.9 [5.5], 10–72.1 | – | – | – | – | – | – | 65206 (47.7) | 0 (0) | 5194 (3.7) | 1329 (1) | 3865 (2.8) |
| Born in Bradford177 | 13,279 | 13,118 | 27.5 (5.6), 15–49 | 25.1 [7.3], 12.9–57 | 4443 (42.4) | 212 (2) | 5380 (51.4) | 0 (0) | 158 (1.5) | 278 (2.7) | 5034 (39.6) | 0 (0) | 334 (2.7) | 0 (0) | 0 (0) |
| Velauthar39 | 1208 | 1206 | – | 23.9 [6], 13.8–46.6 | 402 (35.3) | 90 (7.9) | 565 (49.6) | 0 (0) | 0 (0) | 82 (7.2) | 598 (52.2) | 0 (0) | 26 (2.3) | 3 (0.3) | 23 (2.1) |
| POP161 | 4212 | 4212 | 29.9 (5.1), 16.1–48.4 | 24.1 [5.5], 14.7–54.7 | 3900 (92.6) | 25 (0.6) | 91 (2.2) | 0 (0) | 1 (0) | 195 (4.6) | 4212 (100) | 0 (0) | 273 (6.5) | 10 (0.2) | 263 (6.3) |
| Baschat115 | 1704 | 1704 | 30.2 (6.5), 18–50 | 26.7 [9.1], 15.9–72.9 | 775 (45.5) | 803 (47.1) | 88 (5.2) | 27 (1.6) | 0 (0) | 11 (0.6) | 736 (43.2) | 92 (5.4) | 106 (6.2) | 21 (1.2) | 85 (5) |
| Audibert113 | 893 | 893 | 29.5 (4.5), 18–43 | 22.6 [4.4], 16.9–50 | 720 (80.6) | 62 (6.9) | 14 (1.6) | 26 (2.9) | 44 (4.9) | 27 (3) | 893 (100) | 0 (0) | 40 (4.5) | 9 (1) | 31 (3.5) |
| Caradeux118 | 682 | 682 | 29 (6.4), 14–45.6 | 24.9 [5.4], 15.7–48 | – | – | – | – | – | – | 322 (48.1) | 15 (2.2) | 28 (4.1) | 3 (0.4) | 25 (3.7) |
| Giguère126 | 7866 | 7693 | – | 23 [5.9], 13.9–58.3 | 6422 (96.9) | 37 (0.6) | 23 (0.3) | 47 (0.7) | 47 (0.7) | 49 (0.7) | 3631 (47.2) | 0 (0) | 142 (1.8) | 13 (0.2) | 129 (1.6) |
| Goetzinger128 | 4035 | 4035 | 34.8 (4.4), 16–52 | 24.4 [7], 15.4–62.4 | 3282 (82.6) | 397 (10) | 112 (2.8) | 65 (1.6) | 0 (0) | 116 (2.9) | 751 (20.1) | 0 (0) | 271 (12.1) | 19 (0.8) | 252 (11.3) |
| Antsaklis110 | 3328 | 3328 | 30.9 (4.8), 14–47 | 22.7 [5.2], 14.5–50.1 | 3229 (97.2) | 49 (1.5) | 32 (1) | 0 (0) | 0 (0) | 11 (0.3) | 3328 (100) | 0 (0) | 32 (1.4) | 13 (0.6) | 19 (0.8) |
| Llurba140 | 11,668 | 11,668 | 30.2 (5.9), 14–51 | – | 10,415 (90.6) | 125 (1.1) | 4 (0) | 794 (6.9) | 0 (0) | 163 (1.4) | – | 51 (0.4) | 411 (3.5) | 69 (0.6) | 218 (1.9) |
| WHO175 | 7315 | 7273 | 22.5 (5.8), 11–51 | 22.4 [4.8], 13.1–54.8 | 2222 (30.6) | 756 (10.4) | 1443 (19.9) | 0 (0) | 0 (0) | 2846 (39.2) | 6710 (92.3) | 432 (5.9) | 142 (1.9) | 18 (0.2) | 124 (1.7) |
| Andersen109 | 2161 | 2120 | 30.2 (4.5), 17–45 | 23.4 [5.2], 14.9–50.4 | 1765 (96.6) | 3 (0.2) | 31 (1.7) | 5 (0.3) | 0 (0) | 24 (1.3) | 1193 (56.3) | 0 (0) | 159 (7.5) | 3 (0.1) | 156 (7.4) |
| Arenas111 | 319 | 319 | 30.5 (4.9), 16–43 | 24.2 [4.3], 17.3–39.1 | – | – | – | – | – | – | 193 (60.5) | 2 (0.6) | 11 (3.4) | 1 (0.3) | 10 (3.1) |
| FINNPEC133 | 2506 | 2506 | 30 (5.4), 18–47 | 23.6 [6.1], 16–50.8 | 2498 (99.7) | 4 (0.2) | 4 (0.2) | 0 (0) | 0 (0) | 0 (0) | 1653 (66) | 189 (7.5) | 1440 (57.5) | 260 (10.4) | 1180 (47.1) |
| Verlohren171 | 253 | 235 | 31.9 (5.5), 16–45 | 25.1 [7.1], 17.9–48.8 | 160 (68.4) | 17 (7.3) | 3 (1.3) | 37 (15.8) | 0 (0) | 17 (7.3) | 117 (50.2) | 38 (15.3) | 103 (41.4) | 33 (13.3) | 70 (28.1) |
| Generation R134 | 8727 | 8631 | 29.6 (5.3), 15.3–46.3 | 23.9 [5.3], 15.2–51.2 | 4800 (58.8) | 2117 (26) | 484 (5.9) | 0 (0) | 0 (0) | 756 (9.3) | 4745 (55.7) | 0 (0) | 194 (2.3) | 24 (0.3) | 170 (2) |
| NICHD HR32 | 2539 | 1848 | 27.1 (6.3), 15–43 | 28.3 [11.6], 13.4–68.5 | 612 (33.1) | 1079 (58.4) | 2 (0.1) | 148 (8) | 0 (0) | 7 (0.4) | 430 (23.3) | 76 (5.9) | 485 (19.4) | 108 (4.3) | 377 (15.1) |
| NICHD LR159 | 3134 | 3097 | 20.6 (4.4), 15–39 | 22.7 [5.4], 13.4–51.2 | 548 (17.7) | 1515 (48.9) | 2 (0.1) | 1010 (32.6) | 0 (0) | 22 (0.7) | 3097 (100) | 0 (0) | 163 (5.5) | 13 (0.4) | 150 (5) |
| Placental Health Study176 | 856 | 856 | 32.9 (4.1), 18–45 | 23.3 [4.9], 16.5–41.9 | 593 (71.4) | 29 (3.5) | 66 (7.9) | 28 (3.4) | 0 (0) | 115 (13.8) | 856 (100) | 0 (0) | 69 (8.1) | 3 (0.4) | 66 (7.7) |
| POUCH131 | 3019 | 3019 | 26.4 (5.8), 15–47.3 | 27.7 [8.6], 15.1–66.3 | 2018 (66.8) | 743 (24.6) | 57 (1.9) | 160 (5.3) | 0 (0) | 41 (1.4) | 1293 (42.8) | 106 (3.5) | 44 (3.2) | 12 (0.9) | 32 (2.3) |
| van Kuijk 2011167 | 407 | 407 | 28.8 (3.9), 18.9–40.1 | 24.8 [6.3], 16–42.2 | – | – | – | – | – | – | 0 (0) | 407 (100) | 106 (26) | 24 (5.9) | 82 (20.1) |
| Van Kuijk 2014166 | 230 | 230 | 30.8 (4.8), 19.8–40.8 | 24.7 [6.6], 17.3–48.3 | – | – | – | – | – | – | 0 (0) | 209 (91.7) | 43 (19.8) | 15 (6.9) | 28 (12.9) |
| Odibo147 | 1200 | 1200 | 31.5 (5.6), 17–49 | 25.4 [8.4], 13.6–67.8 | 735 (61.3) | 325 (27.1) | 94 (7.8) | 23 (1.9) | 22 (1.8) | 1 (0.1) | 518 (43.2) | 64 (5.3) | 102 (8.7) | 20 (1.7) | 82 (7) |
| PREDO127 | 1082 | 1082 | – | 25.8 [9.9], 17.2–55 | 1082 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 247 (22.8) | 96 (8.9) | 20 (1.9) | 75 (6.9) |
| Prefumo151 | 273 | 273 | 33.4 (4.2), 21–43 | 23.3 [4.7], 17.4–37.1 | 203 (74.4) | 41 (15) | 29 (10.6) | 0 (0) | 0 (0) | 0 (0) | 108 (39.6) | 56 (20.5) | 12 (4.4) | 3 (1.1) | 9 (3.3) |
| Skråstad160 | 599 | 599 | 27.5 (4), 18–40 | 23.6 [5.1], 17.1–48.9 | 589 (98.3) | 0 (0) | 1 (0.2) | 1 (0.2) | 4 (0.7) | 4 (0.7) | 559 (93.3) | 33 (5.5) | 27 (4.5) | 1 (0.2) | 26 (4.3) |
| Verlohren172 | 566 | 542 | 31 (6.2), 15–55 | 27.6 [8.7], 15.9–57.4 | 503 (93.3) | 15 (2.8) | 12 (2.2) | 0 (0) | 0 (0) | 9 (1.7) | 267 (49.5) | 26 (4.6) | 94 (17.2) | 20 (3.6) | 74 (13.5) |
| Rumbold155 | 1877 | 1877 | 26.4 (5.7), 13–44 | 24.1 [6.2], 13.7–57.6 | 1777 (94.9) | 3 (0.2) | 1 (0.1) | 1 (0.1) | 4 (0.2) | 87 (4.6) | 1877 (100) | 0 (0) | 103 (5.5) | 6 (0.3) | 97 (5.2) |
| Vollebregt174 | 308 | 308 | – | 22.9 [4.3], 17.7–41.2 | 264 (85.7) | 4 (1.3) | 14 (4.5) | 20 (6.5) | 0 (0) | 6 (1.9) | 256 (83.1) | 38 (12.3) | 21 (6.8) | 7 (2.3) | 14 (4.6) |
| JSOG105 | 406,286 | 379,390 | 32.2 (5.4), 10–59 | 20.5 [3.6], 10.5–69.8 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 379,390 (100) | 195,983 (51.9) | 1509 (0.4) | 22,880 (5.6) | 3548 (0.9) | 19,323 (4.8) |
| DOMInO143 | 2399 | 2399 | 28.9 (5.7), 15–47 | 26.2 [7.6], 15.4–60.2 | 2111 (88.1) | 24 (1) | 191 (8) | 4 (0.2) | 7 (0.3) | 60 (2.5) | – | – | 76 (3.2) | 8 (0.3) | 68 (2.9) |
| Danish National Birth Cohort136 | 86,082 | 84,173 | 29 (4.4), 15–46 | 23.9 [4.8], 12.5–65.2 | – | – | – | – | – | – | 0 (0) | – | 2144 (2.5) | 196 (0.2) | 1948 (2.3) |
| Indonesian Cohort157 | 2252 | 2223 | 28.6 (5.9), 10.2–59 | 22.4 [5.7], 12.9–47.3 | 0 (0) | 0 (0) | 2223 (100) | 0 (0) | 0 (0) | 0 (0) | 664 (42.7) | 3 (0.1) | 97 (6.1) | 8 (0.5) | 80 (5) |
| Ohkuchi148 | 288 | 288 | 28.7 (4), 18–42 | – | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 288 (100) | 146 (50.7) | 9 (3.2) | 9 (3.1) | 1 (0.3) | 8 (2.8) |
| Lecarpentier139 | 244 | 243 | 35.9 (4.9), 23–53 | 27.7 [9.7], 15–47.4 | 93 (38.3) | 123 (50.6) | 4 (1.6) | 0 (0) | 0 (0) | 23 (9.5) | 46 (18.9) | 46 (18.9) | 58 (23.8) | 32 (13.1) | 26 (10.7) |
| TEST146 | 557 | 557 | 32 (4.8), 18.2–43.1 | 24 [5.5], 17.4–45.2 | – | – | – | – | – | – | 557 (100) | 0 (0) | 23 (4.1) | 2 (0.4) | 18 (3.2) |
| Massé144 | 1244 | 1244 | 26.1 (4.2), 15.4–41.8 | 21.2 [3.8], 14.8–46.9 | 539 (96.8) | 2 (0.4) | 10 (1.8) | 0 (0) | 6 (1.1) | 0 (0) | 1244 (100) | 0 (0) | 109 (8.8) | 0 (0) | 0 (0) |
| Staff162 | 240 | 240 | 31.1 (4.8), 18–42 | 24.1 [6.2], 17–45.4 | 222 (92.5) | 5 (2.1) | 13 (5.4) | 0 (0) | 0 (0) | 0 (0) | 132 (55) | 0 (0) | 73 (30.4) | 41 (17.1) | 32 (13.3) |
| STORK G135 | 823 | 812 | 29.8 (4.8), 19.3–45.1 | 24.6 [5.7], 16.2–49.8 | 375 (46.2) | 61 (7.5) | 198 (24.4) | 12 (1.5) | 0 (0) | 166 (20.4) | 377 (46.4) | 0 (0) | 46 (5.6) | 1 (0.1) | 45 (5.5) |
| Vatten170 | 736 | 736 | 27.9 (5.3), 17–43 | – | – | – | – | – | – | – | 373 (51) | 0 (0) | 344 (46.7) | 154 (20.9) | 190 (25.8) |
| Vinter173 | 304 | 304 | 29.2 (4.2), 19–43 | 33.6 [5], 28.8–46 | 304 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 160 (52.6) | 1 (0.3) | 19 (6.3) | 2 (0.7) | 17 (5.6) |
| BORN Ontorio104 | 281,466 | 276,329 | – | 23.8 [6.9], 9.8–144 | – | – | – | – | – | – | 116,602 (43.7) | 0 (0) | 2903 (1) | 336 (0.1) | 2379 (0.8) |
| Ghana Cohort165 | 1016 | 1010 | 28 (5.1), 18–49 | 24.8 [6.4], 15.4–64.5 | 0 (0) | 1010 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | – | 34 (3.3) | 16 (2) | 0 (0) | 9 (1.1) |
| MoBa142 | 112,758 | 110,771 | 30.1 (4.7), 14–47 | 24.2 [5], 11–60.1 | – | – | – | – | – | – | 48,457 (43.9) | 0 (0) | 4225 (3.7) | 414 (0.4) | 3785 (3.4) |
| Huang132 | 141,698 | 141,698 | 30.8 (5.1), 13.1–53.3 | – | 89,350 (66.1) | 8708 (6.4) | 35,293 (26.1) | 0 (0) | 0 (0) | 1760 (1.3) | 57,879 (49.1) | 0 (0) | 1036 (0.7) | 0 (0) | 0 (0) |
| Carbillon119 | 42,829 | 42,036 | 29.2 (5.8), 13–57 | 23 [6], 11–68 | – | – | – | – | – | – | 16,809 (40) | 391 (0.9) | 359 (0.8) | 93 (0.2) | 266 (0.6) |
| Goffinet129 | 3317 | 3317 | 28.8 (5.2), 15.4–47.2 | – | – | – | – | – | – | – | 1485 (46.3) | 0 (0) | 33 (1.2) | 1 (0) | 32 (1.2) |
| Rang152 | 42 | 42 | 29.6 (3.7), 22–39 | – | 13,752 (32.7) | 19,602 (46.6) | 2379 (5.7) | 0 (0) | 0 (0) | 6303 (15) | 21 (50) | 21 (50) | 6 (14.3) | 2 (4.8) | 4 (9.5) |
| Cameroni117 | 173 | 173 | 34.5 (4.5), 21–48 | – | 152 (87.9) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 21 (12.1) | 74 (42.8) | 22 (12.7) | 17 (9.8) | 1 (0.6) | 16 (9.2) |
| Conserva123 | 53 | 53 | 33.2 (6), 24–43 | – | 53 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 34 (64.2) | 5 (9.4) | 3 (5.7) | 0 (0) | 3 (5.7) |
| Facchinetti124 | 172 | 172 | 33.2 (5.2), 20–58 | 23.4 [5], 18–49.8 | 172 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 119 (69.2) | 52 (30.4) | 58 (33.7) | 3 (1.7) | 55 (32) |
| Ferrazzani158 | 54 | 54 | 32.5 (5), 21–42 | – | 54 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 54 (100) | 10 (19.6) | 5 (9.8) | 5 (9.8) |
| Figueiró-Filho125 | 81 | 81 | 30 (6.2), 18–46 | – | 59 (72.8) | 4 (4.9) | 0 (0) | 0 (0) | 17 (21) | 1 (1.2) | 0 (0) | 81 (100) | 30 (44.1) | 11 (16.4) | 18 (26.9) |
| Langenveld138 | 208 | 205 | 31.5 (3.9), 22–40 | 24.2 [5.6], 18.4–44.2 | 171 (88.6) | 11 (5.7) | 6 (3.1) | 0 (0) | 0 (0) | 5 (2.6) | 205 (100) | 0 (0) | 54 (26.3) | 21 (10.3) | 32 (15.7) |
| Lykke141 | 536,414 | 536,409 | 29.5 (4.2), 15.9–49.9 | 23.1 [5.3], 12.8–62.9 | – | – | – | – | – | – | – | – | 10,159 (1.9) | 471 (0.1) | 9574 (1.8) |
| Mbah145 | 1,663,167 | 1,601,906 | 25.8 (5.6), 11–53 | 22 [6], 11.1–95.9 | 1,301,530 (83.6) | 236,479 (15.2) | 977 (0.1) | 0 (0) | 161 (0) | 18,611 (1.2) | – | – | 42,608 (2.6) | 4252 (0.3) | 38,305 (2.4) |
| Trogstad164 | 37,738 | 37,738 | 28.1 (4.5), 17–48 | – | 37,738 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 37738 (100) | 4722 (12.6) | 321 (0.9) | 4085 (11) |
| Salim156 | 97 | 97 | 30.3 (5.8), 18–45 | – | – | – | – | – | – | – | – | – | 5 (5.2) | 2 (2.1) | 3 (3.1) |
| Van Oostwaard 2012168 | 425 | 425 | 32 (4.1), 23–42 | 24.3 [6.4], 16.2–41.8 | 288 (84) | 46 (13.4) | 4 (1.2) | 0 (0) | 3 (0.9) | 2 (0.6) | 0 (0) | 208 (48.9) | 22 (11.6) | 4 (2.1) | 18 (9.5) |
| Van Oostwaard 2014169 | 639 | 639 | 32.1 (4.4), 21–43 | 25.9 [8.7], 17.7–56.5 | 360 (71.9) | 119 (23.8) | 17 (3.4) | 0 (0) | 3 (0.6) | 2 (0.4) | 0 (0) | 224 (35.1) | 30 (9.9) | 5 (1.6) | 25 (8.2) |
| Zhang178 | 1639 | 1639 | 22.5 (4), 13–40 | 21.2 [4.1], 12.1–47.7 | 1061 (65.5) | 502 (31) | 0 (0) | 0 (0) | 0 (0) | 57 (3.5) | – | – | 8 (0.5) | 0 (0) | 8 (0.5) |
| Brown 2007116 | 2785 | 2702 | 29.5 (5), 15–46 | – | – | – | – | – | – | – | 1192 (44.1) | 0 (0) | 781 (28) | 79 (2.8) | 702 (25.2) |
| Gurgel Alves 2014130 | 500 | 500 | – | – | – | – | – | – | – | – | – | – | 31 (12.2) | 8 (3.1) | 23 (9.1) |
| Rocha153 | 733 | 733 | 26.1 (6.7), 13–45 | 25 [6], 16–51 | – | – | – | – | – | – | – | 45 (6.1) | 55 (7.5) | 11 (1.5) | 44 (6) |
| Costa 2017107 | 574 | 574 | 34.3 (5.3), 18–50.2 | 25.3 [6], 17.6–51.3 | 431 (75.3) | 21 (3.7) | 96 (16.8) | 0 (0) | 24 (4.2) | 0 (0) | 251 (43.7) | 40 (7) | 29 (5.1) | 3 (0.5) | 26 (4.5) |
| Rocha154 | 733 | 733 | 26.1 (6.7), 13–45 | 25 [6], 16–51 | 128 (18.2) | 20 (2.8) | 556 (79) | 0 (0) | 0 (0) | 0 (0) | 360 (49.1) | 39 (5.3) | 55 (7.5) | 11 (1.5) | 44 (6) |
Appendix 5 Detailed study characteristics of IPPIC data sets
| Study/data set | Study design (randomised, observational) | Data source (trial, cohort, registry) | Country | Data period | Population type [any pregnancy, high risk (women with complications), low risk] | Inclusion criteria | Exclusion criteria |
|---|---|---|---|---|---|---|---|
| IPPIC-UK | |||||||
| SCOPE42 | Observational | Prospective cohort | Multicountry (UK, New Zealand, Australia and Republic of Ireland) | 2004–8 | Low risk | Healthy nulliparous women with singleton pregnancies | Recognised as high risk of pre-eclampsia, small for gestational age fetus or spontaneous preterm birth owing to underlying medical condition such as chronic hypertension requiring antihypertensive drugs, diabetes, renal disease, systemic lupus erythematosus, antiphospholipid syndrome, sickle cell disease or HIV. Previous cervical knife cone biopsy, three or more abortions or miscarriages, current ruptured membranes, known major fetal anomaly or abnormal karyotype, and interventions that can alter the course of pregnancy such as aspirin or cervical suture |
| Allen108 | Observational | Prospective cohort | UK | 2010–14 | Any pregnancy | All pregnant women attending an inner London hospital between 11 and 14 weeks’ gestation | Women with multiple pregnancies and fetal anomalies |
| ALSPAC103 | Observational | Prospective birth cohort | UK | 1991–2 | Any pregnancy | All pregnant women resident in Avon, UK | None |
| Chappell121 | Randomised | Trial | UK | NI | High risk | Pregnant women with an abnormal doppler waveform in either uterine artery at 18–22 weeks’ gestation or pre-eclampsia in a previous pregnancy that led to preterm delivery, eclampsia or HELLP syndrome | Heparin or warfarin treatment, abnormal fetal anomaly scan or multiple pregnancy |
| EMPOWaR122 | Randomised | Trial | UK | 2011–14 | High risk | Women at least 16 years of age at recruitment, between 12 and 16 weeks’ gestation and with a BMI of 30 kg/m2 | Non-white women and those with: history of diabetes, systemic disease at the time of enrolment (requiring either regular drugs or systemic corticosteroids treatment in the past 3 months), previous delivery of a baby smaller than the third centile for weight, history of pre-eclampsia with delivery before 32 weeks’ gestation, known hypersensitivity to metformin hydrochloride or any of the excipients. Known liver or renal failure, acute disorders at the time of trial entry with the potential to change renal function, such as dehydration sufficient to require intravenous infusion, severe infection, shock, intravascular administration of iodinated contrast agents, or acute or chronic diseases that might cause tissue hypoxia (e.g. cardiac or respiratory failure, recent myocardial infarction, hepatic insufficiency, acute alcohol intoxication or alcoholism); lactating women; and women with multiple pregnancy |
| POPPY106 | Observational | Prospective cohort | UK | 2011–13 | Any pregnancy | Pregnant asymptomatic women with a high risk of spontaneous preterm birth, such as previous history of spontaneous preterm delivery, late miscarriage, invasive cervical surgery or a short cervix | NI |
| Poston 2006150 | Randomised | Trial | UK | 2003–5 | High risk | Gestational age of 14–21 weeks plus one or more of the following risk factors: history of pre-eclampsia necessitating preterm delivery, history of HELLP syndrome, eclampsia, essential hypertension requiring medication, maternal DBP of ≥ 90 mmHg before 20 weeks’ gestation in current pregnancy, history of diabetes, antiphospholipid syndrome; chronic renal disease, multiple pregnancy; abnormal uterine artery Doppler waveform, primiparity with BMI at first antenatal appointment of ≥ 30 kg/m2 | Women taking vitamin supplements containing doses of vitamin C of 200 mg or more or of vitamin E of ≥ 40 IU daily. Women treated with warfarin |
| Poston 2015149 | Randomised | Trial | UK | 2009–14 | High risk | Women aged > 16 years with a BMI of ≥ 30 kg/m2 and a singleton pregnancy | Any underlying disorders, including a pre-pregnancy diagnosis of essential hypertension, diabetes, renal disease, systemic lupus erythematosus, antiphospholipid syndrome, sickle cell disease, thalassaemia, coeliac disease, thyroid disease, and current psychosis; or if on metformin |
| Khan137 | Randomised | Trial | UK | NI | High risk | Women identified to be at high risk of adverse pregnancy outcome by uterine arterial waveform analysis | Women with underlying conditions thought likely to compromise renal function, such as diabetes or renal disease |
| St George’s163 | Observational | Prospective registry | UK | 2000–15 | Any pregnancy | All pregnant women attending an inner-London hospital | None |
| PARIS112 | IPD MA of 31 randomised trials | Trial | 33 countries | 1985–2005 | Varied | Varied (dependent on individual study) | Varied (dependent on individual study) |
| AMND114 | Observational | Prospective registry | UK | 1986–2015 | Any pregnancy | Data from every pregnancy event occurring in Aberdeen Maternity Hospital | None |
| Born in Bradford177 | Observational | Prospective birth cohort | UK | 2007–11 | Any pregnancy | All pregnant women attending Bradford Royal Infirmary | None |
| Velauthar39 | Observational | Prospective cohort | UK | NI | Any pregnancy | All pregnant women attending an inner-London hospital | None |
| POP161 | Observational | Prospective cohort | UK | 2008–12 | Any pregnancy | Nulliparous women with singleton pregnancies | None |
| IPPIC international | |||||||
| Baschat115 | Observational | Prospective cohort | USA | 2007–10 | Any pregnancy | All pregnant women attending any of four Baltimore (USA) hospitals for first-trimester screening | None |
| Audibert113 | Observational | Prospective cohort | Canada | 2006–8 | Low risk | Nulliparous women with singleton pregnancies presenting for Down syndrome screening at 11–13 weeks | Pregnancies with a major fetal chromosomal or structural anomaly |
| Caradeux118 | Observational | Prospective cohort | Chile | NI | Any pregnancy | All pregnant women attending an 11- to 14-week ultrasound evaluation | None |
| Giguère126 | Observational | Prospective cohort | Canada | 2005–10 | Any pregnancy | Women at least 18 years old and with a gestational age of at least 10 weeks at their first prenatal visit with no chronic hepatic or renal diseases | Pregnancies with major fetal abnormalities and those ending in termination, miscarriage or fetal death before 24 weeks’ gestation |
| Goetzinger128 | Observational | Retrospective cohort | USA | 2003–8 | Any pregnancy | Women seen for aneuploidy screening | None |
| Antsaklis110 | Observational | Prospective cohort | Greece | 1997–8 | Low risk | All nulliparous women | Women with multiple pregnancies, renal disease, cardiovascular disease and fetal anomalies |
| Llurba140 | Observational | Prospective cohort | Spain | 2002–6 | Any pregnancy | Singleton women attending routine second-trimester anomaly scans | None |
| WHO175 | Observational | Prospective cohort | Multicountry (Argentina, Colombia, India, Italy, Kenya, Peru, Switzerland and Thailand) | 2006–9 | High risk | Women with risk factors for pre-eclampsia | Women with known renal disease or proteinuria |
| Andersen109 | Observational | Prospective cohort | Denmark | 2010–12 | Any pregnancy | Newly pregnant women | Twin pregnancies and early pregnancy fetal losses |
| Arenas111 | Observational | Prospective cohort | Spain | 2000–1 | Any pregnancy | Women attending routine ultrasound scan at 20 weeks | Multiple pregnancies or congenital defects |
| FINNPEC133 | Observational | Prospective/retrospective case–control cohort | Finland | 2008–11 | Any pregnancy | Nulliparous or multiparous women with a singleton pregnancy with or without pre-eclampsia on admission to hospital | Multiple pregnancy, maternal age of < 18 years |
| Verlohren171 | Observational | Prospective case–control cohort | Spain | NI | Any pregnancy | Singleton pregnancies | Multigestation, antiphospholipid antibody syndrome, systemic lupus erythematosus or any other autoimmune disease, as well as chronic corticosteroid or nonsteroidal anti-inflammatory drug use, except low-dose aspirin (< 150 mg/day) |
| Generation R134 | Observational | Prospective birth cohort | Netherlands | 2002–6 | Any pregnancy | Resident mothers delivering during the study period | None |
| NICHD HR32 | Randomised | Trial | USA | 1991–5 | High risk | Women with pre-gestational, insulin-treated diabetes mellitus, women with chronic hypertension, women with multifetal gestations, and women who had had preeclampsia in a previous pregnancy | Women with multifetal gestation if they also had chronic hypertension, renal disease, diabetes, history of pre-eclampsia and current proteinuria |
| NICHD LR159 | Randomised | Trial | USA | NI (early 1990s) | Low risk | Healthy nulliparous women | Women with chronic hypertension, renal disease, diabetes and other illnesses |
| Placental Health Study176 | Observational | Prospective cohort | Canada | 2012–13 | Low risk | Healthy nulliparous women with singleton pregnancies | Chronic hypertension, use of unfractionated or low molecular-weight heparin, pre-gestational diabetes mellitus, major fetal abnormalities, ruptured membranes, vaginal bleeding from 13 0/7 weeks of gestation for greater than 1 day, or a short cervical length on ultrasonography before 20 weeks of gestation (< 2 cm long) |
| POUCH131 | Observational | Prospective cohort | USA | 1998–2004 | Any pregnancy | Women with a singleton pregnancy at 16–27 weeks’ gestation, no known chromosomal abnormality, maternal age of at least 15 years, no pre-pregnancy diabetes mellitus | None |
| van Kuijk 2011167 | Observational | Prospective cohort | Netherlands | 1993–2008 | High risk | Women with preceding singleton pregnancy complicated by pre-eclampsia or HELLP syndrome | NI |
| van Kuijk 2014166 | Observational | Prospective and retrospective cohort | Netherlands | 2008–12 | High risk | Women with preceding singleton pregnancy complicated by pre-eclampsia or HELLP syndrome | Women who had diabetes, autoimmune disease, heart or kidney disease |
| Odibo147 | Observational | Prospective cohort | USA | 2009–11 | Any pregnancy | Women attending first-trimester screening | None |
| PREDO127 | Observational | Prospective case–control cohort | Finland | 2005–9 | Any pregnancy | Pregnant women with known risk factor for pre-eclampsia and intrauterine growth restriction and those without, attending clinics for their first ultrasound screening between 12 and 14 weeks’ gestation | Asthma diagnosed by a physician, allergy to ASA, tobacco smoking during pregnancy, previous peptic ulcer, previous placental ablation, inflammatory bowel diseases (Crohn’s disease, ulcerative colitis), rheumatoid arthritis, haemophilia or thrombophilia (previous venous or pulmonary thrombosis and/or coagulation abnormality), gestational weeks + days < 12 + 0 or > 14 + 0 or multiple pregnancy |
| Prefumo151 | Observational | Prospective cohort | Italy | 2001–5 | Any pregnancy | Women attending routine antenatal care | Known medical condition (e.g. diabetes mellitus, connective tissue disease, essential hypertension) or a history of recurrent miscarriage |
| Skråstad160 | Observational | Prospective cohort | Norway | 2010–12 | High risk | Nulliparous and high-risk parous women with one or more previous pre-eclampsia pregnancies | Use of any anticoagulant medication or acetylsalicylic acid in pregnancy |
| Verlohren172 | Observational | Prospective case–control cohort | Germany | NI | Any pregnancy | Singleton pregnancies | Multigestation, antiphospholipid antibody syndrome, systemic lupus erythematosus or any other autoimmune disease, as well as chronic corticosteroid or nonsteroidal anti-inflammatory drug use except low-dose aspirin (< 150 mg/day) |
| Rumbold155 | Randomised | Trial | Australia | 2001–5 | Low risk | Nulliparous women with a singleton pregnancy between 14 and 22 weeks’ gestation and normal blood pressure | Known multiple pregnancy, known potentially lethal fetal anomaly, known thrombophilia, chronic renal failure, antihypertensive therapy, or specific contraindications to vitamin C or E therapy, such as hemochromatosis or anticoagulant therapy |
| Vollebregt174 | Observational | Prospective cohort | Netherlands | 2004–6 | High risk | Healthy nulliparous women at low risk and women with elevated risk for pre-eclampsia or fetal growth restriction with singleton pregnancies | NI |
| JSOG105 | Observational | Prospective registry | Japan | 2013–14 | Any pregnancy | All women giving birth at participating institutions in Japan | None |
| DOMInO143 | Randomised | Trial | Australia | 2005–8 | Any pregnancy | Singleton pregnancies at less than 21 weeks’ gestation | Already taking a prenatal supplement with DHA, their fetus had a known major abnormality, had a bleeding disorder in which tuna oil was contraindicated, were taking anticoagulant therapy, had a documented history of drug or alcohol abuse, were participating in another fatty acid trial |
| Danish Birth Cohort136 | Observational | Prospective registry | Denmark | 1996–2002 | Any pregnancy | All women in Denmark | None |
| Indonesian Cohort157 | Observational | Prospective cohort | Indonesia | 2012–15 | Any pregnancy | All women attending antenatal care | None |
| Ohkuchi148 | Observational | Prospective cohort | Japan | NI | Any pregnancy | Women with singleton pregnancies attending antenatal care | None |
| Lecarpentier139 | Observational | Retrospective cohort | France | 2004–7 | High risk | Women with chronic hypertension | Multiple pregnancies, women with secondary hypertension, women with proteinuria at < 20 weeks’ gestation, women considered as having chronic hypertension but without any treatment at first prenatal visit, women transferred from other maternities, pregnancies complicated by fetal malformations |
| TEST146 | Randomised | Trial | Ireland | 2014–16 | Low risk | Nulliparous women between 11–14 weeks’ gestation and not already on aspirin | Fetal abnormality or contraindication to aspirin |
| Massé144 | Observational | Prospective cohort | Canada | 1989–91 | Low risk | Nulliparous women attending hospital for routine blood sampling at the start of pregnancy | Diabetes mellitus, cardiovascular disease (including chronic hypertension) or renal disease or women seen after 20 weeks |
| Staff162 | Observational | Prospective case–control cohort | Norway | NI | Low risk | Women with singleton pregnancies | NI |
| STORK G135 | Observational | Prospective cohort | Norway | 2008–10 | Any pregnancy | Healthy pregnant women | Women with diabetes or diseases require intensive hospital follow-up in pregnancy |
| Vatten170 | Observational | Prospective case–control cohort | Norway | 1992–4 | Any pregnancy | Women attending antenatal care | None |
| Vinter173 | Randomised | Trial | Denmark | 2007–10 | High risk | Women aged between 18–40 years with a pre-pregnancy weight of between 30 and 45 kg/m2 | Women with chronic medical disorders (hypertension, diabetes, alcohol or drug use) and serious obstetric complication (multiple pregnancy, congenital malformation, miscarriage) |
| BORN Ontorio104 | Observational | Prospective registry | Canada | 2012–14 | Any pregnancy | Women giving birth during the data period in the Ontario region | None |
| Ghana Cohort165 | Observational | Prospective cohort | Ghana | 2012–14 | Any pregnancy | Women < 17 weeks pregnant, at least 18 years old with no established hypertension at booking | None |
| MoBa142 | Observational | Prospective registry | Norway | 1999–2005 | Any pregnancy | All women giving birth in Norway | None |
| Huang132 | Observational | Retrospective cohort | Canada | 2000–3 | Any pregnancy | All women screened in early pregnancy for Down syndrome | None |
| Carbillion119 | Observational | Prospective registry | France | 1996–2005 | Any pregnancy | Women giving birth during the data period in that region | None |
| Goffinet129 | Randomised | Trial | France | 1994–7 | Low risk | All women attending routine antenatal visit before 24 weeks | Any indications for uterine artery Doppler, such as chronic hypertension, diabetes, previous fetal death, intrauterine growth restriction, hypertensive disorders of pregnancy or contraindication for aspirin |
| Rang152 | Observational | Prospective cohort | Netherlands | NI | High risk | Women with a history of early-onset pre-eclampsia in a previous pregnancy or women who had never been pregnant | None |
| Cameroni 2011117 | Observational | Retrospective cohort | Italy | NI | High risk | Singleton pregnancies at risk of pre-eclampsia or intrauterine growth restriction | NI |
| Conserva 2012123 | Observational | Prospective cohort | Italy | 2001–8 | High risk | Women with previous adverse pregnancy outcomes | Multiple gestation; a previous uneventful pregnancy; a previous pregnancy treated with low-molecular-weight heparin or unfractionated heparin; patients with clinical immune disease and acquired thrombophilia − lupus-like anticoagulant or APL syndrome; patients with positive antinuclear, antimitochondria, anti-smooth muscle antibodies; postnatal or postmortem diagnosis of congenital fetal anomaly or fetal infection; women of non-Caucasian ethnicity; alcohol or illicit drug use; early pregnancy loss was not considered an abnormal pregnancy outcome |
| Facchinetti124 | Observational | Prospective cohort | Italy | 2001–6 | High risk | Previous singleton pregnancies complicated by pre-eclampsia and received evaluation for thrombophilia | History of thromboembolic diseases, renal and/or cardiovascular disorder, systemic lupus erythematosus, diabetes and any ethnic group other than white |
| Ferrazzani158 | Observational | Prospective cohort | Italy | 1990–2001 | High risk | Previous severe preterm pre-eclampsia | Previous HELLP syndrome |
| Figueiró-Filho125 | Observational | Prospective case–control cohort | Brazil | 2007–10 | High risk | Women with severe pre-eclampsia in previous pregnancies | Antiphospholipid antibodies and thrombophilia |
| Langenveld138 | Observational | Retrospective cohort | Netherlands | 1996–2004 | High risk | Women with hypertension (including patients with chronic hypertension), pre-eclampsia or HELLP syndrome, and delivered before 34 weeks of gestation in the study period and primiparous with singleton pregnancy without fetal abnormalities in first pregnancy | NI |
| Lykke141 | Observational | Prospective registry | Denmark | 1978–2007 | Any pregnancy | Singleton deliveries of women with first delivery aged > 15 years and second delivery aged < 50 years | Cardiovascular diagnosis and type 1 or type 2 diabetes |
| Mbah145 | Observational | Prospective registry | USA | 1989–2005 | Any pregnancy | Women with first and second singleton pregnancies within the gestational age range of 20–44 weeks | None |
| Trogstad164 | Observational | Prospective registry | Norway | 1967–98 | Any pregnancy | Women with a first and second delivery | None |
| Salim156 | Observational | Prospective cohort | Israel | 2000–6 | High risk | Previous pregnancy with antepartum complications at ≥ 23 weeks’ gestation | Women who had a previous pregnancy with antepartum complications that could be attributed to multiple gestations, having fetuses with major congenital anomalies or chromosomal abnormalities, fetal infection, chorioamnionitis, hydrops fetalis and diabetes mellitus |
| van Oostwaard 2012168 | Observational | Prospective cohort | Netherlands | 2000–2 | High risk | Women with a hypertensive disorder in the index pregnancy and delivery at 34–37 weeks of gestation | Fetal abnormalities |
| van Oostwaard 2014169 | Observational | Retrospective cohort | Netherlands | 2000–2 | High risk | Women with a hypertensive disorder in the index pregnancy and delivery at 34–37 weeks’ gestation | Fetal abnormalities |
| Zhang178 | Observational | Prospective cohort | USA | 1959–65 | Any pregnancy | Women attending prenatal care | None |
| Brown 2007116 | Observational | Retrospective cohort | Australia | 1988–98 | Any pregnancy | Women referred for management of hypertensive disorders of pregnancy | None |
| Gurgel Alves 2014130 | Observational | Prospective cohort | Brazil | 2009–11 | Any pregnancy | Women attending for first-trimester Down syndrome screening | None |
| Rocha153 | Observational | Prospective cohort | Brazil | 2009–14 | Any pregnancy | Women with singleton pregnancies attending for first-trimester ultrasound scans | Prior maternal renal disease, major fetal malformations or chromosomal abnormalities, miscarriage |
| Costa 2017107 | Observational | Prospective cohort | Australia | 2012–15 | Any pregnancy | Women attending for their second-trimester morphology ultrasound between 19 and 22 weeks | None |
| Rocha154 | Observational | Prospective cohort | Brazil | 2009–14 | Any pregnancy | Singleton pregnancies of women attending routine ultrasound screening | Kidney disease diagnosis in their history or on ultrasound examination, major fetal malformations or chromosomal abnormalities, and fetuses with crown–rump length of > 84 mm |
Appendix 6 Summary of missing data for prioritised predictors and each pre-eclampsia outcome for all pregnancies
| Study | Variable, n (%) | ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Maternal clinical characteristics | Ultrasound markers | Biochemical markers | Outcome | ||||||||||||||||||||||||||
| Ethnicity | Age | Nulliparous | Previous PE | Previous autoimmune disease | Previous heritable thrombophilia | Family history of PE | History of renal disease | History of diabetes | History of hypertension | Smoker | Substance misuse in current pregnancy | Spontaneous conception | Multiple pregnancy | History of SGA | BMI | SBP | DBP | MAP | Urine dipstick | PCR | Uterine PI | Umbilical PI | PAPP-A | PlGF | sFlt-1 | Any-onset PE | Early-onset PE | Late-onset PE | |
| SCOPE42 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 5628 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 4464 (79.3) | 5628 (100) | 3102 (55.1) | 48 (0.9) | 51 (0.9) | 48 (0.9) | 5 (0.1) | 5 (0.1) | 5 (0.1) |
| Allen108 | 2 (0.2) | 1 (0.1) | 0 (0) | 0 (0) | 0 (0) | 1045 (100) | 6 (0.6) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1045 (100) | 1045 (100) | 0 (0) | 1045 (100) | 114 (10.9) | 0 (0) | 1045 (100) | 0 (0) | 0 (0) | 0 (0) |
| ALSPAC103 | 2999 (19.9) | 2078 (13.8) | 2473 (16.4) | 9476 (63) | 15,242 (100) | 15,242 (100) | 15,242 (100) | 2798 (18.4) | 2857 (18.7) | 3049 (20) | 2702 (17.7) | 2334 (15.3) | 2906 (19.1) | 0 (0) | 15,242 (100) | 3147 (20.6) | 2011 (13.2) | 2011 (13.2) | 2011 (13.2) | 2087 (13.7) | 15,242 (100) | 15,242 (100) | 15242 (100) | 15,242 (100) | 15,242 (100) | 15,242 (100) | 1546 (10.1) | 1546 (10.1) | 1546 (10.1) |
| Chappell121 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 316 (100) | 316 (100) | 316 (100) | 0 (0) | 0 (0) | 0 (0) | 316 (100) | 316 (100) | 0 (0) | 316 (100) | 33 (10.4) | 33 (10.4) | 33 (10.4) | 33 (10.4) | 92 (29.1) | 316 (100) | 316 (100) | 316 (100) | 316 (100) | 143 (45.3) | 238 (75.3) | 33 (10.4) | 33 (10.4) | 33 (10.4) |
| EMPOWaR122 | 0 (0) | 0 (0) | 1 (0.2) | 0 (0) | 449 (100) | 449 (100) | 449 (100) | 0 (0) | 0 (0) | 449 (100) | 0 (0) | 0 (0) | 449 (100) | 0 (0) | 0 (0) | 334 (74.4) | 0 (0) | 0 (0) | 0 (0) | 449 (100) | 449 (100) | 449 (100) | 449 (100) | 449 (100) | 449 (100) | 449 (100) | 246 (54.8) | 78 (17.4) | 239 (53.2) |
| POPPY106 | 0 (0) | 0 (0) | 0 (0) | 1010 (83.1) | 1216 (100) | 1216 (100) | 1216 (100) | 1216 (100) | 1216 (100) | 1216 (100) | 1216 (100) | 1216 (100) | 1216 (100) | 0 (0) | 1216 (100) | 0 (0) | 1216 (100) | 1216 (100) | 1216 (100) | 1216 (100) | 1216 (100) | 1216 (100) | 1216 (100) | 1216 (100) | 1216 (100) | 1216 (100) | 12 (1) | 12 (1) | 12 (1) |
| Poston 2006150 | 2422 (100) | 22 (1.1) | 2422 (100) | 0 (0) | 0 (0) | 2422 (100) | 2422 (100) | 0 (0) | 0 (0) | 0 (0) | 2422 (100) | 2422 (100) | 2422 (100) | 0 (0) | 2422 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 2214 (91.4) | 2249 (92.9) | 2422 (100) | 2422 (100) | 2249 (92.9) | 2249 (92.9) | 0 (0) | 0 (0) | 0 (0) |
| Poston 2015149 | 0 (0) | 0 (0) | 0 (0) | 7 (0.5) | 0 (0) | 1554 (100) | 61 (3.9) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1554 (100) | 4 (0.3) | 0 (0) | 1554 (100) | 0 (0) | 8 (0.5) | 8 (0.5) | 8 (0.5) | 357 (23) | 1547 (99.5) | 1554 (100) | 1554 (100) | 1554 (100) | 395 (25.4) | 1554 (100) | 47 (3) | 47 (3) | 47 (3) |
| Khan137 | 222 (100) | 222 (100) | 0 (0) | 76 (34.2) | 222 (100) | 222 (100) | 222 (100) | 0 (0) | 0 (0) | 222 (100) | 0 (0) | 222 (100) | 222 (100) | 0 (0) | 222 (100) | 0 (0) | 2 (0.9) | 2 (0.9) | 2 (0.9) | 222 (100) | 222 (100) | 222 (100) | 43 (19.4) | 222 (100) | 222 (100) | 222 (100) | 0 (0) | 0 (0) | 0 (0) |
| St George’s163 | 1086 (2) | 0 (0) | 104 (0.2) | 25,326 (46.4) | 54,656 (100) | 54,656 (100) | 54,656 (100) | 54,656 (100) | 54,656 (100) | 0 (0) | 4054 (7.4) | 54,656 (100) | 1427 (2.6) | 0 (0) | 54,656 (100) | 9151 (16.7) | 54,656 (100) | 54,656 (100) | 54,656 (100) | 54,656 (100) | 54,656 (100) | 20,069 (36.7) | 54,656 (100) | 54,656 (100) | 54,656 (100) | 54,656 (100) | 0 (0) | 0 (0) | 0 (0) |
| PARIS Collaborative Group112 | 35,955 (100) | 35,955 (100) | 190 (0.5) | 3073 (8.9) | 27,798 (77.3) | 35,955 (100) | 35,395 (98.4) | 10,741 (29.9) | 7599 (21.1) | 19,304 (53.7) | 35,955 (100) | 35,955 (100) | 35,955 (100) | 0 (0) | 4814 (13.4) | 35,955 (100) | 5255 (14.6) | 5246 (14.6) | 5258 (14.6) | 35,955 (100) | 35,955 (100) | 35,955 (100) | 35,955 (100) | 35,955 (100) | 35,955 (100) | 35,955 (100) | 2603 (7.2) | 2630 (7.3) | 2630 (7.3) |
| AMND114 | 138,967 (100) | 19 (0) | 7 (0) | 72,684 (53.2) | 0 (0) | 138,967 (100) | 138,967 (100) | 0 (0) | 138,967 (100) | 0 (0) | 2961 (2.1) | 0 (0) | 138,967 (100) | 0 (0) | 138,967 (100) | 26,080 (18.8) | 138,967 (100) | 138,967 (100) | 138,967 (100) | 138,967 (100) | 138,967 (100) | 138,967 (100) | 138,967 (100) | 138,967 (100) | 138,967 (100) | 138,967 (100) | 0 (0) | 0 (0) | 0 (0) |
| Born in Bradford177 | 2676 (20.4) | 405 (3.1) | 405 (3.1) | 8178 (62.3) | 13,279 (100) | 13,279 (100) | 13,279 (100) | 13,279 (100) | 925 (7) | 936 (7) | 2679 (20.2) | 4038 (30.4) | 13,279 (100) | 0 (0) | 13,279 (100) | 3360 (25.3) | 2747 (20.7) | 2747 (20.7) | 2747 (20.7) | 6122 (46.1) | 13,279 (100) | 13,279 (100) | 13,279 (100) | 13,279 (100) | 13,279 (100) | 13,279 (100) | 955 (7.2) | 1289 (9.7) | 1289 (9.7) |
| Velauthar39 | 67 (5.6) | 1208 (100.2) | 61 (5.1) | 608 (50.4) | 1208 (100) | 1208 (100) | 1208 (100) | 1208 (100) | 62 (5.1) | 62 (5.1) | 62 (5.1) | 1208 (100) | 1208 (100) | 0 (0) | 1208 (100) | 67 (5.5) | 69 (5.7) | 69 (5.7) | 69 (5.7) | 1208 (100) | 600 (49.7) | 63 (5.2) | 1208 (100) | 1208 (100) | 1208 (100) | 1208 (100) | 93 (7.7) | 93 (7.7) | 93 (7.7) |
| POP161 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1704 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1704 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1704 (100) | 1704 (100) | 266 (15.6) | 1704 (100) | 704 (41.3) | 704 (41.3) | 1704 (100) | 0 (0) | 0 (0) | 0 (0) |
| Baschat115 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 893 (100) | 893 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 893 (100) | 893 (100) | 0 (0) | 893 (100) | 0 (0) | 893 (100) | 893 (100) | 893 (100) | 893 (100) | 893 (100) | 28 (3.1) | 893 (100) | 4 (0.4) | 362 (40.5) | 893 (100) | 0 (0) | 0 (0) | 0 (0) |
| Audibert113 | 682 (100) | 3 (0.4) | 12 (1.8) | 0 (0) | 0 (0) | 0 (0) | 682 (100) | 682 (100) | 0 (0) | 0 (0) | 0 (0) | 682 (100) | 682 (100) | 0 (0) | 682 (100) | 6 (0.9) | 3 (0.4) | 2 (0.3) | 3 (0.4) | 682 (100) | 682 (100) | 38 (5.6) | 370 (54.3) | 682 (100) | 682 (100) | 682 (100) | 0 (0) | 0 (0) | 0 (0) |
| Caradeux118 | 63 (14.7) | 7866 (102.2) | 12 (0.2) | 4149 (53.9) | 5553 (70.6) | 5515 (70.1) | 7866 (100) | 426 (5.4) | 313 (4) | 256 (3.3) | 872 (11.1) | 6307 (80.2) | 7866 (100) | 0 (0) | 7866 (100) | 1956 (24.9) | 539 (6.9) | 540 (6.9) | 540 (6.9) | 7866 (100) | 7866 (100) | 7866 (100) | 7866 (100) | 7212 (91.7) | 7866 (100) | 7212 (91.7) | 0 (0) | 0 (0) | 0 (0) |
| Giguère126 | 63 (1.6) | 72 (1.8) | 302 (7.5) | 3284 (81.4) | 0 (0) | 4035 (100) | 4035 (100) | 0 (0) | 3406 (84.4) | 281 (7) | 244 (6) | 281 (7) | 92 (2.3) | 0 (0) | 4035 (100) | 606 (15) | 4035 (100) | 4035 (100) | 4035 (100) | 4035 (100) | 4035 (100) | 4035 (100) | 4035 (100) | 2119 (52.5) | 4035 (100) | 4035 (100) | 1798 (44.6) | 1798 (44.6) | 1798 (44.6) |
| Goetzinger128 | 7 (0.2) | 11 (0.3) | 0 (0) | 0 (0) | 0 (0) | 3328 (100) | 3328 (100) | 0 (0) | 0 (0) | 0 (0) | 6 (0.2) | 3328 (100) | 3328 (100) | 0 (0) | 3328 (100) | 9 (0.3) | 2898 (87.1) | 2905 (87.3) | 2905 (87.3) | 3328 (100) | 3328 (100) | 2770 (83.2) | 3328 (100) | 3328 (100) | 3328 (100) | 3328 (100) | 985 (29.6) | 985 (29.6) | 985 (29.6) |
| Antsaklis110 | 167 (1.4) | 11,668 (100) | 11,668 (100) | 39 (0.3) | 11,668 (100) | 61 (0.5) | 11,668 (100) | 11,668 (100) | 61 (0.5) | 59 (0.5) | 1229 (10.5) | 11,668 (100) | 11,668 (100) | 0 (0) | 0 (0) | 11,668 (100) | 11,668 (100) | 11,668 (100) | 11,668 (100) | 11,668 (100) | 11,668 (100) | 6299 (54) | 11,668 (100) | 11,668 (100) | 11,668 (100) | 11,668 (100) | 0 (0) | 0 (0) | 0 (0) |
| Llurba140 | 6 (0.1) | 1 (0) | 0 (0) | 0 (0) | 2 (0) | 2 (0) | 7315 (100) | 0 (0) | 1 (0) | 1 (0) | 2 (0) | 7315 (100) | 7315 (100) | 0 (0) | 7315 (100) | 9 (0.1) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 7315 (100) | 7315 (100) | 7315 (100) | 7315 (100) | 3180 (43.5) | 3428 (46.9) | 0 (0) | 0 (0) | 0 (0) |
| WHO175 | 299 (14.1) | 0 (0) | 0 (0) | 939 (44.3) | 2161 (100) | 2161 (100) | 2161 (100) | 2161 (100) | 198 (9.2) | 197 (9.1) | 2 (0.1) | 2161 (100) | 2161 (100) | 0 (0) | 2161 (100) | 3 (0.1) | 2161 (100) | 2161 (100) | 2161 (100) | 2161 (100) | 2161 (100) | 2161 (100) | 2161 (100) | 2161 (100) | 1028 (47.6) | 1028 (47.6) | 47 (2.2) | 47 (2.2) | 47 (2.2) |
| Andersen109 | 319 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 319 (100) | 319 (100) | 319 (100) | 319 (100) | 0 (0) | 0 (0) | 0 (0) | 319 (100) | 0 (0) | 319 (100) | 0 (0) | 319 (100) | 319 (100) | 319 (100) | 319 (100) | 319 (100) | 2 (0.6) | 2 (0.6) | 319 (100) | 319 (100) | 319 (100) | 0 (0) | 0 (0) | 0 (0) |
| Arenas111 | 0 (0) | 1 (0) | 0 (0) | 0 (0) | 0 (0) | 2506 (100) | 2506 (100) | 0 (0) | 0 (0) | 0 (0) | 2506 (100) | 2506 (100) | 2506 (100) | 0 (0) | 2506 (100) | 5 (0.2) | 10 (0.4) | 10 (0.4) | 10 (0.4) | 0 (0) | 2506 (100) | 2506 (100) | 2506 (100) | 2506 (100) | 1930 (77) | 1929 (77) | 0 (0) | 0 (0) | 0 (0) |
| FINNPEC133 | 1 (0.4) | 0 (0) | 3 (1.3) | 5 (2.1) | 0 (0) | 253 (100) | 253 (100) | 7 (2.8) | 3 (1.2) | 8 (3.2) | 1 (0.4) | 253 (100) | 253 (100) | 0 (0) | 253 (100) | 15 (5.9) | 2 (0.8) | 2 (0.8) | 2 (0.8) | 253 (100) | 253 (100) | 253 (100) | 78 (30.8) | 253 (100) | 3 (1.2) | 3 (1.2) | 4 (1.6) | 4 (1.6) | 4 (1.6) |
| Galindo171 | 478 (5.5) | 2 (0) | 108 (1.3) | 3938 (45.6) | 2573 (29.5) | 8727 (100) | 8727 (100) | 8727 (100) | 1397 (16) | 1253 (14.4) | 1112 (12.7) | 0 (0) | 592 (6.8) | 0 (0) | 8727 (100) | 38 (0.4) | 18 (0.2) | 18 (0.2) | 18 (0.2) | 8727 (100) | 8727 (100) | 3549 (40.7) | 8727 (100) | 8727 (100) | 635 (7.3) | 635 (7.3) | 255 (2.9) | 10 (0.1) | 245 (2.8) |
| Generation R134 | 0 (0) | 19 (1) | 0 (0) | 1250 (67.6) | 2539 (100) | 2539 (100) | 2539 (100) | 2539 (100) | 0 (0) | 0 (0) | 0 (0) | 2539 (100) | 2539 (100) | 0 (0) | 2539 (100) | 25 (1) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 2539 (100) | 2539 (100) | 2539 (100) | 2539 (100) | 1571 (61.9) | 2539 (100) | 36 (1.4) | 36 (1.4) | 36 (1.4) |
| NICHD HR32 | 0 (0) | 99 (3.2) | 0 (0) | 0 (0) | 0 (0) | 3134 (100) | 3134 (100) | 0 (0) | 0 (0) | 0 (0) | 6 (0.2) | 3134 (100) | 3134 (100) | 0 (0) | 3134 (100) | 94 (3) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 3134 (100) | 3134 (100) | 3134 (100) | 3134 (100) | 3134 (100) | 3134 (100) | 150 (4.8) | 150 (4.8) | 150 (4.8) |
| NICHD LR159 | 25 (2.9) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 36 (4.2) | 856 (100) | 856 (100) | 0 (0) | 0 (0) | 0 (0) | 856 (100) | 1 (0.1) | 0 (0) | 856 (100) | 2 (0.2) | 2 (0.2) | 2 (0.2) | 2 (0.2) | 0 (0) | 754 (88.1) | 1 (0.1) | 3 (0.4) | 856 (100) | 856 (100) | 856 (100) | 0 (0) | 0 (0) | 0 (0) |
| Placental Health Study176 | 0 (0) | 0 (0) | 1 (0) | 1 (0) | 2458 (81.4) | 1706 (56.5) | 3019 (100) | 1921 (63.6) | 0 (0) | 1 (0) | 6 (0.2) | 7 (0.2) | 3019 (100) | 0 (0) | 3019 (100) | 1 (0) | 1648 (54.6) | 1648 (54.6) | 1648 (54.6) | 0 (0) | 3019 (100) | 3019 (100) | 3019 (100) | 3019 (100) | 1715 (56.8) | 1712 (56.7) | 1648 (54.6) | 1648 (54.6) | 1648 (54.6) |
| POUCH131 | 407 (100) | 0 (0) | 0 (0) | 0 (0) | 407 (100) | 407 (100) | 407 (100) | 407 (100) | 407 (100) | 0 (0) | 407 (100) | 407 (100) | 407 (100) | 0 (0) | 407 (100) | 0 (0) | 407 (100) | 407 (100) | 407 (100) | 407 (100) | 407 (100) | 407 (100) | 407 (100) | 407 (100) | 407 (100) | 407 (100) | 0 (0) | 0 (0) | 0 (0) |
| van Kuijk 2011167 | 230 (100) | 0 (0) | 0 (0) | 2 (0.9) | 0 (0) | 230 (100) | 230 (100) | 0 (0) | 0 (0) | 0 (0) | 230 (100) | 230 (100) | 20 (8.7) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 230 (100) | 230 (100) | 230 (100) | 230 (100) | 230 (100) | 230 (100) | 230 (100) | 13 (5.7) | 13 (5.7) | 13 (5.7) |
| van Kuijk 2014166 | 0 (0) | 1 (0.1) | 1 (0.1) | 0 (0) | 1200 (100) | 1200 (100) | 1200 (100) | 7 (0.6) | 0 (0) | 0 (0) | 9 (0.8) | 11 (0.9) | 5 (0.4) | 0 (0) | 6 (0.5) | 29 (2.4) | 81 (6.8) | 81 (6.8) | 82 (6.8) | 1200 (100) | 1200 (100) | 10 (0.8) | 1200 (100) | 1200 (100) | 697 (58.1) | 889 (74.1) | 23 (1.9) | 23 (1.9) | 23 (1.9) |
| Odibo147 | 0 (0) | 1082 (100) | 0 (0) | 0 (0) | 0 (0) | 1082 (100) | 1082 (100) | 0 (0) | 0 (0) | 0 (0) | 1082 (100) | 1082 (100) | 125 (11.6) | 0 (0) | 1082 (100) | 16 (1.5) | 670 (61.9) | 670 (61.9) | 670 (61.9) | 1004 (92.8) | 1082 (100) | 1082 (100) | 1082 (100) | 661 (61.1) | 976 (90.2) | 976 (90.2) | 0 (0) | 1 (0.1) | 1 (0.1) |
| PREDO127 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 273 (100) | 273 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 273 (100) | 273 (100) | 0 (0) | 273 (100) | 60 (22) | 273 (100) | 273 (100) | 273 (100) | 273 (100) | 273 (100) | 62 (22.7) | 273 (100) | 273 (100) | 273 (100) | 273 (100) | 0 (0) | 0 (0) | 0 (0) |
| Prefumo151 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 599 (100) | 599 (100) | 599 (100) | 599 (100) | 599 (100) | 0 (0) | 0 (0) | 599 (100) | 0 (0) | 0 (0) | 599 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 599 (100) | 599 (100) | 98 (16.4) | 599 (100) | 0 (0) | 0 (0) | 599 (100) | 0 (0) | 0 (0) | 0 (0) |
| Skråstad160 | 3 (0.6) | 0 (0) | 3 (0.6) | 0 (0) | 235 (41.5) | 0 (0) | 566 (100) | 2 (0.4) | 2 (0.4) | 2 (0.4) | 21 (3.7) | 566 (100) | 566 (100) | 0 (0) | 0 (0) | 12 (2.1) | 3 (0.5) | 3 (0.5) | 3 (0.5) | 28 (4.9) | 566 (100) | 477 (84.3) | 166 (29.3) | 566 (100) | 103 (18.2) | 103 (18.2) | 18 (3.2) | 18 (3.2) | 18 (3.2) |
| Verlohren172 | 4 (0.2) | 0 (0) | 0 (0) | 0 (0) | 1877 (100) | 0 (0) | 1877 (100) | 0 (0) | 0 (0) | 0 (0) | 39 (2.1) | 1877 (100) | 39 (2.1) | 0 (0) | 0 (0) | 152 (8.1) | 26 (1.4) | 26 (1.4) | 26 (1.4) | 357 (19) | 1877 (100) | 1877 (100) | 1877 (100) | 1877 (100) | 1877 (100) | 1877 (100) | 0 (0) | 0 (0) | 0 (0) |
| Rumbold155 | 0 (0) | 308 (100) | 0 (0) | 0 (0) | 308 (100) | 308 (100) | 308 (100) | 2 (0.6) | 2 (0.6) | 2 (0.6) | 308 (100) | 308 (100) | 308 (100) | 0 (0) | 308 (100) | 0 (0) | 308 (100) | 308 (100) | 308 (100) | 308 (100) | 308 (100) | 26 (8.4) | 308 (100) | 308 (100) | 308 (100) | 308 (100) | 1 (0.3) | 1 (0.3) | 1 (0.3) |
| Vollebregt174 | 0 (0) | 1147 (0.3) | 1586 (0.4) | 0 (0) | 0 (0) | 406,286 (100) | 406,286 (100) | 0 (0) | 0 (0) | 0 (0) | 84,755 (20.9) | 406,286 (100) | 0 (0) | 0 (0) | 406,286 (100) | 57,224 (14.1) | 406,286 (100) | 406,286 (100) | 406,286 (100) | 406,286 (100) | 406,286 (100) | 406,286 (100) | 406,286 (100) | 406,286 (100) | 406,286 (100) | 406,286 (100) | 0 (0) | 9 (0) | 9 (0) |
| JSOG105 | 2 (0.1) | 0 (0) | 2399 (100) | 2399 (100) | 2399 (100) | 2399 (100) | 2399 (100) | 2399 (100) | 2399 (100) | 2399 (100) | 3 (0.1) | 2399 (100) | 3 (0.1) | 0 (0) | 2399 (100) | 27 (1.1) | 2399 (100) | 2399 (100) | 2399 (100) | 2399 (100) | 2399 (100) | 2399 (100) | 2399 (100) | 2399 (100) | 2399 (100) | 2399 (100) | 34 (1.4) | 34 (1.4) | 32 (1.3) |
| DOMInO143 | 86,082 (100) | 0 (0) | 0 (0) | 86,082 (100) | 0 (0) | 86,082 (100) | 86,082 (100) | 0 (0) | 0 (0) | 0 (0) | 3164 (3.7) | 86,082 (100) | 86,082 (100) | 0 (0) | 86,082 (100) | 9656 (11.2) | 86,082 (100) | 86,082 (100) | 86,082 (100) | 86,082 (100) | 86,082 (100) | 86,082 (100) | 86,082 (100) | 86,082 (100) | 86,082 (100) | 86,082 (100) | 0 (0) | 0 (0) | 0 (0) |
| Danish Birth Cohort136 | 0 (0) | 75 (3.4) | 674 (30.3) | 0 (0) | 2252 (100) | 2252 (100) | 2252 (100) | 2252 (100) | 1 (0) | 18 (0.8) | 1264 (56.1) | 2252 (100) | 2252 (100) | 0 (0) | 0 (0) | 177 (7.9) | 470 (20.9) | 470 (20.9) | 471 (20.9) | 2248 (99.8) | 2252 (100) | 2252 (100) | 2252 (100) | 2252 (100) | 2252 (100) | 2252 (100) | 655 (29.1) | 664 (29.5) | 664 (29.5) |
| Indonesian Cohort157 | 0 (0) | 288 (100) | 0 (0) | 4 (1.4) | 288 (100) | 288 (100) | 288 (100) | 288 (100) | 288 (100) | 288 (100) | 288 (100) | 288 (100) | 288 (100) | 0 (0) | 288 (100) | 288 (100) | 288 (100) | 288 (100) | 288 (100) | 288 (100) | 288 (100) | 288 (100) | 288 (100) | 288 (100) | 288 (100) | 288 (100) | 0 (0) | 0 (0) | 0 (0) |
| Ohkuchi148 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 244 (100) | 244 (100) | 244 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 244 (100) | 244 (100) | 0 (0) | 244 (100) | 20 (8.2) | 33 (13.5) | 33 (13.5) | 33 (13.5) | 244 (100) | 244 (100) | 244 (100) | 244 (100) | 244 (100) | 244 (100) | 244 (100) | 0 (0) | 0 (0) | 0 (0) |
| Lecarpentier139 | 557 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 557 (100) | 557 (100) | 0 (0) | 557 (100) | 0 (0) | 0 (0) | 557 (100) | 0 (0) | 0 (0) | 557 (100) | 1 (0.2) | 0 (0) | 0 (0) | 0 (0) | 557 (100) | 557 (100) | 3 (0.5) | 557 (100) | 0 (0) | 0 (0) | 557 (100) | 0 (0) | 0 (0) | 0 (0) |
| TEST146 | 1244 (100) | 0 (0) | 0 (0) | 0 (0) | 1244 (100) | 1244 (100) | 1244 (100) | 0 (0) | 0 (0) | 0 (0) | 1244 (100) | 1244 (100) | 1244 (100) | 0 (0) | 1244 (100) | 2 (0.2) | 724 (58.2) | 724 (58.2) | 724 (58.2) | 1244 (100) | 1244 (100) | 1244 (100) | 1244 (100) | 1244 (100) | 1244 (100) | 1244 (100) | 0 (0) | 109 (8.8) | 109 (8.8) |
| Massé144 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 240 (100) | 240 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 240 (100) | 240 (100) | 0 (0) | 240 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 240 (100) | 240 (100) | 240 (100) | 240 (100) | 240 (100) | 29 (12.1) | 8 (3.3) | 0 (0) | 0 (0) | 0 (0) |
| Staff162 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 823 (100) | 823 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 823 (100) | 0 (0) | 0 (0) | 0 (0) | 3 (0.4) | 15 (1.8) | 15 (1.8) | 15 (1.8) | 823 (100) | 823 (100) | 823 (100) | 71 (8.6) | 823 (100) | 823 (100) | 823 (100) | 0 (0) | 0 (0) | 0 (0) |
| STORK G135 | 736 (100) | 736 (100) | 4 (0.5) | 363 (49.3) | 736 (100) | 736 (100) | 736 (100) | 736 (100) | 736 (100) | 736 (100) | 736 (100) | 736 (100) | 736 (100) | 0 (0) | 736 (100) | 736 (100) | 736 (100) | 736 (100) | 736 (100) | 736 (100) | 736 (100) | 736 (100) | 736 (100) | 736 (100) | 17 (2.3) | 56 (7.6) | 0 (0) | 0 (0) | 0 (0) |
| Vatten170 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 304 (100) | 304 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 304 (100) | 304 (100) | 0 (0) | 304 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 304 (100) | 304 (100) | 304 (100) | 304 (100) | 304 (100) | 304 (100) | 304 (100) | 0 (0) | 0 (0) | 0 (0) |
| Vinter173 | 281,466 (100) | 281,466 (101.9) | 10,027 (3.6) | 162,627 (58.9) | 15,312 (5.4) | 281,466 (100) | 281,466 (100) | 15,312 (5.4) | 0 (0) | 15,312 (5.4) | 0 (0) | 21,145 (7.5) | 0 (0) | 0 (0) | 281,466 (100) | 48,947 (17.4) | 281,466 (100) | 281,466 (100) | 281,466 (100) | 281,466 (100) | 281,466 (100) | 281,466 (100) | 281,466 (100) | 281,466 (100) | 281,466 (100) | 281,466 (100) | 0 (0) | 188 (0.1) | 188 (0.1) |
| BORN Ontorio104 | 0 (0) | 0 (0) | 1016 (100) | 0 (0) | 1016 (100) | 1016 (100) | 1016 (100) | 1016 (100) | 1016 (100) | 0 (0) | 1016 (100) | 1016 (100) | 1016 (100) | 0 (0) | 1016 (100) | 8 (0.8) | 526 (51.8) | 526 (51.8) | 526 (51.8) | 532 (52.4) | 1016 (100) | 1016 (100) | 1016 (100) | 71 (7) | 71 (7) | 1016 (100) | 221 (21.8) | 228 (22.4) | 228 (22.4) |
| Ghana Cohort165 | 112,758 (100) | 496 (0.4) | 496 (0.4) | 63,352 (57.2) | 112,758 (100) | 112,758 (100) | 112,758 (100) | 496 (0.4) | 0 (0) | 496 (0.4) | 18,757 (16.6) | 0 (0) | 0 (0) | 0 (0) | 112,758 (100) | 15,779 (14) | 23,192 (20.6) | 23,348 (20.7) | 23,373 (20.7) | 112,758 (100) | 112,758 (100) | 112,758 (100) | 112,758 (100) | 112,758 (100) | 112,758 (100) | 112,758 (100) | 0 (0) | 26 (0) | 26 (0) |
| MoBA142 | 6587 (4.6) | 141,698 (100) | 23,866 (16.8) | 83,819 (59.2) | 141,698 (100) | 141,698 (100) | 141,698 (100) | 141,698 (100) | 0 (0) | 0 (0) | 141,698 (100) | 141,698 (100) | 141,698 (100) | 0 (0) | 141,698 (100) | 141,698 (100) | 141,698 (100) | 141,698 (100) | 141,698 (100) | 141,698 (100) | 141,698 (100) | 141,698 (100) | 141,698 (100) | 134,442 (94.9) | 141,698 (100) | 141,698 (100) | 0 (0) | 1036 (0.7) | 1036 (0.7) |
| Huang132 | 42,829 (100) | 0 (0) | 53 (0.1) | 0 (0) | 0 (0) | 42,829 (100) | 42,829 (100) | 42,829 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 451 (1.1) | 2722 (6.4) | 122 (0.3) | 118 (0.3) | 123 (0.3) | 5752 (13.4) | 42,829 (100) | 42,829 (100) | 42,829 (100) | 42,829 (100) | 42,829 (100) | 42,829 (100) | 0 (0) | 0 (0) | 0 (0) |
| Carbillion119 | 3317 (100) | 3317 (100) | 112 (3.4) | 0 (0) | 0 (0) | 3317 (100) | 3317 (100) | 0 (0) | 0 (0) | 592 (17.8) | 3317 (100) | 3317 (100) | 3317 (100) | 0 (0) | 3317 (100) | 3317 (100) | 3317 (100) | 3317 (100) | 3317 (100) | 3317 (100) | 3317 (100) | 3317 (100) | 3317 (100) | 3317 (100) | 3317 (100) | 3317 (100) | 602 (18.1) | 602 (18.1) | 602 (18.1) |
| Goffinet129 | 42 (100) | 42 (100) | 0 (0) | 0 (0) | 42 (100) | 42 (100) | 42 (100) | 42 (100) | 42 (100) | 0 (0) | 0 (0) | 42 (100) | 42 (100) | 0 (0) | 42 (100) | 42 (100) | 0 (0) | 0 (0) | 0 (0) | 42 (100) | 42 (100) | 0 (0) | 42 (100) | 42 (100) | 42 (100) | 42 (100) | 0 (0) | 0 (0) | 0 (0) |
| Rang152 | 0 (0) | 173 (100) | 0 (0) | 0 (0) | 173 (100) | 173 (100) | 173 (100) | 173 (100) | 173 (100) | 0 (0) | 173 (100) | 173 (100) | 173 (100) | 0 (0) | 0 (0) | 173 (100) | 173 (100) | 173 (100) | 173 (100) | 173 (100) | 173 (100) | 173 (100) | 173 (100) | 173 (100) | 173 (100) | 173 (100) | 0 (0) | 0 (0) | 0 (0) |
| Cameroni 2011117 | 0 (0) | 53 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 53 (100) | 0 (0) | 0 (0) | 0 (0) | 53 (100) | 53 (100) | 53 (100) | 0 (0) | 0 (0) | 53 (100) | 53 (100) | 53 (100) | 53 (100) | 53 (100) | 53 (100) | 53 (100) | 53 (100) | 53 (100) | 53 (100) | 53 (100) | 0 (0) | 0 (0) | 0 (0) |
| Conserva 2012123 | 0 (0) | 2 (1.2) | 0 (0) | 1 (0.6) | 0 (0) | 0 (0) | 172 (100) | 0 (0) | 0 (0) | 0 (0) | 23 (13.4) | 172 (100) | 172 (100) | 0 (0) | 172 (100) | 90 (52.3) | 172 (100) | 172 (100) | 172 (100) | 172 (100) | 172 (100) | 172 (100) | 172 (100) | 172 (100) | 172 (100) | 172 (100) | 0 (0) | 0 (0) | 0 (0) |
| Facchinetti124 | 0 (0) | 54 (100) | 0 (0) | 0 (0) | 0 (0) | 54 (100) | 54 (100) | 0 (0) | 0 (0) | 0 (0) | 54 (100) | 54 (100) | 54 (100) | 0 (0) | 54 (100) | 54 (100) | 3 (5.6) | 3 (5.6) | 3 (5.6) | 54 (100) | 54 (100) | 54 (100) | 54 (100) | 54 (100) | 54 (100) | 54 (100) | 3 (5.6) | 3 (5.6) | 3 (5.6) |
| Ferrazzani158 | 0 (0) | 81 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 81 (100) | 81 (100) | 0 (0) | 0 (0) | 81 (100) | 81 (100) | 81 (100) | 0 (0) | 0 (0) | 81 (100) | 81 (100) | 81 (100) | 81 (100) | 81 (100) | 81 (100) | 81 (100) | 81 (100) | 81 (100) | 81 (100) | 81 (100) | 13 (16) | 14 (17.3) | 14 (17.3) |
| Figueiró-Filho125 | 12 (5.9) | 2 (1) | 0 (0) | 0 (0) | 19 (9.1) | 19 (9.1) | 208 (100) | 0 (0) | 19 (9.1) | 16 (7.7) | 23 (11.1) | 208 (100) | 208 (100) | 0 (0) | 208 (100) | 111 (53.4) | 102 (49) | 89 (42.8) | 102 (49) | 208 (100) | 208 (100) | 208 (100) | 208 (100) | 208 (100) | 208 (100) | 208 (100) | 3 (1.4) | 4 (1.9) | 4 (1.9) |
| Langenveld138 | 536,414 (100) | 0 (0) | 536,414 (100) | 536,414 (100) | 536,414 (100) | 536,414 (100) | 536,414 (100) | 536,414 (100) | 0 (0) | 536,414 (100) | 189,726 (35.4) | 536,414 (100) | 536,414 (100) | 0 (0) | 536,414 (100) | 467,141 (87.1) | 536,414 (100) | 536,414 (100) | 536,414 (100) | 536,414 (100) | 536,414 (100) | 536,414 (100) | 536,414 (100) | 536,414 (100) | 536,414 (100) | 536,414 (100) | 0 (0) | 114 (0) | 114 (0) |
| Lykke141 | 46,177 (2.9) | 44,301 (2.8) | 1,663,167 (100) | 1,663,167 (100) | 1,663,167 (100) | 1,663,167 (100) | 1,663,167 (100) | 45,547 (2.7) | 45,547 (2.7) | 45,547 (2.7) | 54,336 (3.3) | 1,663,167 (100) | 1,663,167 (100) | 0 (0) | 1,663,167 (100) | 82,947 (5) | 1,663,167 (100) | 1,663,167 (100) | 1,663,167 (100) | 1,663,167 (100) | 1,663,167 (100) | 1,663,167 (100) | 1,663,167 (100) | 1,663,167 (100) | 1,663,167 (100) | 1,663,167 (100) | 45,547 (2.7) | 45,598 (2.7) | 45,598 (2.7) |
| Mbah145 | 0 (0) | 37,738 (100) | 0 (0) | 0 (0) | 37,738 (100) | 37,738 (100) | 37,738 (100) | 0 (0) | 0 (0) | 0 (0) | 30,152 (79.9) | 37,738 (100) | 37,738 (100) | 0 (0) | 37,738 (100) | 37,738 (100) | 37,738 (100) | 37,738 (100) | 37,738 (100) | 37,738 (100) | 37,738 (100) | 3,7738 (100) | 37,738 (100) | 37,738 (100) | 37,738 (100) | 37,738 (100) | 299 (0.8) | 615 (1.6) | 615 (1.6) |
| Trogstad164 | 97 (100) | 97 (100) | 97 (100) | 97 (100) | 97 (100) | 97 (100) | 97 (100) | 97 (100) | 97 (100) | 97 (100) | 97 (100) | 97 (100) | 97 (100) | 0 (0) | 97 (100) | 97 (100) | 97 (100) | 97 (100) | 97 (100) | 97 (100) | 97 (100) | 97 (100) | 97 (100) | 97 (100) | 97 (100) | 97 (100) | 0 (0) | 0 (0) | 0 (0) |
| Salim156 | 82 (19.3) | 232 (54.6) | 0 (0) | 0 (0) | 278 (65.4) | 278 (65.4) | 425 (100) | 278 (65.4) | 278 (65.4) | 273 (64.2) | 242 (56.9) | 425 (100) | 425 (100) | 0 (0) | 0 (0) | 265 (62.4) | 251 (59.1) | 247 (58.1) | 251 (59.1) | 425 (100) | 425 (100) | 425 (100) | 425 (100) | 425 (100) | 425 (100) | 425 (100) | 235 (55.3) | 235 (55.3) | 235 (55.3) |
| van Oostwaard 2012168 | 138 (21.6) | 329 (51.5) | 0 (0) | 0 (0) | 407 (63.7) | 407 (63.7) | 639 (100) | 408 (63.8) | 406 (63.5) | 404 (63.2) | 350 (54.8) | 639 (100) | 639 (100) | 0 (0) | 0 (0) | 388 (60.7) | 373 (58.4) | 373 (58.4) | 373 (58.4) | 639 (100) | 639 (100) | 639 (100) | 639 (100) | 639 (100) | 639 (100) | 639 (100) | 335 (52.4) | 335 (52.4) | 335 (52.4) |
| van Oostwaard 2014169 | 19 (1.2) | 19 (1.2) | 1639 (100) | 1639 (100) | 1639 (100) | 33 (2) | 1639 (100) | 30 (1.8) | 19 (1.2) | 17 (1) | 26 (1.6) | 1639 (100) | 1639 (100) | 0 (0) | 17 (1) | 78 (4.8) | 1639 (100) | 1639 (100) | 1639 (100) | 1639 (100) | 1639 (100) | 1639 (100) | 1639 (100) | 1639 (100) | 1639 (100) | 1639 (100) | 0 (0) | 0 (0) | 0 (0) |
| Zhang178 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 4212 (100) | 4212 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 4212 (100) | 0 (0) | 0 (0) | 4212 (100) | 6 (0.1) | 143 (3.4) | 143 (3.4) | 144 (3.4) | 4212 (100) | 4212 (100) | 133 (3.2) | 70 (1.7) | 134 (3.2) | 8 (0.2) | 8 (0.2) | 5 (0.1) | 5 (0.1) | 5 (0.1) |
| Brown 2007116 | 2785 (100) | 2785 (100) | 0 (0) | 1551 (57.4) | 2785 (100) | 2785 (100) | 2785 (100) | 2785 (100) | 0 (0) | 0 (0) | 1368 (49.1) | 2785 (100) | 2785 (100) | 0 (0) | 2785 (100) | 2785 (100) | 165 (5.9) | 165 (5.9) | 165 (5.9) | 2785 (100) | 2785 (100) | 2785 (100) | 2785 (100) | 2785 (100) | 2785 (100) | 2785 (100) | 0 (0) | 0 (0) | 0 (0) |
| Gurgel Alves130 | 500 (100) | 500 (100) | 500 (100) | 500 (100) | 500 (100) | 500 (100) | 500 (100) | 500 (100) | 500 (100) | 0 (0) | 500 (100) | 500 (100) | 500 (100) | 0 (0) | 500 (100) | 500 (100) | 500 (100) | 500 (100) | 500 (100) | 500 (100) | 500 (100) | 7 (1.4) | 500 (100) | 500 (100) | 500 (100) | 500 (100) | 246 (49.2) | 246 (49.2) | 246 (49.2) |
| Rocha153 | 733 (100) | 0 (0) | 733 (100) | 0 (0) | 733 (100) | 733 (100) | 733 (100) | 0 (0) | 733 (100) | 733 (100) | 733 (100) | 733 (100) | 733 (100) | 0 (0) | 733 (100) | 0 (0) | 733 (100) | 733 (100) | 0 (0) | 733 (100) | 733 (100) | 733 (100) | 733 (100) | 733 (100) | 733 (100) | 733 (100) | 0 (0) | 0 (0) | 0 (0) |
| Costa 2017107 | 2 (0.3) | 0 (0) | 0 (0) | 0 (0) | 574 (100) | 0 (0) | 574 (100) | 574 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 574 (100) | 574 (100) | 0 (0) | 574 (100) | 574 (100) | 32 (5.6) | 574 (100) | 574 (100) | 574 (100) | 574 (100) | 0 (0) | 0 (0) | 0 (0) |
| Rocha154 | 29 (4) | 0 (0) | 0 (0) | 0 (0) | 733 (100) | 733 (100) | 733 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 733 (100) | 733 (100) | 0 (0) | 733 (100) | 0 (0) | 733 (100) | 733 (100) | 0 (0) | 733 (100) | 733 (100) | 733 (100) | 733 (100) | 733 (100) | 733 (100) | 733 (100) | 0 (0) | 0 (0) | 0 (0) |
Appendix 7 International Prediction of Pregnancy Complications Collaborators pre-eclampsia predictors prioritisation survey
Appendix 8 Risk-of-bias assessment of data sets on the IPPIC project
| Study/data set | Domain: participant selection | ||||
|---|---|---|---|---|---|
| Appropriate data sources | Appropriate inclusion and exclusion of participants | Participant selection similar to model development study | Risk | Rationale of rating | |
| SCOPE42 | Yes | Yes | N/A | Low | Responses to relevant signalling questions are yes |
| Allen108 | Yes | Yes | N/A | Low | Responses to relevant signalling questions are yes |
| ALSPAC103 | Yes | Yes | N/A | Low | Responses to relevant signalling questions are yes |
| Chappell121 | Yes | No | N/A | High | Selected high-risk population |
| EMPOWaR122 | Yes | No | N/A | High | Selected high-risk population |
| POPPY106 | Yes | Yes | N/A | Low | Responses to relevant signalling questions are yes |
| Poston 2006150 | Yes | No | N/A | High | Selected high-risk population |
| Poston 2015149 | Yes | No | N/A | High | Selected high-risk population |
| Khan137 | Yes | No | N/A | High | Selected high-risk population |
| St George’s163 | Yes | Yes | N/A | Low | Responses to relevant signalling questions are yes |
| PARIS Collaborative Group112 | Yes | Probably no | N/A | High | Likely to be a mix of high- and low-risk population |
| AMND114 | Yes | Yes | N/A | Low | Responses to relevant signalling questions are yes |
| Born in Bradford177 | Yes | Yes | N/A | Low | Responses to relevant signalling questions are yes |
| Velauthar39 | Yes | Yes | N/A | Low | Responses to relevant signalling questions are yes |
| POP161 | Yes | Yes | N/A | Low | Responses to relevant signalling questions are yes |
| Baschat115 | Yes | Yes | N/A | Low | Responses to relevant signalling questions are yes |
| Audibert113 | Yes | Yes | N/A | Low | Responses to relevant signalling questions are yes |
| Caradeux118 | Yes | Yes | N/A | Low | Responses to relevant signalling questions are yes |
| Giguère126 | Yes | Yes | N/A | Low | Responses to relevant signalling questions are yes |
| Goetzinger128 | Yes | Yes | N/A | Low | Responses to relevant signalling questions are yes |
| Antsaklis110 | Yes | Yes | N/A | Low | Responses to relevant signalling questions are yes |
| Llurba140 | Yes | Yes | N/A | Low | Responses to relevant signalling questions are yes |
| WHO175 | Yes | No | N/A | High | Selected high-risk population |
| Andersen109 | Yes | Yes | N/A | Low | Responses to relevant signalling questions are yes |
| Arenas111 | Yes | Yes | N/A | Low | Responses to relevant signalling questions are yes |
| FINNPEC133 | Yes | Yes | N/A | Low | Responses to relevant signalling questions are yes |
| Galindo171 | Yes | Yes | N/A | Low | Responses to relevant signalling questions are yes |
| Generation R134 | Yes | Yes | N/A | Low | Responses to relevant signalling questions are yes |
| NICHD HR32 | Yes | No | N/A | High | Selected high-risk population |
| NICHD LR159 | Yes | Yes | N/A | Low | Responses to relevant signalling questions are yes |
| Placental Health Study176 | Yes | Yes | N/A | Low | Responses to relevant signalling questions are yes |
| POUCH131 | Yes | Yes | N/A | Low | Responses to relevant signalling questions are yes |
| van Kuijk 2011167 | Yes | No | N/A | High | Selected high-risk population |
| van Kuijk 2014166 | Yes | No | N/A | High | Selected high-risk population |
| Odibo147 | Yes | Yes | N/A | Low | Responses to relevant signalling questions are yes |
| PREDO127 | Yes | Yes | N/A | Low | Responses to relevant signalling questions are yes |
| Prefumo151 | Yes | Yes | N/A | Low | Responses to relevant signalling questions are yes |
| Skråstad160 | Yes | No | N/A | High | Selected high-risk population |
| Verlohren172 | Yes | Yes | N/A | Low | Responses to relevant signalling questions are yes |
| Rumbold155 | Yes | Yes | N/A | Low | Responses to relevant signalling questions are yes |
| Vollebregt174 | Yes | No | N/A | High | Selected high-risk population |
| JSOG105 | Yes | Yes | N/A | Low | Responses to relevant signalling questions are yes |
| DOMInO143 | Yes | Yes | N/A | Low | Responses to relevant signalling questions are yes |
| Danish Birth Cohort136 | Yes | Yes | N/A | Low | Responses to relevant signalling questions are yes |
| Indonesian Cohort157 | Yes | Yes | N/A | Low | Responses to relevant signalling questions are yes |
| Ohkuchi148 | Yes | Yes | N/A | Low | Responses to relevant signalling questions are yes |
| Lecarpentier139 | Yes | No | N/A | High | Selected high-risk population |
| TEST146 | Yes | Yes | N/A | Low | Responses to relevant signalling questions are yes |
| Massé144 | Yes | Yes | N/A | Low | Responses to relevant signalling questions are yes |
| Staff162 | Yes | Yes | N/A | Low | Responses to relevant signalling questions are yes |
| STORK G135 | Yes | Yes | N/A | Low | Responses to relevant signalling questions are yes |
| Vatten170 | Yes | Yes | N/A | Low | Responses to relevant signalling questions are yes |
| Vinter173 | Yes | No | N/A | High | Selected high-risk population |
| BORN Ontorio104 | Yes | Yes | N/A | Low | Responses to relevant signalling questions are yes |
| Ghana Cohort165 | Yes | Yes | N/A | Low | Responses to relevant signalling questions are yes |
| MoBa142 | Yes | Yes | N/A | Low | Responses to relevant signalling questions are yes |
| Huang132 | Yes | Yes | N/A | Low | Responses to relevant signalling questions are yes |
| Carbillion119 | Yes | Yes | N/A | Low | Responses to relevant signalling questions are yes |
| Goffinet129 | Yes | Yes | N/A | Low | Responses to relevant signalling questions are yes |
| Rang152 | Yes | No | N/A | High | Selected high-risk population |
| Cameroni117 | Yes | No | N/A | High | Selected high-risk population |
| Conserva123 | Yes | No | N/A | High | Selected high-risk population |
| Facchinetti124 | Yes | No | N/A | High | Selected high-risk population |
| Ferrazzani158 | Yes | No | N/A | High | Selected high-risk population |
| Figueiro-Filho125 | Yes | No | N/A | High | Selected high-risk population |
| Langenveld138 | Yes | No | N/A | High | Selected high-risk population |
| Lykke141 | Yes | Yes | N/A | Low | Responses to relevant signalling questions are yes |
| Mbah145 | Yes | Yes | N/A | Low | Responses to relevant signalling questions are yes |
| Trogstad164 | Yes | Yes | N/A | Low | Responses to relevant signalling questions are yes |
| Salim156 | Yes | No | N/A | High | Selected high-risk population |
| van Oostwaard 2012168 | Yes | No | N/A | High | Selected high-risk population |
| van Oostwaard 2014169 | Yes | No | N/A | High | Selected high-risk population |
| Zhang178 | Yes | Yes | N/A | Low | Responses to relevant signalling questions are yes |
| Brown116 | Yes | Yes | N/A | Low | Responses to relevant signalling questions are yes |
| Gurgel Alves130 | Yes | Yes | N/A | Low | Responses to relevant signalling questions are yes |
| Rocha153 | Yes | Yes | N/A | Low | Responses to relevant signalling questions are yes |
| Costa 2017107 | Yes | Yes | N/A | Low | Responses to relevant signalling questions are yes |
| Rocha154 | Yes | Yes | N/A | Low | Responses to relevant signalling questions are yes |
| Study/data set | Domain: predictors | |||||
|---|---|---|---|---|---|---|
| Predictors defined in a similar way for participants | Predictors defined in a similar way to model development study | Predictors assessed without knowledge of outcome data | All predictors available at the time model is to be used | Risk | Rationale of rating | |
| SCOPE42 | Yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| Allen108 | Yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| ALSPAC103 | Yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| Chappell121 | Yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| EMPOWaR122 | Yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| POPPY106 | NI | N/A | NI | Yes | Unclear | No information to make assessment |
| Poston 2006150 | Yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| Poston 2015149 | Yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| Khan137 | Yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| St George’s163 | Probably yes | N/A | Probably yes | Yes | Low | Responses to relevant signalling questions are yes |
| PARIS Collaborative Group112 | Probably no | N/A | Probably yes | Yes | High | Definition of predictor may differ across studies |
| AMND114 | Probably yes | N/A | Probably yes | Yes | Low | Responses to relevant signalling questions are yes |
| Born in Bradford177 | Yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| Velauthar39 | NI | N/A | NI | Yes | Unclear | No information to make assessment |
| POP161 | Yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| Baschat115 | Yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| Audibert113 | Yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| Caradeux118 | Yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| Giguère126 | Yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| Goetzinger128 | Yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| Antsaklis110 | Yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| Llurba140 | Yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| WHO175 | Yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| Andersen109 | Yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| Arenas111 | Yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| FINNPEC133 | Yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| Galindo171 | Yes | N/A | Probably yes | Yes | Low | Responses to relevant signalling questions are yes |
| Generation R134 | Yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| NICHD HR32 | Yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| NICHD LR159 | Yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| Placental Health Study176 | Yes | N/A | Probably yes | Yes | Low | Responses to relevant signalling questions are yes |
| POUCH131 | Yes | N/A | Probably yes | Yes | Low | Responses to relevant signalling questions are yes |
| van Kuijk 2011167 | Yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| van Kuijk 2014166 | Yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| Odibo147 | Yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| PREDO127 | Yes | N/A | Probably yes | Yes | Low | Responses to relevant signalling questions are yes |
| Prefumo151 | Yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| Skråstad160 | Yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| Verlohren172 | Yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| Rumbold155 | Yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| Vollebregt174 | Yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| JSOG105 | NI | N/A | NI | Yes | Unclear | No information to make assessment |
| DOMInO143 | Yes | N/A | Probably yes | Yes | Low | Responses to relevant signalling questions are yes |
| Danish Birth Cohort136 | Probably yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| Indonesian Cohort157 | Yes | N/A | Probably yes | Yes | Low | Responses to relevant signalling questions are yes |
| Ohkuchi148 | Yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| Lecarpentier139 | Yes | N/A | Probably yes | Yes | Low | Responses to relevant signalling questions are yes |
| TEST146 | Yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| Massé144 | Probably yes | N/A | Probably yes | Yes | Low | Responses to relevant signalling questions are yes |
| Staff162 | Yes | N/A | Probably yes | Yes | Low | Responses to relevant signalling questions are yes |
| STORK G135 | Yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| Vatten170 | Yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| Vinter173 | Yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| BORN Ontorio104 | Yes | N/A | Probably yes | Yes | Low | Responses to relevant signalling questions are yes |
| Ghana Cohort165 | Yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| MoBa142 | Probably yes | N/A | Probably yes | Yes | Low | Responses to relevant signalling questions are yes |
| Huang132 | Probably yes | N/A | Probably yes | Yes | Low | Responses to relevant signalling questions are yes |
| Carbillion119 | Probably yes | N/A | NI | Yes | Unclear | No information to make assessment |
| Goffinet129 | Yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| Rang152 | Yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| Cameroni117 | Yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| Conserva123 | Yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| Facchinetti124 | Yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| Ferrazzani158 | Yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| Figueiró-Filho125 | Yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| Langenveld138 | Yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| Lykke141 | Probably yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| Mbah145 | Probably yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| Trogstad164 | Probably yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| Salim156 | Yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| van Oostwaard 2012168 | Yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| van Oostwaard 2014169 | Yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| Zhang178 | Yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| Brown 2007116 | Yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| Gurgel Alves130 | Yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| Rocha153 | Yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| Costa 2017107 | Yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| Rocha154 | Yes | N/A | Yes | Yes | Low | Responses to relevant signalling questions are yes |
| Study/data set | Domain: outcome | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Outcome appropriately determined | Prespecified or standard definition used | Predictors excluded from outcome definition | Outcome defined and determined in the same way for all participants | Outcome defined and determined in the same way to model development study | Outcome determined without knowledge of predictor information | Time interval between predictor and outcome assessment appropriate | Risk | Rationale of rating | |
| SCOPE42 | Yes | Yes | No | Yes | N/A | No | Yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| Allen108 | NI | NI | Probably no | Probably yes | N/A | Probably no | Probably yes | Unclear | No information to make assessment |
| ALSPAC103 | NI | NI | Probably no | Probably yes | N/A | Probably no | Probably yes | Unclear | No information to make assessment |
| Chappell121 | Yes | Yes | No | Yes | N/A | No | Yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| EMPOWaR122 | NI | NI | Probably no | Probably yes | N/A | Probably no | Probably yes | Unclear | No information to make assessment |
| POPPY106 | NI | NI | Probably no | Probably yes | N/A | Probably no | Probably yes | Unclear | No information to make assessment |
| Poston 2006150 | Yes | Yes | No | Yes | N/A | No | Yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| Poston 2015149 | Yes | Yes | No | Yes | N/A | No | Yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| Khan137 | Yes | Yes | No | Yes | N/A | No | Yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| St George’s163 | NI | NI | Probably no | Probably yes | N/A | Probably no | Probably yes | Unclear | No information to make assessment |
| PARIS Collaborative Group112 | Yes | Yes | No | Yes | N/A | No | Yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| AMND114 | NI | NI | Probably no | Probably yes | N/A | Probably no | Probably yes | Unclear | No information to make assessment |
| Born in Bradford177 | NI | NI | Probably no | Probably yes | N/A | Probably no | Probably yes | Unclear | No information to make assessment |
| Velauthar39 | NI | NI | Probably no | Probably yes | N/A | Probably no | Probably yes | Unclear | No information to make assessment |
| POP161 | Yes | Yes | No | Yes | N/A | No | Yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| Baschat115 | Yes | Yes | No | Yes | N/A | No | Yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| Audibert113 | Yes | Yes | No | Yes | N/A | No | Yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| Caradeux118 | Yes | Yes | No | Yes | N/A | No | Yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| Giguère126 | Yes | Yes | No | Yes | N/A | No | Yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| Goetzinger128 | Yes | Yes | No | Yes | N/A | No | Yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| Antsaklis110 | Yes | Yes | No | Yes | N/A | No | Yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| Llurba140 | Yes | Yes | No | Yes | N/A | No | Yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| WHO175 | Yes | Yes | No | Probably yes | N/A | Probably no | Yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| Andersen109 | Yes | Yes | No | Yes | N/A | No | Yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| Arenas111 | Yes | Yes | No | Yes | N/A | No | Yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| FINNPEC133 | Yes | Yes | No | Yes | N/A | No | Yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| Galindo171 | Yes | Yes | No | Yes | N/A | No | Yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| Generation R134 | Yes | Probably yes | No | Probably yes | N/A | Probably no | Probably yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| NICHD HR32 | Yes | Yes | No | Yes | N/A | No | Yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| NICHD LR159 | Yes | Yes | No | Yes | N/A | No | Yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| Placental Health Study176 | Yes | Yes | No | Yes | N/A | No | Yes | Unclear | No information to make assessment |
| POUCH131 | NI | NI | Probably no | Probably yes | N/A | Probably no | Probably yes | Unclear | No information to make assessment |
| van Kuijk 2011167 | Yes | Yes | No | Yes | N/A | No | Yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| van Kuijk 2014166 | Yes | Yes | No | Yes | N/A | No | Yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| Odibo147 | Yes | Yes | No | Yes | N/A | No | Yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| PREDO127 | Yes | Yes | No | Yes | N/A | No | Yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| Prefumo151 | Yes | Yes | No | Yes | N/A | No | Yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| Skråstad160 | Yes | Yes | No | Yes | N/A | No | Yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| Verlohren172 | Yes | Yes | No | Yes | N/A | No | Yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| Rumbold155 | Yes | Yes | No | Yes | N/A | No | Yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| Vollebregt174 | Yes | Yes | No | Yes | N/A | No | Yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| JSOG105 | NI | NI | Probably no | Probably yes | N/A | Probably no | Probably yes | Unclear | No information to make assessment |
| DOMInO143 | Yes | Yes | No | Yes | N/A | No | Yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| Danish Birth Cohort136 | Yes | Probably yes | No | Probably yes | N/A | Probably no | Probably yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| Indonesian cohort157 | Yes | Yes | No | Yes | N/A | No | Yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| Ohkuchi148 | Yes | Yes | No | Yes | N/A | No | Yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| Lecarpentier139 | Yes | Yes | No | Yes | N/A | No | Yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| TEST146 | Yes | Yes | No | Yes | N/A | No | Yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| Massé144 | NI | NI | Probably no | Probably yes | N/A | Probably no | Probably yes | Unclear | No information to make assessment |
| Staff162 | Yes | Yes | No | Yes | N/A | No | Yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| STORK G135 | NI | NI | Probably no | Probably yes | N/A | Probably no | Probably yes | Unclear | No information to make assessment |
| Vatten170 | Yes | Yes | No | Yes | N/A | No | Yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| Vinter173 | Yes | Yes | No | Yes | N/A | No | Yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| BORN Ontorio104 | NI | NI | Probably no | Probably yes | N/A | Probably no | Probably yes | Unclear | Unable to confirm definition of outcome used |
| Ghana cohort165 | Yes | Yes | No | Yes | N/A | No | Yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| MoBA142 | NI | NI | Probably no | Probably yes | N/A | Probably no | Probably yes | Unclear | No information to make assessment |
| Huang132 | NI | NI | Probably no | Probably yes | N/A | Probably no | Probably yes | Unclear | No information to make assessment |
| Carbillion119 | NI | NI | Probably no | Probably yes | N/A | Probably no | Probably yes | Unclear | No information to make assessment |
| Goffinet129 | Yes | Yes | No | Yes | N/A | No | Yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| Rang152 | Yes | Yes | No | Yes | N/A | No | Yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| Cameroni 2011117 | Yes | Yes | No | Yes | N/A | No | Yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| Conserva 2012123 | Yes | Yes | No | Yes | N/A | No | Yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| Facchinetti124 | Yes | Yes | No | Yes | N/A | No | Yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| Ferrazzani158 | Yes | Yes | No | Yes | N/A | No | Yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| Figueiró-Filho125 | Yes | Yes | No | Yes | N/A | No | Yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| Langenveld138 | Yes | Yes | No | Probably yes | N/A | Probably no | Probably yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| Lykke141 | Yes | Yes | No | Probably yes | N/A | Probably no | Probably yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| Mbah145 | Yes | Yes | No | Probably yes | N/A | Probably no | Probably yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| Trogstad164 | Yes | Yes | No | Probably yes | N/A | Probably no | Probably yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| Salim156 | Yes | Yes | No | Yes | N/A | No | Yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| van Oostwaard 2012168 | Yes | Yes | No | Yes | N/A | No | Yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| van Oostwaard 2014169 | Yes | Yes | No | Yes | N/A | No | Yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| Zhang178 | Yes | Yes | No | Yes | N/A | No | Yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| Brown 2007116 | Yes | Yes | No | Yes | N/A | No | Yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| Gurgel Alves130 | Yes | Yes | No | Yes | N/A | No | Yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| Rocha153 | Yes | Yes | No | Yes | N/A | No | Yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| Costa 2017107 | Yes | Yes | No | Yes | N/A | No | Yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
| Rocha154 | Yes | Yes | No | Yes | N/A | No | Yes | Low | Blood pressure and proteinuria are components of predictor and need to be known for diagnosis |
Appendix 9 Prediction models and equations identified from the literature search
For more information, see published reviews. 37,42 Model equations were extracted from the original manuscripts, which may not incorporate later corrections to the equation by the authors, if any.
| First author, year | Predictor category | Trimester of measurement of variables | Equation(s)a | Included in validation | If no, reason for exclusion |
|---|---|---|---|---|---|
| Any-onset pre-eclampsia | |||||
| Baschat, 2014115 | C + B | First trimester | LP: –8.72 + 0.157 (if nulliparous) + 0.341 (if history of hypertension) + 0.635 (if history of PE) + 0.064 (MAP) – 0.186 (PAPP-A, Ln MoM) | Yes | |
| North, 201142 | C | 15 weeks’ gestation | LP1: –6.8855 – 0.0393 (age, years) + 0.0659 (MAP) + 0.0483 (BMI) + 0.6861 (if family history of PE) + 0.6232 (if family history CHD) – 0.3881 (woman’s birthweight, kg) + 0.7129 (if vaginal bleeding ≥ 5 days) – 0.8033 (if one miscarriage ≤ 10 weeks, same partner) – 0.9070 (if ≥ 12 months to conceive) – 0.3733 (if high fruit intake at 15 weeks) – 0.508 (if alcohol consumed in first trimester) – 0.063 (number of cigarettes/day at 15 weeks) | No | Predictor not available in IPPIC-UK data set |
| C + U | LP2: –9.1113 + 0.0634 (MAP) + 0.0485 (BMI) + 0.6539 (if family history of PE) + 0.6093 (if family history of CHD) – 0.3787 (participant’s birthweight, kg) + 0.6493 (if vaginal bleeding ≥ 5 days) + 0.5008 (if months in sexual relationship ≤ 6 months) + 0.5084 (if bilateral notches) + 2.0802 (mean Ut RI) – 0.8248 (one miscarriage ≤ 10 weeks, same partner) – 0.8983 (≥ 12 months to conceive) – 0.4389 (high fruit intake at 15 weeks) – 0.5573 (if alcohol consumed in first trimester) | No | Predictor not available in IPPIC-UK data set | ||
| Odibo, 2011147 | C + B | First trimester | LP1: –3.389 – 0.716 (PAPP-A, MoM) + 0.05 (BMI) + 0.319 (if black ethnicity) + 1.57 (if history of chronic hypertension) | Yes | |
| C + B | LP2: –2.607 – 0.502 (PP13, MoM) + 0.759 (if pre-gestational diabetic) + 0.777 (if black ethnicity) + 1.268 (if history of chronic hypertension) | No | Predictor not available in IPPIC-UK data set | ||
| C + U | LP3: –3.895 – 0.593 (mean uterine artery PI) + 0.944 (if pre-gestational diabetes) + 0.059 (BMI) + 1.532 (if history of chronic hypertension) | Yes | |||
| C + B + U | LP4: –1.308 – 0.574 (PP13, MoM) – 0.502 (PAPP-A, MoM) – 0.643 (mean uterine artery PI) + 0.799 (if pre-gestational diabetic) + 0.664 (if black ethnicity) + 1.340 (if history of chronic hypertension) | No | Predictor not available in IPPIC-UK data set | ||
| Seed, 2011 | C | Second trimester | LP: –1.2422 + 0.4061 (if chronic hypertension) + 0.5071 (if DBP > 70) – 0.3846 (if DBP > 90) + 0.8890 (if SBP > 120) + 0.7040 (if previous PE) – 0.4043 (if on folates) + 0.8311 (if mother is Indian, Bangladeshi, Pakistani, African or African Caribbean) | No | Predictor not available in IPPIC-UK data set |
| Goetzinger, 2010128 | C + B | First trimester | LP: –3.25 + 0.51 (if PAPP-A < 10th centile) + 0.93 (if BMI > 25) + 0.94 (if chronic hypertension) + 0.97 (if diabetes) + 0.61 (if African American ethnicity) | Yes | |
| Emonts, 2008 | C | Post pregnancy | LP: –3.72 + 0.030 (age, years) – 0.50 (parity) + 0.15 (gestation) + 1.89 (if chronic hypertension in patient’s mother) + 0.14 (BMI) + 0.079 (SBP) – 0.13 (DBP) | No | Predictor not available in IPPIC-UK data set |
| Poon, 2008233 | C | First trimester | LP: –6.311 + 1.299 (if African Caribbean ethnicity) + 0.092 (BMI) + 0.855 (if woman’s mother had PE) – 1.481 (if parous without previous PE) + 0.933 (if parous with previous PE) | Yes | |
| Plasencia, 2007231 | C | First trimester | LP: –6.253 + 1.432 (if African Caribbean ethnicity) + 1.465 (if mixed ethnicity) + 0.084 (BMI) + 0.81 (if patient’s mother had PE) – 1.539 (if parous without previous PE) + 1.049 (if parous with previous PE) | Yes | |
| Yu, 2005238 | C + U | Second trimester | LP: 1.8552 + 5.9228 (mean uterine artery PI)–2 – 14.4474 (mean uterine artery PI)–1 – 0.5478 (if smoker) + 0.6719 (bilateral notch) + 0.0372 (age) + 0.4949 (if black ethnicity) + 1.5033 (if history of PE) – 1.2217 (if previous term live birth) + 0.0367 (BMI) | Yes | |
| Kenny, 2014 | C + B + U | First trimester | LP: −12.200−0.655 (high fruit intake) + 0.054 (BMI) + 0.065 (MAP) + 2.569 (mean uterine artery RI)−0.311 (PlGF artery RI) + 1.232 (cystatin C/PlGF) | No | Predictor not available in IPPIC-UK data set |
| Direkvand-Moghadam, 2013 | C | First trimester | LP: 0.74 – 1.016 prior infertility + 0.72 chronic hypertension + 1.69 prior PE | No | Predictor not available in IPPIC-UK data set |
| Teixeira, 2014 | C + B | First trimester | LP: –5.723 + 0.870 (if chronic hypertension) + 1.428 (if diabetic) – 0.787 (if multiparous) + 3.952 (if history of PE) + 0.039 × (maternal age) + 6.159 × (maternal weight MoM, log) + 0.027 × (CRL) + (–0.483) × NT + 0.766 × (Free B-HCG MoM, log) | No | Predictor not available in IPPIC-UK data set |
| Early-onset pre-eclampsia | |||||
| Crovetto, 2015229 | C | First trimester | LP1: –5.177 + 2.383 (if black ethnicity) – 1.105 (if nulliparous) + 3.543 (if parous with previous PE) + 2.229 (if chronic hypertension) + 2.201 (if renal disease) a priori risk = exp(LP1)/(1 + exp(LP1)) | Yes | |
| C + U | LP2: –21.99 + 12.25 (a priori risk, log10) + 11.516 (MAP, MoM) + 3.784 (mean uterine artery PI, MoM) | No | Predictor not available in IPPIC-UK data set | ||
| C + B + U | LP3: 21.515 + 12.884 (a priori risk, log10) + 11.219 (MAP, MoM) + 3.325 (mean uterine PI, MoM) – 7.346 (PlGF, log10) + 3.559 (sFlt-1, log10) | No | Predictor not available in IPPIC-UK data set | ||
| Baschat, 2014115 | C | First trimester | LP: –5.803 + 0.302 (if history of diabetes) + 0.767 (if history of hypertension) + 0.00948 (MAP) | Yes | |
| Parra-cordero, 2013 | C + B + U | First trimester | LP: –6.942 + 0.074 (BMI) + 1.878 (if smoker) + 2.1116 (lowest uterine artery PI, Ln MoM) – 0.671 (PlGF, Ln MoM) | No | Predictor not available in IPPIC-UK data set |
| Scazzocchio, 2013235 | C | First trimester | LP1: –7.703 + 0.086 (BMI) + 1.708 (if chronic hypertension) + 4.033 (if renal disease) + 1.931 (if parous with previous PE) + 0.005 (if parous with no previous PE) a priori risk = exp(LP1)/(1 + exp(LP1)) | Yes | |
| C + U | LP2: –0.32 + 2.681 (a priori risk, Ln) + 13.12 (mean uterine artery PI, Ln MoM) + 25.733 (MAP, Ln MoM) | No | Predictor not available in IPPIC-UK data set | ||
| Kuc, 2013230 | C | First trimester | LP: –6.790 – 0.119 (maternal height, cm) + 4.8565 (maternal weight, Ln) + 1.845 (if nulliparous) + 0.086 (maternal age, years) + 1.353 (if smoker) | Yes | |
| Odibo, 2011 | C + B + U | First trimester | LP: –4.678 – 0.443 (PP13, MoM) – 0.009 (PAPP-A, MoM) + 0.347 (mean uterine artery PI) + 3.059 (if history of chronic hypertension) | No | Predictor not available in IPPIC-UK data set |
| Seed, 2011 | C | Second trimester | LP: –2.693 + 1.735 (if SBP > 140) + 1.004 (if on antihypertensive therapy) – 0.9790 (if previous PE in most recent pregnancy) + 2.121 (if previous PE with delivery < 34 weeks) + 1.285 (if mother is Indian, Bangladeshi, Pakistani, African) | No | Predictor not available in IPPIC-UK data set |
| Poon, 2010 (1) | C + B | First trimester | LP1: 3.022 + 2.652 (maternal factor-derived a priori risk for early-PE, Ln) – 6.056 (PlGF, Ln MoM) + 3.103 (inhibin-A, Ln MoM) + 9.753 (TNF-R1, Ln MoM) | No | Predictor not available in IPPIC-UK data set |
| C + B | LP2: 2.547 + 2.518 (combined a priori risk for early-onset PE, Ln) – 6.012 (PlGF, Ln MoM) | No | Predictor not available in IPPIC-UK data set | ||
| Poon, 2010 (2)232 | C | First trimester | LP: –5.674 + 1.267 (if black ethnicity) + 2.193 (if history of chronic hypertension) – 1.184 (if parous without previous PE) + 1.362 (if parous with previous PE) + 1.537 (if conceived with ovulation induction) | Yes | |
| Poon, 2009 (1) | C + B | First trimester | LP: –8.776 + 14.177 (uterine artery PI, Ln MoM) + 42.960 (MAP, Ln MoM) – 2.249 (PAPP-A, Ln MoM) – 3.529 (PlGF, Ln MoM) + 0.120 (BMI) – 1.472 (if parous with no previous PE) | No | Predictor not available in IPPIC-UK data set |
| Poon, 2009 (2) | C + U | First trimester | LP: –3.657 + 1.592 (maternal factor-derived a priori risk for early PE, Ln) + 31.396 (MAP, Ln MoM) + 13.322 (lowest uterine artery PI, Ln MoM) | No | Predictor not available in IPPIC-UK data set |
| Poon, 2009 (3)234 | C + B | First trimester | LP: –6.413 – 3.612 (PAPP-A, Ln MoM) + 1.803 (if history of chronic hypertension) + 1.564 (if black ethnicity) – 1.005 (if parous without previous PE) + 1.491 (if parous with previous PE) | Yes | |
| Akolekar, 2008 | C + B + U | First trimester | LP: –5.620 – 4.717 (PlGF, Ln MoM) – 1.865 (PAPP-A, Ln MoM) + 14.519 (uterine artery PI, MoM) + 5.471 (if history of chronic hypertension) + 1.159 (if black ethnicity) | No | Predictor not available in IPPIC-UK data set |
| Onwudiwe, 2008 | C + U | First and second trimester | LP: –11.4487 + 31.2443 (uterine artery PI, Ln MoM) + 40.1105 (MAP, Ln MoM) + 1.5442 (if African Caribbean ethnicity) | No | Predictor not available in IPPIC-UK data set |
| Plasencia, 2008 | C + U | First trimester | LP: –6.546 + 3.769 (if chronic hypertension) + 15.692 (uterine artery PI, Ln MoM) | No | Predictor not available in IPPIC-UK data set |
| Plasencia, 2007231 | C | First trimester | LP: –6.431 + 1.680 (if African Caribbean ethnicity) + 1.889 (if mixed ethnicity) + 2.822 (if parous with previous PE) | Yes | |
| Yu, 2005238 | U | Second trimester | LP: –9.81223 + 2.10910 (mean uterine artery PI)3 – 1.79921 (mean uterine artery PI)3 + 1.059463 (if bilateral notch) | Yes | |
| Wright, 2015236 | C | First trimester | LP: mean gestational age at delivery with PE = 54.3637 – 0.0206886 (age, years – 35, if age ≥ 35) + 0.11711 (height, cm – 164) – 2.6786 (if Afro-Caribbean ethnicity) – 1.129 (if South Asian ethnicity) – 7.2897 (if chronic hypertension) – 3.0519 (if systemic lupus erythematosus or antiphospholipid syndrome) – 1.6327 (if conception by in vitro fertilisation) – 8.1667 (if parous with previous PE) + 0.0271988 (if parous with previous PE, previous gestation in weeks – 24)2 – 4.335 (if parous with no previous PE) – 4.15137651 (if parous with no previous PE, interval between pregnancies in years)–1 + 9.21473572 (if parous with no previous PE, interval between pregnancies in years)–0.5 – 0.0694096 (if no chronic hypertension, weight in kg – 69) – 1.7154 (if no chronic hypertension and family history of PE) – 3.3899 (if no chronic hypertension and diabetes mellitus type 1 or 2) | Yes | |
| Kenny, 2014 | C + B + U | First trimester | LP: −14.164 + 0.075 (MAP) + 6.1782 (mean uterine artery RI) + 0.649 (interleukin-1 receptor antagonist/PlGF); early-onset PE: −34.347 + 0.109 (MAP) + 7.679 (mean uterine) | No | Predictor not available in IPPIC-UK data set |
| Teixeira, 2014 | C | First trimester | LP: –4.951 + 1.519 (if chronic hypertension) – 1.201 (if multiparous) + 3.201 (if history of PE) + 7.108 (maternal weight MoM, log) | No | Predictor not available in IPPIC-UK data set |
| Keikkala, 2013 | C + B | First trimester | LP: 1.75 – 3.27 (%hCG-H MoM) –3.63 (PAPP-AMoM) + 1.49 [parity (1 = nulliparous, 0 = multiparous)] + 0.03 (MAP) | No | Predictor not available in IPPIC-UK data set |
| Myers, 2013 | C | Second trimester | LP1: –8.4093 + 0.9037 (fertility treatment) + 0.7999 (any sister with PE) + 0.1030 (MAP) | No | Predictor not available in IPPIC-UK data set |
| C + B | LP2: –7.7769 + 0.7307 (fertility treatment) + 0.1047 (MAP) – 1.7269 (PlGF MoM) | No | Predictor not available in IPPIC-UK data set | ||
| C + U | LP3: –13.5946 + 0.8402 (fertility treatment) + 0.1039 (MAP) + 7.0938 (20-week mean uterine artery RI) | No | Predictor not available in IPPIC-UK data set | ||
| C + B + U | LP4: –12.5382 + 0.1078 (MAP) –1.5658 (PlGF MoM) + 6.1087 (20-week mean uterine artery RI) | No | Predictor not available in IPPIC-UK data set | ||
| C + B + U | LP5: –10.4272 + 0.0994 (MAP) –1.1787 (PlGF MoM) + 0.0344 (endoglin 20-week) + 0.5285 (20-week bilateral notches of uterine arteries) | No | Predictor not available in IPPIC-UK data set | ||
| Late-onset pre-eclampsia | |||||
| Crovetto, 2015229 | C | First trimester | LP1: –5.873 – 0.462 (if white ethnicity) + 0.109 (BMI) – 0.825 (if nulliparous) + 2.726 (if parous with previous PE) + 1.956 (if chronic hypertension) – 0.575 (if smoker) a priori risk = exp(LP1)/(1 + exp(LP1)) | Yes | |
| C + U | LP2: –14.315 + 8.864 (a priori risk, log10) + 7.429 (MAP, MoM) + 2.447 (mean uterine artery PI, MoM) | No | Predictor not available in IPPIC-UK data set | ||
| C + B + U | LP3: 25.921 + 9.652 (a priori risk, log10) + 6.89 (MAP, MoM) + 2.343 (mean uterine artery PI, MoM) – 5.618 (PlGF, log10) + 6.579 (sFlt-1, log10) | No | Predictor not available in IPPIC-UK data set | ||
| Parra-cordero, 2013 | C + B + U | First trimester | LP: –5.584 + 0.137 (BMI) + 0.822 (lowest uterine artery PI, Ln MoM) – 0.533671 (PlGF, Ln MoM) | No | Predictor not available in IPPIC-UK data set |
| Scazzocchio, 2013235 | C | First trimester | LP1: 6.135 + 2.124 (if previous PE) + 1.571 (if chronic hypertension) + 0.958 (if diabetes) + 1.416 (if thrombophilic condition) – 0.487 (if multiparous) + 0.093 (BMI) a priori risk = exp(LP1)/(1 + exp(LP1)) | Yes | |
| C + B | LP2: 0.328 + 2.205 (a priori risk, Ln) – 1.307 (PAPP-A, Ln MoM) | No | Predictor not available in IPPIC-UK data set | ||
| Kuc, 2013230 | C | First trimester | LP: –14.374 + 2.300 (maternal weight, Ln) + 1.303 (if nulliparous) + 0.068 (maternal age, years) | Yes | |
| Poon, 2010 (1) | C + B | First trimester | LP1: 3.810 + 2.898 (maternal factor-derived a priori risk for late PE, Ln) – 3.171 (PlGF, Ln MoM) + 3.792 (activin-A, Ln MoM) + 2.013 (MMP-9, Ln MoM) + 5.242 (P-selectin, Ln MoM) | No | Predictor not available in IPPIC-UK data set |
| LP2: 3.490 + 2.717 (combined a priori risk for late PE, Ln) – 2.966 (PlGF, Ln MoM) + 3.937 (activin-A, Ln MoM) + 4.190 (P-selectin, Ln MoM) | No | Predictor not available in IPPIC-UK data set | |||
| Poon, 2010 (2)232 | C | First trimester | LP: –7.860 + 0.034 (age, years) + 0.096(BMI) + 1.089 (if black ethnicity) + 0.980 (if Indian or Pakistani ethnicity) + 1.196 (if mixed ethnicity) + 1.070 (if woman’s mother had PE) – 1.413 (if parous without previous PE) + 0.780 (if parous with previous PE) | Yes | |
| Poon, 2009 (1) | C + B + U | First trimester | LP: –5.324 + 2.233 (uterine artery PI, Ln) + 23.134 (MAP, Ln MoM) – 2.408 (PlGF, Ln MoM) + 0.123 (BMI) + 1.019 (if black ethnicity) + 2.028 (if mixed ethnicity) + 1.298 (if family history of PE) – 1.443 (if parous with no previous PE) | No | Predictor not available in IPPIC-UK data set |
| Poon, 2009 (2) | C + U | First trimester | LP: –0.468 + 2.272 (maternal factor-derived a priori risk for late PE, Ln) + 21.147 (MAP, Ln MoM) + 3.537 (lowest uterine artery PI, Ln MoM) | No | Predictor not available in IPPIC-UK data set |
| Poon, 2009 (3)234 | C + B | First trimester | LP: –6.652 – 0.884 (PAPP-A, Ln MoM) + 1.127 (if family history of PE) + 1.222 (if black ethnicity) + 0.936 (if Indian or Pakistani ethnicity) + 1.335 (if mixed ethnicity) + 0.084 (BMI) – 1.255 (if parous without previous PE) + 0.818 (if parous with previous PE) | Yes | |
| Akolekar, 2008 | C + B + U | First trimester | LP: –5.136 – 2.400 (PlGF, Ln MoM) + 2.641 (uterine artery PI, Ln MoM) + 0.108 (BMI) + 1.441 (if patient’s mother had PE) + 1.366 (if black ethnicity) + 1.083 (if Indian or Pakistani ethnicity) + 1.549 (if mixed ethnicity) – 1.281 (if parous and no previous PE) | No | Predictor not available in IPPIC-UK data set |
| Onwudiwe, 2008 | C + U | First and second trimester | LP: –7.4924 + 6.2361 (uterine artery PI, Ln MoM) + 23.1953 (MAP, Ln MoM) + 0.6003 (if African Caribbean ethnicity) + 0.1197 (BMI) – 1.1058 (if parous without previous PE) | No | Predictor not available in IPPIC-UK data set |
| Plasencia, 2008 | C + U | First trimester | LP: –6.140 + 0.082 (BMI) + 0.813 (if African Caribbean ethnicity) – 1.234 (if parous with no previous PE) + 0.922 (if parous with a previous PE) + 1.049 (if patient’s mother had PE) + 2.198 (uterine artery PI, Ln MoM) | No | Predictor not available in IPPIC-UK data set |
| Plasencia, 2007231 | C | First trimester | LP: –6.585 + 1.368 (if African Caribbean ethnicity) + 1.311 (if mixed ethnicity) + 0.091 (BMI) + 0.960 (if patient’s mother had PE) – 1.663 (if parous without previous PE) | Yes | |
| Yu, 2005238 | C + U | Second trimester | LP: 0.7901 + 5.1473 (mean uterine artery PI)–2 – 12.5152 (mean uterine artery PI)–1 – 0.5575 (if smoker) + 0.5333 (if bilateral notch) + 0.0328 (age) + 0.4958 (if black ethnicity) + 1.5109 (if history of PE) + 1.1556 (if previous term live birth) + 0.0378 (BMI) | Yes | |
| Kenny, 2014 | C + B + U | First trimester | LP: −9.504 − 0.577 high fruit intake + 0.058 MAP + 0.058 BMI + 0.550 tissue inhibitor of metalloproteinase 1 | No | Predictor not available in IPPIC-UK data set |
| Teixeira, 2014 | C + B | First trimester | LP: –8.248 + 0.050 (if chronic hypertension) + 1.649 (if diabetic) – 0.623 (if multiparous) + 3.668 (if history of PE) + 5.834 × (maternal weight MoM, log) + 0.036 × (CRL) + (–0.592) × NT + 1.046 × (free B-HCG MoM, log) | No | Predictor not available in IPPIC-UK data set |
Appendix 10 Patient characteristics of IPPIC-UK individual participant data sets
| Maternal characteristics and outcomes | SCOPE UK42 (N = 658) | Allen et al.108 (N = 1045) | ALSPAC103 (N = 14344) | Chappell et al.121 (N = 316) | EMPOWaR122 (N = 449) | Poston et al. 2006150 (N = 2422) | Poston et al. 2015149 (N = 1554) | St George’s163 (N = 54635) | AMND114 (N = 136,635) | Velauthar et al.39 (N = 1145) | POP161 (N = 4212) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Age (years), mean (SD), range | 28.5 (5.6), 15–42 | 29.9 (5.1), 15–48 | 27.7 (4.9), 13–46 | 29.6 (5.9), 16–43 | 28.7 (5.4), 17–43 | 31.0 (5.8), 16–48 | 30.5 (5.5), 16–45 | 30.4 (5.6), 13–54 | 28.4 (5.6), 13–56 | NR | 29.9 (5.1), 16–48 |
| BMI (kg/m2), median [IQR], range | 24.1 [21.8–26.8], 16.5–50.0 | 23.6 [21.1–26.8], 14.8–51.1 | 21.5 [19.7–23.7], 11.7–61.3 | 24.2 [21.7–27.0], 16.4–56.5 | 38.2 [35.2–41.8], 30.8–56.3 | 30.1 [24.6–34.7], 16.6–65.4 | 35.1 [32.8–38.5], 30.0–66.0 | 23.5 [21.3–26.8], 11.9–79.8 | 28.0 [24.0–32.0], 10.0–72.1 | 23.9 [21.3–27.3], 13.8–46.6 | 24.1 [21.8–27.3], 14.7–54.7 |
| Ethnicity, n (%) | |||||||||||
| White | 554 (84) | 398 (38) | 11,769 (97) | 215 (68) | 449 (100) | NR | 954 (61) | 33,257 (62) | NR | 402 (35) | 3900 (93) |
| Black | 49 (7) | 108 (10) | 127 (1) | 91 (29) | 0 (0) | 395 (25) | 7820 (15) | 90 (8) | 25 (< 1) | ||
| Asian | 47 (7) | 495 (47) | 113 (< 1) | 6 (2) | 0 (0) | 97 (6) | 10,388 (19) | 565 (50) | 91 (2) | ||
| Hispanic | 1 (< 1) | 0 (0) | 0 (0) | 3 (< 1) | 0 (0) | 0 (0) | 5 (< 1) | 0 (0) | 0 (0) | ||
| Mixed | 0 (0) | 12 (1) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1528 (3) | 0 (0) | 1 (< 1) | ||
| Other | 7 (1) | 30 (3) | 76 (< 1) | 1 (< 1) | 0 (0) | 108 (7) | 555 (1) | 82 (7) | 195 (5) | ||
| Nulliparous, n (%) | 658 (100) | 584 (56) | 5704 (45) | 202 (64) | 291 (65) | NR | 674 (43) | 29,319 (54) | 65,206 (48) | 598 (52) | 4212 (100) |
| Previous PE, n (%) | 0 (0) | 17 (2) | NR | 56 (18) | 0 (0) | 657 (27) | 69 (4) | NR | NR | NR | 0 (0) |
| Outcome, n (%) | |||||||||||
| Any-onset PE | 32 (5) | 14 (1) | 288 (2) | 35 (12) | 10 (5) | 371 (15) | 54 (4) | 1487 (3) | 4970 (4) | 26 (2) | 273 (6) |
| Early-onset PE | 6 (1) | 1 (< 1) | 37 (< 1) | 6 (2) | 0 (0) | 144 (6) | 5 (< 1) | 151 (< 1) | 1237 (< 1) | 3 (< 1) | 10 (< 1) |
| Late-onset PE | 26 (4) | 13 (1) | 251 (2) | 29 (10) | 10 (5) | 227 (9) | 49 (3) | 1336 (2) | 3733 (3) | 23 (2) | 263 (6) |
Appendix 11 Number and proportion missing (or not recorded) for each predictor in each data set used for external validation
| Variable | Missing or not recorded, n (%) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SCOPE UK42 (N = 658) | Allen et al.108 (N = 1045) | ALSPAC103 (N = 14344) | Chappell et al.121 (N = 316) | EMPOWaR122 (N = 449) | Poston et al. 2006150 (N = 2422) | Poston et al. 2015149 (N = 1554) | St George’s163 (N = 54635) | AMND (N = 136635)114 | Velauthar et al.39 (N = 1145) | POP161 (N = 4212) | |
| Predictor | |||||||||||
| Maternal age | 0 (0) | 1 (< 1) | 1353 (9) | 0 (0) | 0 (0) | 22 (< 1) | 0 (0) | 0 (0) | 19 (< 1) | 1145 (100) | 0 (0) |
| Ethnicity | 0 (0) | 2 (< 1) | 2259 (16) | 0 (0) | 0 (0) | 2422 (100) | 0 (0) | 1082 (2) | 136,635 (100) | 6 (< 1) | 0 (0) |
| Nulliparous | 0 (0) | 0 (0) | 1745 (12) | 0 (0) | 1 (< 1) | 2422 (100) | 0 (0) | 104 (< 1) | 6 (< 1) | 0 (0) | 0 (0) |
| Parity and previous PE | 0 (0) | 0 (0) | 14,344 (100) | 0 (0) | 1 (< 1) | 2422 (100) | 8 (< 1) | 54,635 (100) | 136,635 (100) | 1145 (100) | 0 (0) |
| Family history of PE | 0 (0) | 6 (< 1) | 14,344 (100) | 316 (100) | 449 (100) | 2422 (100) | 61 (4) | 54,635 (100) | 136,635 (100) | 1145 (100) | 4212 (100) |
| Family history of PE in mother | 0 (0) | 6 (< 1) | 14,344 (100) | 316 (100) | 449 (100) | 2422 (100) | 101 (6) | 54,635 (100) | 136,635 (100) | 1145 (100) | 4212 (100) |
| Spontaneous conception | 0 (0) | 0 (0) | 2167 (15) | 316 (100) | 449 (100) | 2422 (100) | 4 (< 1) | 1427 (3) | 136,635 (100) | 1145 (100) | 0 (0) |
| History of hypertension | 0 (0) | 0 (0) | 2307 (16) | 0 (0) | 449 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (< 1) | 0 (0) |
| History of renal disease | 0 (0) | 0 (0) | 2060 (14) | 316 (100) | 0 (0) | 0 (0) | 0 (0) | 54,635 (100) | 0 (0) | 1145 (100) | 0 (0) |
| History of diabetes | 0 (0) | 0 (0) | 2119 (15) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 54,635 (100) | 136,635 (100) | 1 (< 1) | 0 (0) |
| Previous heritable thrombophilia | 0 (0) | 1045 (100) | 14,344 (100) | 316 (100) | 449 (100) | 2422 (100) | 1554 (100) | 54,635 (100) | 136,635 (100) | 1145 (100) | 4212 (100) |
| Smoker | 0 (0) | 0 (0) | 1972 (14) | 0 (0) | 0 (0) | 2422 (100) | 0 (0) | 4053 (7) | 2902 (2) | 1 (< 1) | 0 (0) |
| Maternal height | 0 (0) | 0 (0) | 8769 (61) | 316 (100) | 0 (0) | 0 (0) | 0 (0) | 8454 (15) | 3867 (3) | 5 (< 1) | 6 (< 1) |
| Maternal weight (trimester 1) | 0 (0) | 5 (< 1) | 9190 (64) | 316 (100) | 152 (34) | 0 (0) | 0 (0) | 7992 (15) | 23,926 (18) | 6 (< 1) | 146 (3) |
| Trimester 1 BMI | 0 (0) | 5 (< 1) | 2409 (17) | 33 (10) | 152 (34) | 0 (0) | 0 (0) | 9151 (17) | 25,650 (19) | 6 (< 1) | 152 (4) |
| Trimester 2 BMI | 0 (0) | 1040 (100) | 14,344 (100) | 316 (100) | 297 (66) | 0 (0) | 0 (0) | 25,183 (46) | 136,635 (100) | 1145 (100) | 57 (1) |
| Trimester 1 MAP | 0 (0) | 5 (< 1) | 3618 (25) | 316 (100) | 152 (34) | 0 (0) | 1536 (99) | 54,635 (100) | 136,635 (100) | 8 (< 1) | 280 (7) |
| Trimester 1 PAPP-A (MoM) | 658 (100) | 119 (11) | 14,344 (100) | 316 (100) | 449 (100) | 2422 (100) | 1554 (100) | 30,919 (57) | 136,635 (100) | 1145 (100) | 171 (4) |
| Trimester 1 PAPP-A | 658 (100) | 119 (11) | 14,344 (100) | 316 (100) | 449 (100) | 2422 (100) | 526 (34) | 54,635 (100) | 136,635 (100) | 1145 (100) | 134 (3) |
| Trimester 1 UtPI | 658 (100) | 1045 (100) | 14,344 (100) | 316 (100) | 449 (100) | 2422 (100) | 1554 (100) | 32,595 (60) | 136,635 (100) | 2 (< 1) | 4212 (100) |
| Trimester 2 UtPI | 658 (100) | 1045 (100) | 14,344 (100) | 316 (100) | 449 (100) | 2257 (93) | 1554 (100) | 28,109 (51) | 136,635 (100) | 1145 (100) | 133 (3) |
| Trimester 2 bilateral notching | 3 (< 1) | 1040 (100) | 14,344 (100) | 316 (100) | 449 (100) | 27 (1) | 1554 (100) | 21,596 (40) | 136,635 (100) | 1145 (100) | 133 (3) |
| Outcome | |||||||||||
| Any-onset PE | 4 (< 1) | 0 (0) | 832 (5) | 33 (10) | 246 (55) | 0 (0) | 47 (3) | 0 (0) | 0 (0) | 32 (3) | 5 (< 1) |
| Early-onset PE | 4 (< 1) | 0 (0) | 832 (5) | 33 (10) | 78 (17) | 0 (0) | 47 (3) | 0 (0) | 0 (0) | 32 (3) | 5 (< 1) |
| Late-onset PE | 4 (< 1) | 0 (0) | 832 (5) | 33 (10) | 239 (53) | 0 (0) | 47 (3) | 0 (0) | 0 (0) | 32 (3) | 5 (< 1) |
Appendix 12 Summary of linear predictor and predicted probability values from external validation
| Model number | Authors, year | Type of predictors | Study | N | Events,a n (%) | LPa | Predicted probabilitya | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Median | IQR | Range (minimum to maximum) | Median | IQR | Range (minimum to maximum) | ||||||
| First-trimester any-onset pre-eclampsia models | |||||||||||
| 1 | Plasencia et al., 2007231 | C | SCOPE UK42 | 658 | 33 (5.0) | –4.186 | –4.408 to –3.868 | –4.865 to –1.052 | 0.015 | 0.012 to 0.020 | 0.008 to 0.259 |
| Allen et al.108 | 1045 | 14 (1.3) | –4.508 | –5.576 to –4.139 | –6.464 to –1.023 | 0.011 | 0.004 to 0.016 | 0.002 to 0.264 | |||
| Poston et al. 2015149 | 1554 | 56 (3.6) | –3.488 | –4.531 to –2.943 | –5.269 to 0.337 | 0.03 | 0.011 to 0.050 | 0.005 to 0.584 | |||
| 2 | Poon et al., 2008233 | C | SCOPE UK42 | 658 | 33 (5.0) | –4.047 | –4.290 to –3.700 | –4.790 to –0.884 | 0.017 | 0.014 to 0.024 | 0.008 to 0.292 |
| Allen et al.108 | 1045 | 14 (1.3) | –4.406 | –5.386 to –4.002 | –6.337 to –0.885 | 0.012 | 0.005 to 0.018 | 0.002 to 0.292 | |||
| Poston et al. 2015149 | 1554 | 56 (3.6) | –3.292 | –4.220 to –2.700 | –5.028 to 0.691 | 0.036 | 0.014 to 0.063 | 0.007 to 0.666 | |||
| 3 | Wright et al., 2015236 | C | SCOPE UK42 | 658 | 33 (5.0) | –3.272 | –3.513 to –2.929 | –4.179 to –0.758 | 0.037 | 0.029 to 0.051 | 0.015 to 0.319 |
| Allen et al.108 | 584 | 8 (1.4) | –3.093 | –3.436 to –2.729 | –4.182 to –0.962 | 0.043 | 0.031 to 0.061 | 0.015 to 0.277 | |||
| Poston et al. 2015149 | 674 | 36 (5.3) | –2.084 | –2.300 to –1.710 | –3.090 to –0.437 | 0.111 | 0.091 to 0.153 | 0.044 to 0.393 | |||
| 4 | Baschat et al., 2014115 | C + B | Allen et al.108 | 1045 | 14 (1.3) | –3.041 | –3.440 to –2.615 | –4.705 to 0.283 | 0.046 | 0.031 to 0.068 | 0.009 to 0.570 |
| POP161 | 4212 | 273 (6.5) | –3.549 | –3.885 to –3.107 | –5.801 to 0.185 | 0.028 | 0.020 to 0.043 | 0.003 to 0.546 | |||
| 5 | Goetzinger et al., 2010128 | C + B | Allen et al.108 | 1045 | 14 (1.3) | –3.25 | –3.250 to –2.320 | –3.250 to –0.260 | 0.037 | 0.037 to 0.089 | 0.037 to 0.435 |
| Poston et al. 2015149 | 1554 | 56 (3.6) | –2.32 | –2.320 to –1.710 | –2.320 to –1.200 | 0.089 | 0.089 to 0.153 | 0.089 to 0.231 | |||
| POP161 | 4212 | 273 (6.5) | –3.25 | –3.250 to –2.320 | –3.250 to 0.100 | 0.037 | 0.037 to 0.089 | 0.037 to 0.525 | |||
| 6 | Odibo et al., 2011147 | C + B | Allen et al.108 | 1045 | 14 (1.3) | –2.92 | –3.223 to –2.614 | –5.367 to –0.301 | 0.051 | 0.038 to 0.068 | 0.005 to 0.425 |
| St George’s163 | 54,635 | 1487 (2.7) | –2.866 | –3.200 to –2.573 | –36.527 to 0.882 | 0.054 | 0.039 to 0.071 | 0.000 to 0.707 | |||
| POP161 | 4212 | 273 (6.5) | –2.874 | –3.287 to –2.517 | –21.418 to 0.230 | 0.053 | 0.036 to 0.075 | 0.000 to 0.557 | |||
| 7 | Odibo et al., 2011147 | C + U | Velauthar et al.39 | 1145 | 28 (2.4) | –3.331 | –3.543 to –3.080 | –4.586 to –0.194 | 0.035 | 0.028 to 0.044 | 0.010 to 0.452 |
| First-trimester early-onset pre-eclampsia models | |||||||||||
| 8 | Baschat et al., 2014115 | C | SCOPE UK42 | 658 | 6 (0.9) | –5.070 | –5.114 to –5.019 | –5.234 to –4.069 | 0.006 | 0.006 to 0.007 | 0.005 to 0.017 |
| ALSPAC103 | 14,344 | 40 (0.3) | –5.013 | –5.076 to –4.949 | –5.362 to –3.502 | 0.007 | 0.006 to 0.007 | 0.005 to 0.029 | |||
| Poston et al. 2006150 | 2422 | 144 (6.0) | –4.891 | –5.013 to –4.151 | –5.297 to –3.631 | 0.007 | 0.007 to 0.016 | 0.005 to 0.026 | |||
| Velauthar et al.39 | 1145 | 4 (0.3) | –5.029 | –5.090 to –4.975 | –5.272 to –3.867 | 0.007 | 0.006 to 0.007 | 0.005 to 0.020 | |||
| POP161 | 4212 | 10 (0.2) | –5.059 | –5.108 to –4.992 | –5.361 to –3.761 | 0.006 | 0.006 to 0.007 | 0.005 to 0.023 | |||
| 9 | Crovetto et al., 2015229 | C | SCOPE UK42 | 658 | 6 (0.9) | –6.282 | –6.282 to –6.282 | –6.282 to –1.670 | 0.002 | 0.002 to 0.002 | 0.002 to 0.158 |
| Poston et al. 2015149 | 1554 | 6 (0.4) | –5.177 | –6.282 to –3.899 | –6.282 to 0.749 | 0.006 | 0.002 to 0.020 | 0.002 to 0.679 | |||
| POP161 | 4212 | 10 (0.2) | –6.282 | –6.282 to –6.282 | –6.282 to –1.670 | 0.002 | 0.002 to 0.002 | 0.002 to 0.158 | |||
| 10 | Kuc et al., 2013230 | C | SCOPE UK42 | 658 | 6 (0.9) | –1.679 | –2.24 to –0.955 | –4.285 to 2.445 | 0.157 | 0.096 to 0.278 | 0.014 to 0.920 |
| ALSPAC103 | 14,344 | 40 (0.3) | –3.339 | –4.242 to –2.413 | –8.414 to 2.483 | 0.034 | 0.014 to 0.082 | 0.000 to 0.919 | |||
| Poston et al. 2015149 | 1554 | 6 (0.4) | –0.543 | –1.413 to 0.373 | –3.744 to 3.982 | 0.367 | 0.196 to 0.592 | 0.023 to 0.982 | |||
| St George’s163 | 54,635 | 151 (0.3) | –2.351 | –3.236 to –1.488 | –11.455 to 7.092 | 0.087 | 0.038 to 0.184 | 0.000 to 0.999 | |||
| AMND114 | 136,635 | 1237 (0.9) | –2.304 | –3.223 to –1.371 | –7.660 to 4.857 | 0.091 | 0.038 to 0.202 | 0.000 to 0.992 | |||
| POP161 | 4212 | 10 (0.2) | –1.599 | –2.268 to –0.842 | –4.585 to 3.497 | 0.168 | 0.094 to 0.301 | 0.010 to 0.970 | |||
| 11 | Plasencia et al., 2007231 | C | SCOPE UK42 | 658 | 6 (0.9) | –6.431 | –6.431 to –6.431 | –6.431 to –4.751 | 0.002 | 0.002 to 0.002 | 0.002 to 0.009 |
| Chappell et al.121 | 316 | 7 (2.1) | –6.431 | –6.431 to –4.751 | –6.431 to –1.929 | 0.002 | 0.002 to 0.009 | 0.002 to 0.127 | |||
| Poston et al. 2015149 | 1554 | 6 (0.4) | –6.431 | –6.431 to –4.751 | –6.431 to –1.929 | 0.002 | 0.002 to 0.009 | 0.002 to 0.127 | |||
| POP161 | 4212 | 10 (0.2) | –6.431 | –6.431 to –6.431 | –6.431 to –4.542 | 0.002 | 0.002 to 0.002 | 0.002 to 0.011 | |||
| 12 | Poon et al., 2010232 | C | SCOPE UK42 | 658 | 6 (0.9) | –5.674 | –5.674 to –5.674 | –5.674 to –2.214 | 0.003 | 0.003 to 0.003 | 0.003 to 0.099 |
| Poston et al. 2015149 | 1554 | 6 (0.4) | –4.137 | –5.321 to –4.054 | –6.858 to –1.508 | 0.016 | 0.005 to 0.017 | 0.001 to 0.181 | |||
| POP161 | 4212 | 10 (0.2) | –4.137 | –4.137 to –4.137 | –5.674 to –0.677 | 0.016 | 0.016 to 0.016 | 0.003 to 0.337 | |||
| 13 | Scazzocchio et al., 2013235 | C | SCOPE UK42 | 658 | 6 (0.9) | –5.631 | –5.832 to –5.397 | –6.282 to –3.145 | 0.004 | 0.003 to 0.005 | 0.002 to 0.041 |
| Poston et al. 2015149 | 1554 | 6 (0.4) | –4.662 | –4.877 to –4.333 | –5.127 to –0.099 | 0.009 | 0.008 to 0.013 | 0.006 to 0.475 | |||
| POP161 | 4212 | 10 (0.2) | –5.617 | –5.822 to –5.303 | –6.448 to 1.688 | 0.004 | 0.003 to 0.005 | 0.002 to 0.844 | |||
| 14 | Wright et al., 2015236 | C | SCOPE UK42 | 658 | 6 (0.9) | –6.461 | –6.804 to –5.972 | –7.744 to –2.932 | 0.002 | 0.001 to 0.003 | 0.000 to 0.051 |
| Poston et al. 2015149 | 674 | 6 (0.4) | –4.761 | –5.069 to –4.231 | –6.201 to –2.521 | 0.008 | 0.006 to 0.014 | 0.002 to 0.074 | |||
| 15 | Poon et al., 2009234 | C + B | POP161 | 4212 | 10 (0.2) | –6.321 | –7.824 to –4.693 | –18.294 to 3.971 | 0.002 | 0.000 to 0.009 | 0.000 to 0.981 |
| First-trimester late-onset pre-eclampsia models | |||||||||||
| 16 | Crovetto et al., 2015229 | C | SCOPE UK42 | 658 | 26 (4.0) | –4.504 | –4.789 to –4.149 | –5.703 to –1.592 | 0.011 | 0.008 to 0.016 | 0.003 to 0.169 |
| Allen et al.108 | 1045 | 13 (1.2) | –3.914 | –4.494 to –3.298 | –5.654 to 1.509 | 0.020 | 0.011 to 0.036 | 0.003 to 0.819 | |||
| Chappell et al.121 | 316 | 32 (10.0) | –4.084 | –4.648 to –2.645 | –5.903 to 2.098 | 0.017 | 0.009 to 0.066 | 0.003 to 0.891 | |||
| Poston et al. 2015149 | 1554 | 51 (3.3) | –2.658 | –3.207 to –2.114 | –4.468 to 3.007 | 0.066 | 0.039 to 0.108 | 0.011 to 0.953 | |||
| POP161 | 4212 | 263 (6.2) | –4.496 | –4.775 to –4.109 | –6.133 to –0.074 | 0.011 | 0.008 to 0.016 | 0.002 to 0.481 | |||
| 17 | Kuc et al., 2013230 | C | SCOPE UK42 | 658 | 26 (4.0) | –1.464 | –1.871 to –1.062 | –3.016 to 0.191 | 0.188 | 0.133 to 0.257 | 0.047 to 0.548 |
| Allen et al.108 | 1045 | 13 (1.2) | –2.104 | –2.653 to –1.526 | –4.520 to 0.558 | 0.109 | 0.066 to 0.179 | 0.011 to 0.636 | |||
| ALSPAC103 | 14,344 | 266 (1.9) | –2.698 | –3.226 to –2.070 | –5.045 to 0.134 | 0.063 | 0.038 to 0.112 | 0.006 to 0.533 | |||
| EMPOWaR122 | 449 | 28 (6.3) | –0.801 | –1.395 to –0.281 | –3.032 to 1.118 | 0.310 | 0.199 to 0.430 | 0.047 to 0.751 | |||
| Poston et al. 2015149 | 1554 | 51 (3.3) | –1.286 | –1.801 to –0.641 | –3.074 to 1.077 | 0.217 | 0.142 to 0.345 | 0.044 to 0.746 | |||
| St George’s163 | 54,635 | 1336 (2.4) | –1.992 | –2.562 to –1.437 | –4.741 to 1.303 | 0.120 | 0.072 to 0.192 | 0.009 to 0.786 | |||
| AMND114 | 136,635 | 3733 (2.7) | –2.238 | –2.791 to –1.652 | –4.987 to 1.023 | 0.096 | 0.058 to 0.161 | 0.007 to 0.735 | |||
| POP161 | 4212 | 263 (6.2) | –1.367 | –1.723 to –1.011 | –3.233 to 0.874 | 0.203 | 0.151 to 0.267 | 0.038 to 0.705 | |||
| 18 | Plasencia et al., 2007231 | C | SCOPE UK42 | 658 | 26 (4.0) | –4.346 | –4.586 to –4.001 | –5.081 to –1.134 | 0.013 | 0.010 to 0.018 | 0.006 to 0.243 |
| Allen et al.108 | 1045 | 13 (1.2) | –4.702 | –5.848 to –4.318 | –6.809 to –1.020 | 0.009 | 0.003 to 0.013 | 0.001 to 0.265 | |||
| Poston et al. 2015149 | 1554 | 51 (3.3) | –3.636 | –4.715 to –3.103 | –5.514 to 0.141 | 0.026 | 0.009 to 0.043 | 0.004 to 0.535 | |||
| 19 | Poon et al., 2010232 | C | SCOPE UK42 | 658 | 26 (4.0) | –4.460 | –4.746 to –3.925 | –5.459 to –1.342 | 0.011 | 0.009 to 0.019 | 0.004 to 0.207 |
| Allen et al.108 | 1045 | 13 (1.2) | –4.542 | –5.007 to –3.860 | –6.748 to –1.142 | 0.011 | 0.007 to 0.021 | 0.001 to 0.242 | |||
| Poston et al. 2015149 | 1554 | 51 (3.3) | –3.656 | –4.321 to –2.968 | –5.612 to 0.285 | 0.025 | 0.013 to 0.049 | 0.004 to 0.571 | |||
| 20 | Scazzocchio et al., 2013235 | C | SCOPE UK42 | 658 | 26 (4.0) | –3.894 | –4.111 to –3.641 | –4.598 to –1.482 | 0.020 | 0.016 to 0.026 | 0.010 to 0.185 |
| 21 | Poon et al., 2009234 | C + B | Allen et al.108 | 1045 | 13 (1.2) | –4.505 | –5.108 to –3.883 | –7.179 to –0.850 | 0.011 | 0.006 to 0.020 | 0.001 to 0.305 |
| Second-trimester any-onset pre-eclampsia models | |||||||||||
| 22 | Yu et al., 2005238 | C + U | POP161 | 4212 | 273 (6.5) | –4.470 | –4.806 to –3.860 | –6.206 to 65.058 | 0.011 | 0.008 to 0.021 | 0.002 to 1.000 |
| Second-trimester early-onset pre-eclampsia models | |||||||||||
| 23 | Yu et al., 2005238 | C + U | POP161 | 4212 | 10 (0.2) | –9.601 | –9.694 to –9.412 | –9.809 to 0.374 | 0.000 | 0.000 to 0.000 | 0.000 to 0.591 |
| Second-trimester late-onset pre-eclampsia models | |||||||||||
| 24 | Yu et al., 2005238 | C + U | POP161 | 4212 | 263 (6.2) | –4.488 | –4.789 to –3.967 | –6.145 to 56.090 | 0.011 | 0.008 to 0.019 | 0.002 to 1.000 |
FIGURE 18.
Median (dot) and range (bar) of values for (a) LP and (b) predicted probabilities across validation data sets for each model being externally validated. This figure contains information from several sources. 115,128,147,229–236,238 Reproduced from Snell et al. 52 Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
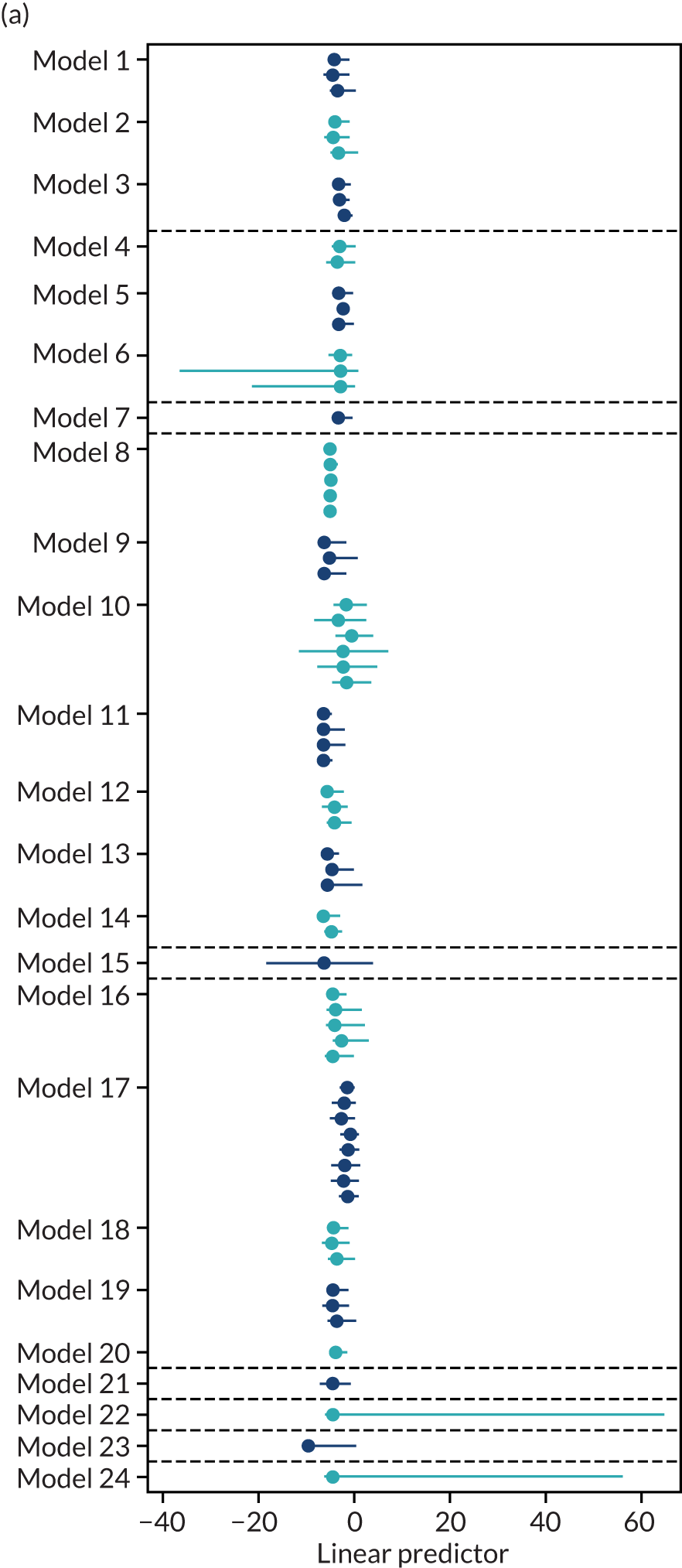
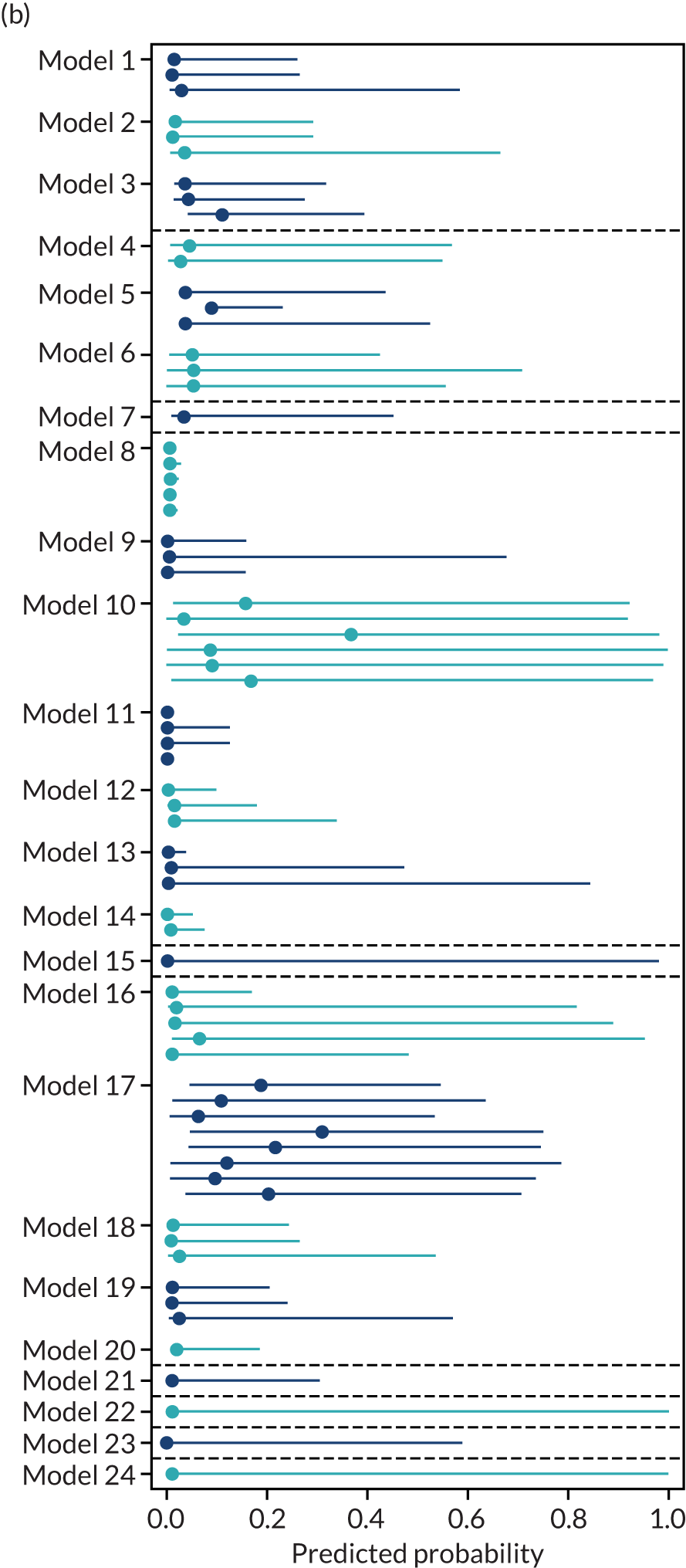
Appendix 13 Predictive performance statistics for models in the individual IPPIC-UK data sets
| Model number | Authors, year | Type of predictors | Validation cohort | N | Events,a n (%) | Performance statistic (95% CI) | ||
|---|---|---|---|---|---|---|---|---|
| C-statistic | Calibration slope | Calibration-in-the-large | ||||||
| First-trimester any-onset pre-eclampsia models | ||||||||
| 1 | Plasencia et al., 2007231 | C | SCOPE UK42 | 658 | 33 (5.0) | 0.636 (0.532 to 0.729) | 0.521 (0.025 to 1.017) | 0.865 (0.502 to 1.228) |
| Allen et al.108 | 1045 | 14 (1.3) | 0.782 (0.638 to 0.880) | 1.110 (0.600 to 1.619) | –0.117 (–0.651 to 0.417) | |||
| Poston et al. 2015149 | 1554 | 56 (3.6) | 0.692 (0.617 to 0.758) | 0.580 (0.346 to 0.813) | –0.326 (–0.610 to –0.042) | |||
| 2 | Poon et al., 2008233 | C | SCOPE UK42 | 658 | 33 (5.0) | 0.637 (0.532 to 0.730) | 0.546 (0.051 to 1.040) | 0.731 (0.368 to 1.093) |
| Allen et al.108 | 1045 | 14 (1.3) | 0.782 (0.638 to 0.880) | 1.155 (0.622 to 1.689) | –0.209 (–0.743 to 0.324) | |||
| Poston et al. 2015149 | 1554 | 56 (3.6) | 0.694 (0.620 to 0.759) | 0.597 (0.357 to 0.837) | –0.518 (–0.801 to –0.234) | |||
| 3 | Wright et al., 2015236 | C | SCOPE UK42 | 658 | 33 (5.0) | 0.647 (0.552 to 0.732) | 0.638 (0.097 to 1.179) | 1.880 (1.519 to 2.240) |
| Allen et al.108 | 584 | 8 (1.4) | 0.680 (0.453 to 0.845) | 0.840 (–0.100 to 1.781) | 0.367 (–0.334 to 1.069) | |||
| Poston et al. 2015149 | 674 | 36 (5.3) | 0.592 (0.497 to 0.681) | 0.560 (–0.067 to 1.187) | 0.544 (0.200 to 0.888) | |||
| 4 | Baschat et al., 2014115 | C + B | Allen et al.108 | 1045 | 14 (1.3) | 0.758 (0.620 to 0.857) | 1.246 (0.667 to 1.824) | –1.543 (–2.074 to –1.011) |
| POP161 | 4212 | 273 (6.5) | 0.704 (0.670 to 0.737) | 1.237 (1.034 to 1.439) | 0.658 (0.533 to 0.782) | |||
| 5 | Goetzinger et al., 2010128 | C + B | Allen et al.108 | 1045 | 14 (1.3) | 0.670 (0.502 to 0.804) | 0.910 (0.092 to 1.729) | –1.691 (–2.221 to –1.162) |
| Poston et al. 2015149 | 1554 | 56 (3.6) | 0.521 (0.451 to 0.590) | 0.409 (–0.567 to 1.386) | –1.206 (–1.480 to –0.933) | |||
| POP161 | 4212 | 273 (6.5) | 0.764 (0.730 to 0.795) | 1.706 (1.499 to 1.913) | –0.070 (–0.195 to 0.054) | |||
| 6 | Odibo et al., 2011147 | C + B | Allen et al.108 | 1045 | 14 (1.3) | 0.660 (0.491 to 0.796) | 0.847 (–0.022 to 1.716) | –1.512 (–2.041 to –0.982) |
| St George’s163 | 54,635 | 1487 (2.7) | 0.672 (0.654 to 0.690) | 0.962 (0.885 to 1.039) | –0.897 (–0.950 to –0.845) | |||
| POP161 | 4212 | 273 (6.5) | 0.779 (0.744 to 0.812) | 1.490 (1.329 to 1.651) | –0.033 (–0.159 to 0.094) | |||
| 7 | Odibo et al., 2011147 | C + U | Velauthar et al.39 | 1145 | 28 (2.4) | 0.526 (0.390 to 0.658) | 0.277 (–0.642 to 1.195) | –0.520 (–0.907 to –0.134) |
| First-trimester early-onset pre-eclampsia models | ||||||||
| 8 | Baschat et al., 2014115 | C | SCOPE UK42 | 658 | 6 (0.9) | 0.513 (0.291 to 0.73) | 0.212 (–8.468 to 8.892) | 0.399 (–0.407 to 1.205) |
| ALSPAC103 | 14,344 | 40 (0.3) | 0.706 (0.600 to 0.794) | 2.070 (1.234 to 2.905) | –1.029 (–1.352 to –0.705) | |||
| Poston et al. 2006150 | 2422 | 144 (6.0) | 0.672 (0.626 to 0.716) | 1.281 (0.898 to 1.664) | 1.797 (1.628 to 1.967) | |||
| Velauthar et al.39 | 1145 | 4 (0.3) | 0.698 (0.434 to 0.876) | 1.639 (–4.040 to 7.319) | –0.755 (–1.875 to 0.365) | |||
| POP161 | 4212 | 10 (0.2) | 0.739 (0.494 to 0.891) | 3.403 (2.017 to 4.789) | –1.054 (–1.674 to –0.433) | |||
| 9 | Crovetto et al., 2015229 | C | SCOPE UK42 | 658 | 6 (0.9) | 0.464 (0.428 to 0.500) | NE | 1.039 (0.218 to 1.861) |
| Poston et al. 2015149 | 1554 | 6 (0.4) | 0.596 (0.278 to 0.850) | 0.264 (–0.216 to 0.743) | –2.484 (–3.401 to –1.567) | |||
| POP161 | 4212 | 10 (0.2) | 0.721 (0.532 to 0.855) | 0.996 (0.596 to 1.395) | –0.317 (–0.943 to 0.309) | |||
| 10 | Kuc et al., 2013230 | C | SCOPE UK42 | 658 | 6 (0.9) | 0.762 (0.520 to 0.905) | 0.682 (0.050 to 1.314) | –3.709 (–4.530 to –2.889) |
| ALSPAC103 | 14,344 | 40 (0.3) | 0.664 (0.484 to 0.812) | 0.442 (–0.025 to 0.909) | –3.458 (–3.786 to –3.130) | |||
| Poston et al. 2015149 | 1554 | 6 (0.4) | 0.519 (0.323 to 0.710) | 0.010 (–0.705 to 0.726) | –5.978 (–6.861 to –5.094) | |||
| St George’s163 | 54,635 | 151 (0.3) | 0.635 (0.587 to 0.679) | 0.343 (0.227 to 0.460) | –4.510 (–4.674 to –4.347) | |||
| AMND114 | 136,635 | 1237 (0.9) | 0.681 (0.665 to 0.696) | 0.470 (0.430 to 0.511) | –3.387 (–3.445 to –3.330) | |||
| POP161 | 4212 | 10 (0.2) | 0.656 (0.518 to 0.772) | 0.450 (–0.069 to 0.969) | –5.191 (–5.815 to –4.567) | |||
| 11 | Plasencia et al., 2007231 | C | SCOPE UK42 | 658 | 6 (0.9) | 0.465 (0.429 to 0.501) | NE | 1.500 (0.691 to 2.309) |
| Chappell et al.121 | 316 | 7 (2.1) | 0.706 (0.463 to 0.872) | 0.510 (0.013 to 1.007) | 0.462 (–0.402 to 1.327) | |||
| Poston et al. 2015149 | 1554 | 6 (0.4) | 0.667 (0.391 to 0.863) | 0.517 (–0.135 to 1.170) | –0.478 (–1.367 to 0.411) | |||
| POP161 | 4212 | 10 (0.2) | 0.497 (0.496 to 0.498) | NE | 0.365 (–0.256 to 0.985) | |||
| 12 | Poon et al., 2010232 | C | SCOPE UK42 | 658 | 6 (0.9) | 0.524 (0.355 to 0.687) | 0.236 (–1.154 to 1.627) | 0.648 (–0.163 to 1.458) |
| Poston et al. 2015149 | 1554 | 6 (0.4) | 0.745 (0.512 to 0.892) | 0.846 (0.004 to 1.688) | –1.667 (–2.548 to –0.787) | |||
| POP161 | 4212 | 10 (0.2) | 0.687 (0.455 to 0.852) | 1.176 (0.617 to 1.734) | –2.228 (–2.849 to –1.606) | |||
| 13 | Scazzocchio et al., 2013235 | C | SCOPE UK42 | 658 | 6 (0.9) | 0.728 (0.448 to 0.898) | 1.131 (–0.130 to 2.392) | 0.788 (–0.020 to 1.596) |
| ALSPAC103 | 1554 | 6 (0.4) | 0.612 (0.333 to 0.834) | 0.586 (–0.376 to 1.548) | –1.434 (–2.316 to –0.551) | |||
| Poston et al. 2006150 | 4212 | 10 (0.2) | 0.844 (0.640 to 0.943) | 0.746 (0.449 to 1.042) | –1.439 (–2.092 to –0.787) | |||
| 14 | Wright et al., 2015236 | C | Velauthar et al.39 | 658 | 6 (0.9) | 0.763 (0.614 to 0.867) | 0.907 (–0.027 to 1.842) | 1.396 (0.585 to 2.207) |
| POP161 | 674 | 6 (0.4) | 0.591 (0.207 to 0.890) | 0.958 (–0.727 to 2.642) | –0.906 (–2.048 to 0.236) | |||
| 15 | Poon et al., 2009234 | C + B | SCOPE UK42 | 4212 | 10 (0.2) | 0.741 (0.507 to 0.888) | 0.452 (0.210 to 0.693) | –2.671 (–3.35 to –1.991) |
| First-trimester late-onset pre-eclampsia models | ||||||||
| 16 | Crovetto et al., 2015229 | C | SCOPE UK42 | 658 | 26 (4.0) | 0.569 (0.448 to 0.683) | 0.379 (–0.231 to 0.988) | 1.074 (0.675 to 1.472) |
| Allen et al.108 | 1045 | 13 (1.2) | 0.544 (0.352 to 0.723) | 0.516 (0.108 to 0.924) | –1.205 (–1.779 to –0.630) | |||
| Chappell et al.121 | 316 | 32 (10.0) | 0.696 (0.586 to 0.788) | 0.345 (0.152 to 0.538) | 0.037 (–0.451 to 0.525) | |||
| Poston et al. 2015149 | 1554 | 51 (3.3) | 0.504 (0.412 to 0.596) | 0.210 (–0.035 to 0.456) | –1.502 (–1.812 to –1.192) | |||
| POP161 | 4212 | 263 (6.2) | 0.781 (0.745 to 0.813) | 1.248 (1.120 to 1.376) | 1.309 (1.177 to 1.441) | |||
| 17 | Kuc et al., 2013230 | C | SCOPE UK42 | 658 | 26 (4.0) | 0.544 (0.421 to 0.663) | 0.224 (–0.445 to 0.893) | –1.867 (–2.262 to –1.471) |
| Allen et al.108 | 1045 | 13 (1.2) | 0.583 (0.425 to 0.726) | 0.340 (–0.356 to 1.036) | –2.570 (–3.119 to –2.020) | |||
| ALSPAC103 | 14,344 | 266 (1.9) | 0.657 (0.616 to 0.696) | 0.761 (0.550 to 0.973) | –1.574 (–1.699 to –1.448) | |||
| EMPOWaR122 | 449 | 28 (6.3) | 0.337 (0.162 to 0.558) | –0.755 (–1.808 to 0.299) | –2.127 (–2.795 to –1.459) | |||
| Poston et al. 2015149 | 1554 | 51 (3.3) | 0.592 (0.512 to 0.668) | 0.390 (0.019 to 0.762) | –2.435 (–2.724 to –2.147) | |||
| St George’s163 | 54,635 | 1336 (2.4) | 0.636 (0.621 to 0.651) | 0.632 (0.560 to 0.704) | –1.970 (–2.025 to –1.915) | |||
| AMND114 | 136,635 | 3733 (2.7) | 0.844 (0.640 to 0.943) | 0.746 (0.449 to 1.042) | –1.439 (–2.092 to –0.787) | |||
| POP161 | 4212 | 263 (6.2) | 0.599 (0.561 to 0.636) | 0.673 (0.452 to 0.894) | –1.487 (–1.613 to –1.361) | |||
| 18 | Plasencia et al., 2007231 | C | SCOPE UK42 | 658 | 26 (4.0) | 0.627 (0.509 to 0.732) | 0.523 (–0.003 to 1.049) | 0.778 (0.377 to 1.178) |
| Allen et al.108 | 1045 | 13 (1.2) | 0.751 (0.600 to 0.858) | 1.008 (0.459 to 1.556) | 0.043 (–0.510 to 0.596) | |||
| Poston et al. 2015149 | 1554 | 51 (3.3) | 0.673 (0.596 to 0.742) | 0.523 (0.264 to 0.782) | –0.204 (–0.498 to 0.090) | |||
| 19 | Poon et al., 2010232 | C | SCOPE UK42 | 658 | 26 (4.0) | 0.570 (0.405 to 0.682) | 0.369 (–0.199 to 0.937) | 0.909 (0.511 to 1.308) |
| Allen et al.108 | 1045 | 13 (1.2) | 0.716 (0.556 to 0.836) | 0.913 (0.312 to 1.514) | –0.289 (–0.841 to 0.262) | |||
| Poston et al. 2015149 | 1554 | 51 (3.3) | 0.666 (0.591 to 0.734) | 0.541 (0.280 to 0.802) | –0.271 (–0.566 to 0.025) | |||
| 20 | Scazzocchio et al., 2013235 | C | SCOPE UK42 | 658 | 26 (4.0) | 0.597 (0.478 to 0.705) | 0.562 (–0.168 to 1.291) | 0.524 (0.128 to 0.920) |
| 21 | Poon et al., 2009234 | C + B | Allen et al.108 | 1045 | 13 (1.2) | 0.684 (0.550 to 0.792) | 0.799 (0.257 to 1.341) | –0.349 (–0.902 to 0.205) |
| Second-trimester any-onset pre-eclampsia models | ||||||||
| 22 | Yu et al., 2005238 | C + U | POP161 | 4212 | 273 (6.5) | 0.610 (0.574 to 0.645) | 0.075 (0.007 to 0.144) | NE |
| Second-trimester early-onset pre-eclampsia models | ||||||||
| 23 | Yu et al., 2005238 | C + U | POP161 | 4212 | 10 (0.2) | 0.908 (0.826 to 0.954) | 0.557 (0.293 to 0.821) | 2.473 (1.716 to 3.229) |
| Second-trimester late-onset pre-eclampsia models | ||||||||
| 24 | Yu et al., 2005238 | C + U | POP161 | 4212 | 263 (6.2) | 0.607 (0.570 to 0.642) | 0.077 (0.005 to 0.148) | NE |
Appendix 14 Decision curves for prediction models of early-onset pre-eclampsia in (a) SCOPE UK,42 (b) Poston et al. 2015149 and (c) POP161
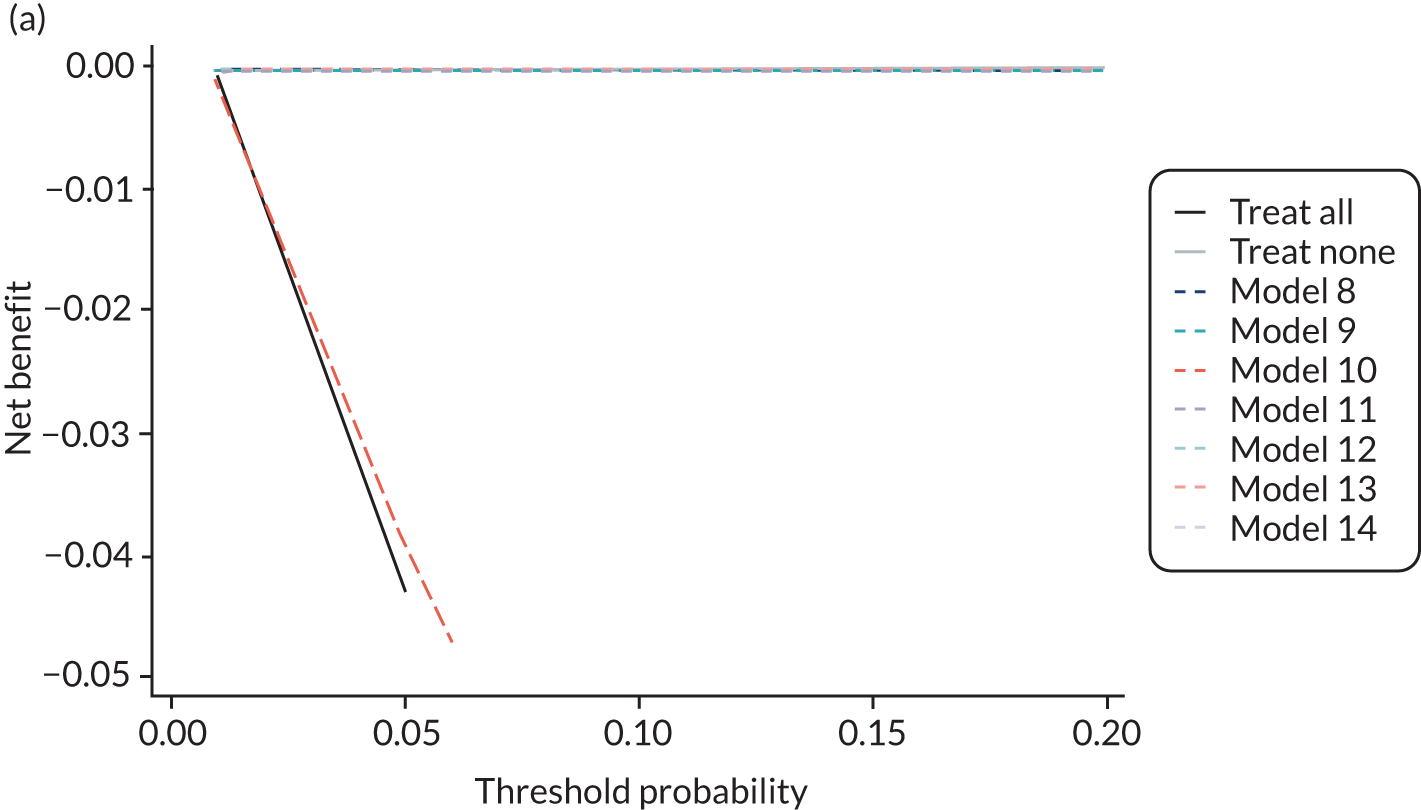
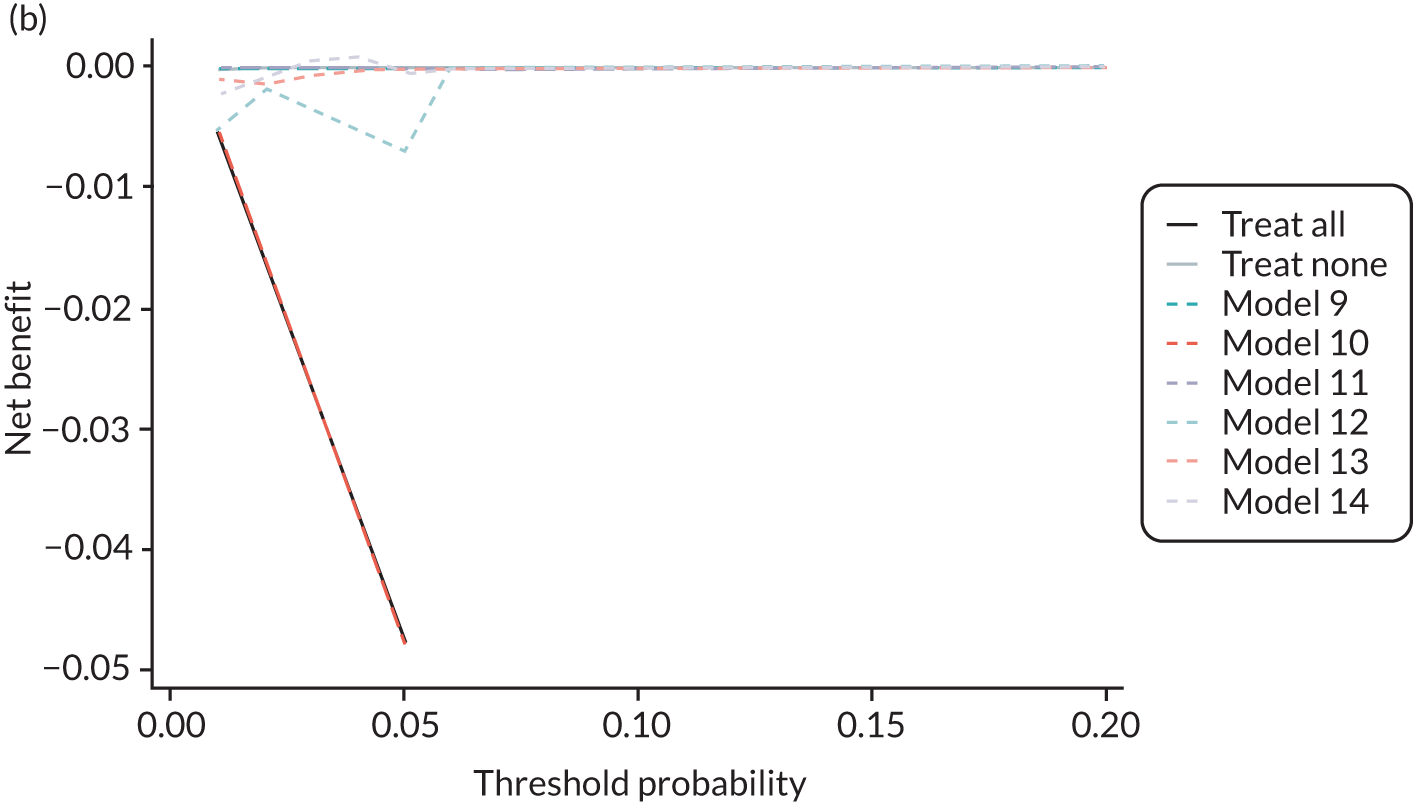
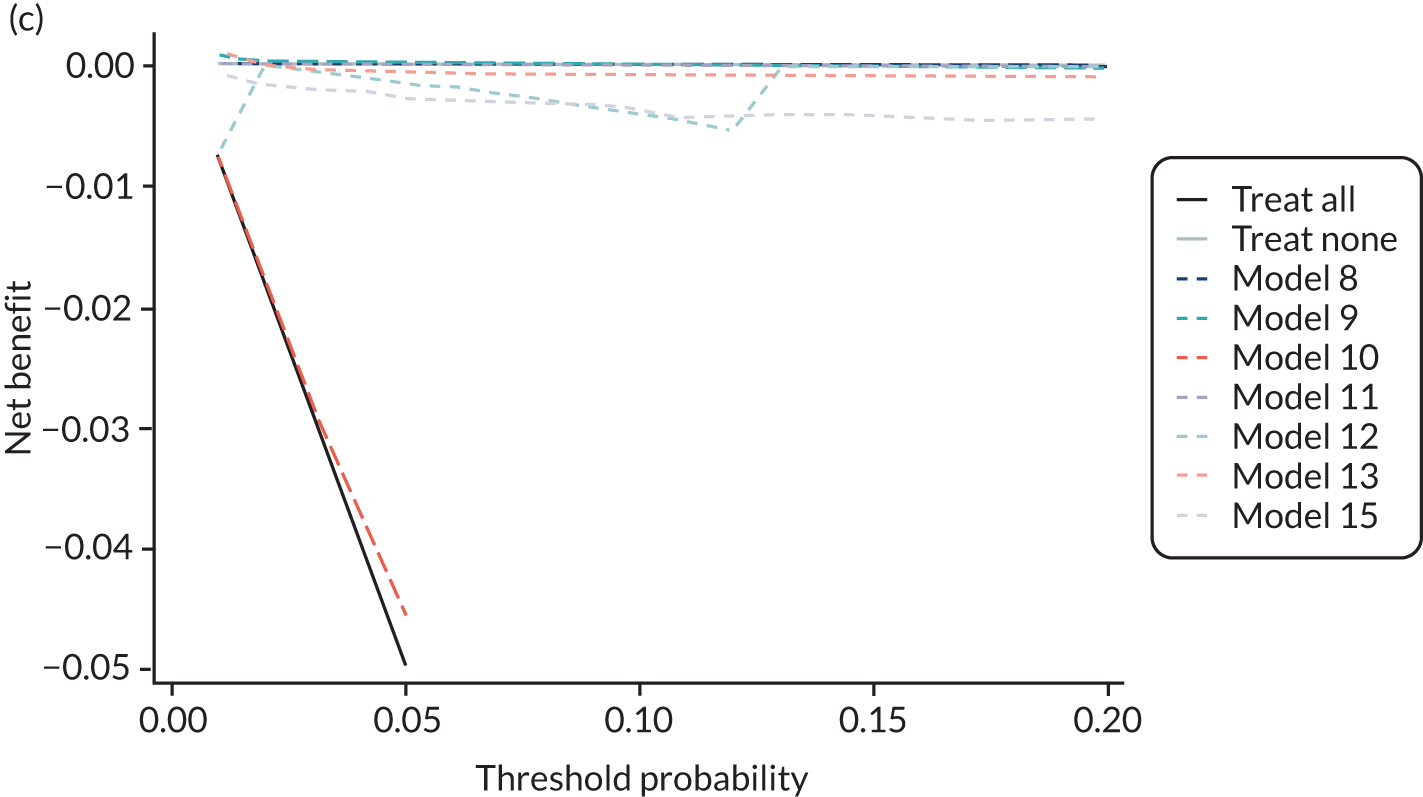
Appendix 15 Imputation checking for model development
Imputation checking for clinical characteristics models
Trimester 1 predictors
BMI values were not imputed reliably in POUCH, illustrated by the distribution of imputed values getting wider in later imputations, particularly for transformations of BMI.
FIGURE 19.
Median and range of BMI values in imputed data set for cohorts included in the development of models with trimester 1 clinical characteristics.
FIGURE 20.
Median and range of ln(BMI) values in imputed data set for cohorts included in the development of models with trimester 1 clinical characteristics.
FIGURE 21.
Median and range of (BMI/10)–2 values in imputed data set for cohorts included in the development of models with trimester 1 clinical characteristics.
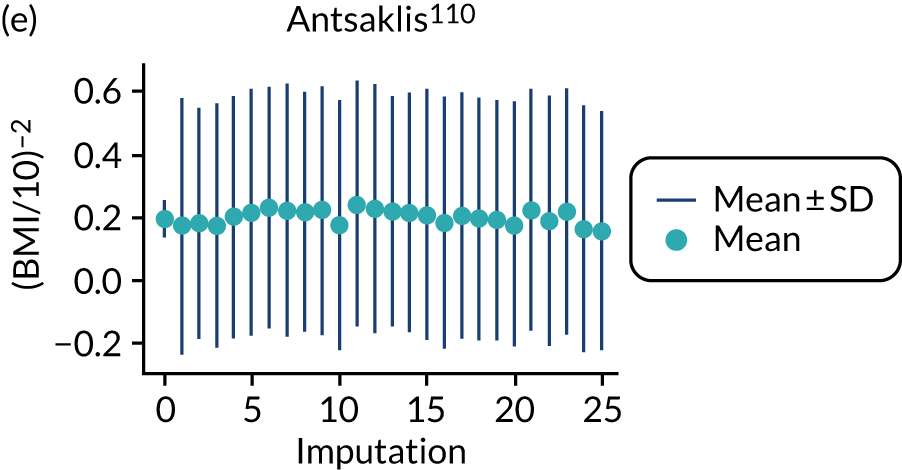
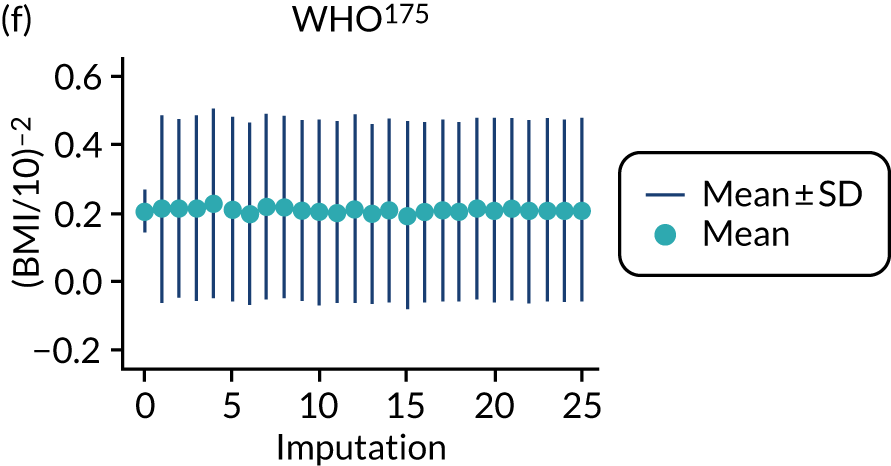
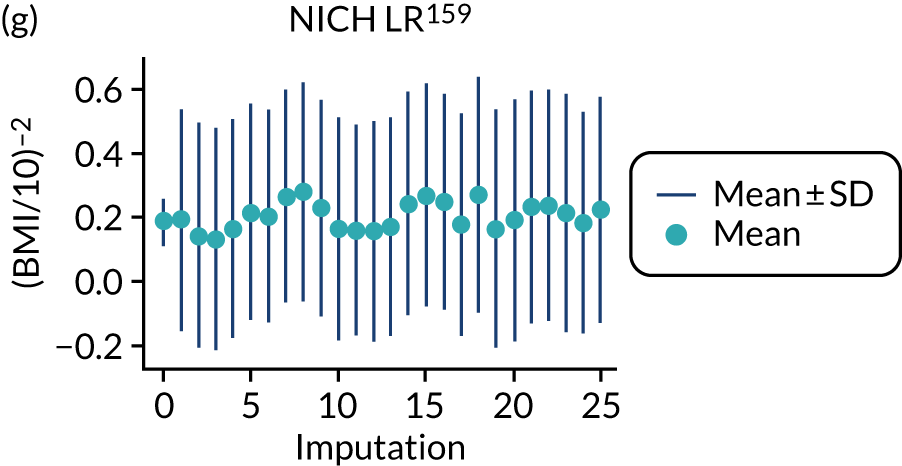
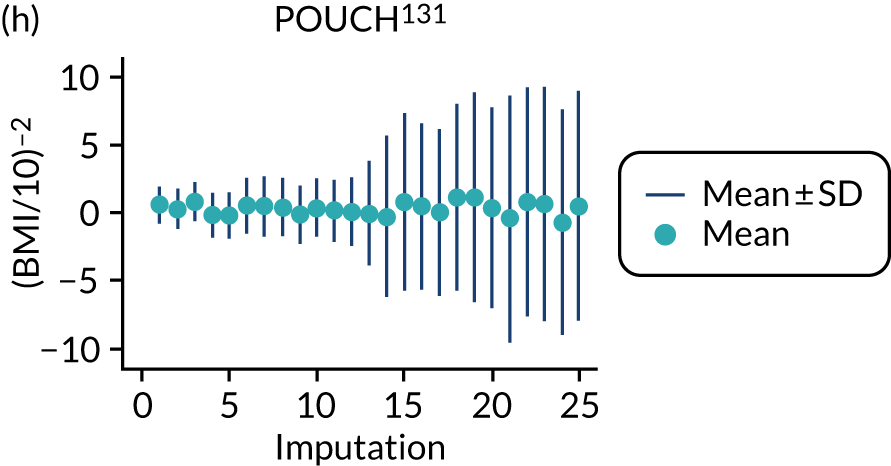
Trimester 2 predictors
SBP, DBP and BMI values were not imputed reliably in Allen, Baschat and Vinter, as illustrated by extreme imputed values for later imputations (particularly for Allen and Vinter).
FIGURE 22.
Median and range of DBP values in imputed data set for cohorts included in the development of models with trimester 2 clinical characteristics.
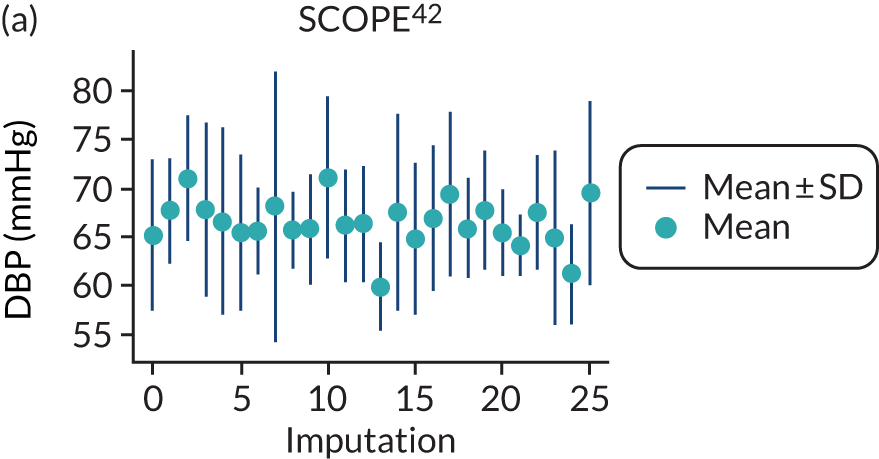
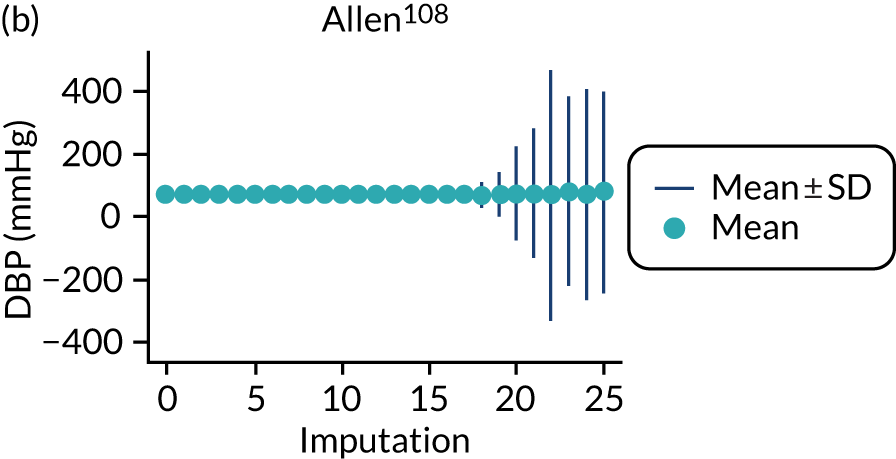
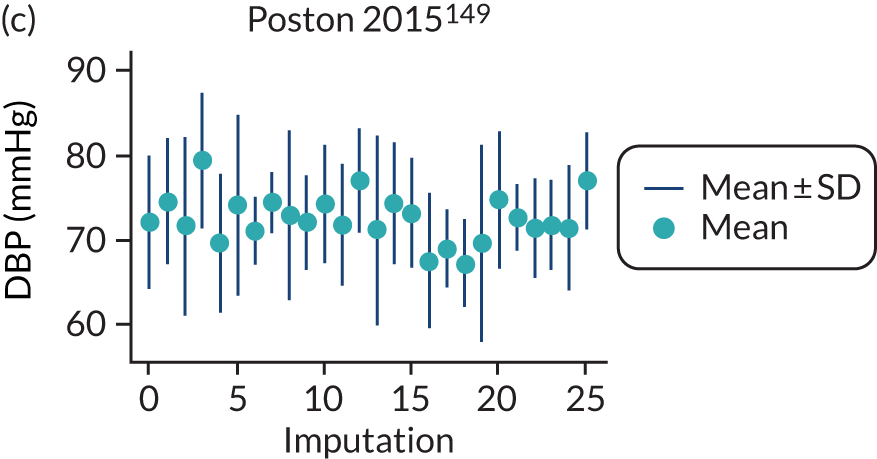
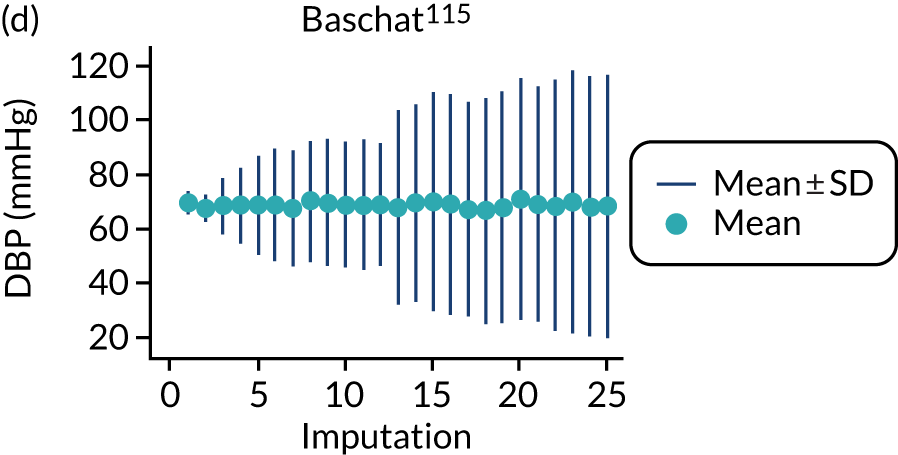
FIGURE 23.
Median and range of SBP values in imputed data set for cohorts included in the development of models with trimester 2 clinical characteristics.
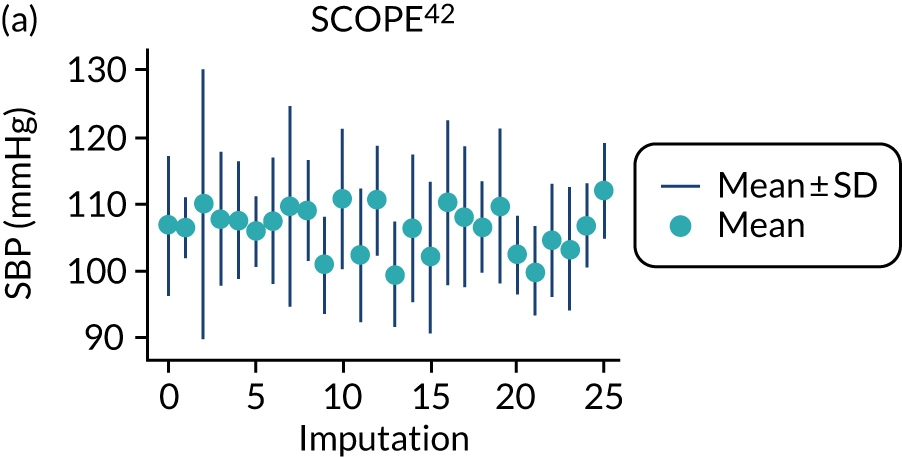
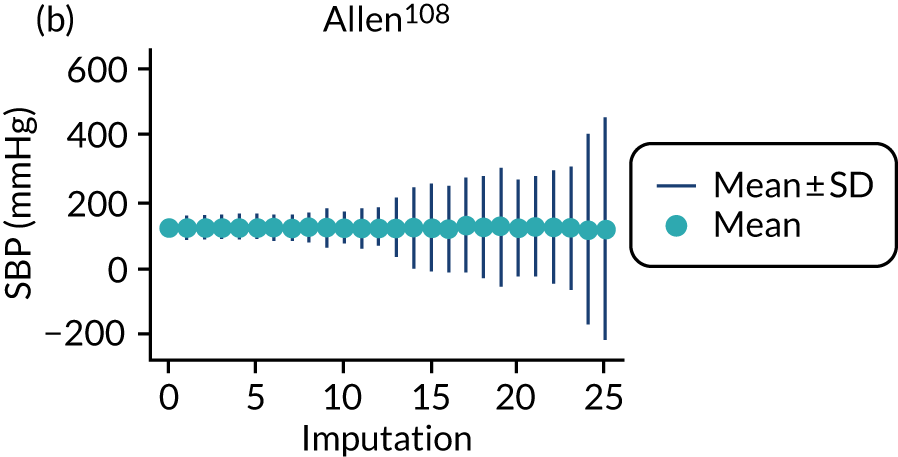
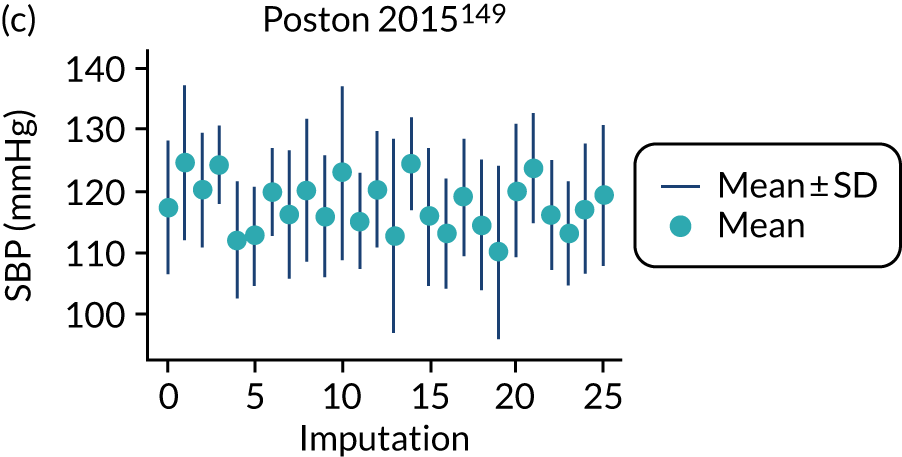
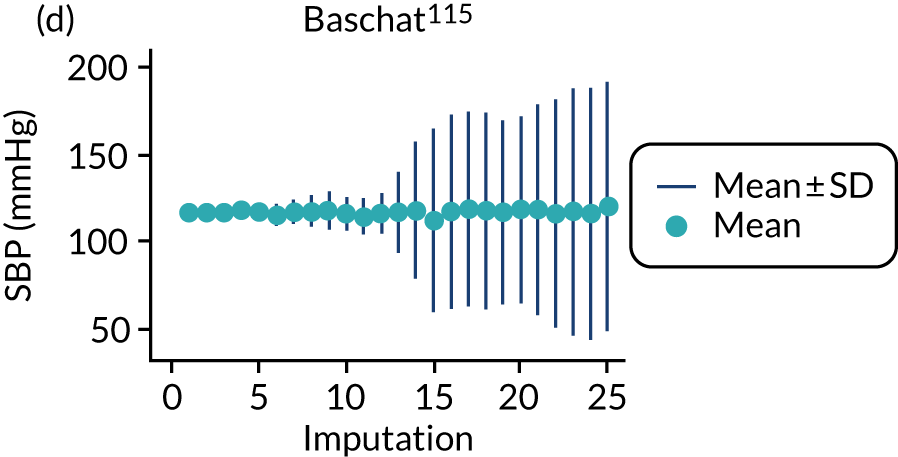
FIGURE 24.
Median and range of BMI values in imputed data set for cohorts included in the development of models with trimester 2 clinical characteristics.

FIGURE 25.
Median and range of ln(BMI) values in imputed data set for cohorts included in the development of models with trimester 2 clinical characteristics.
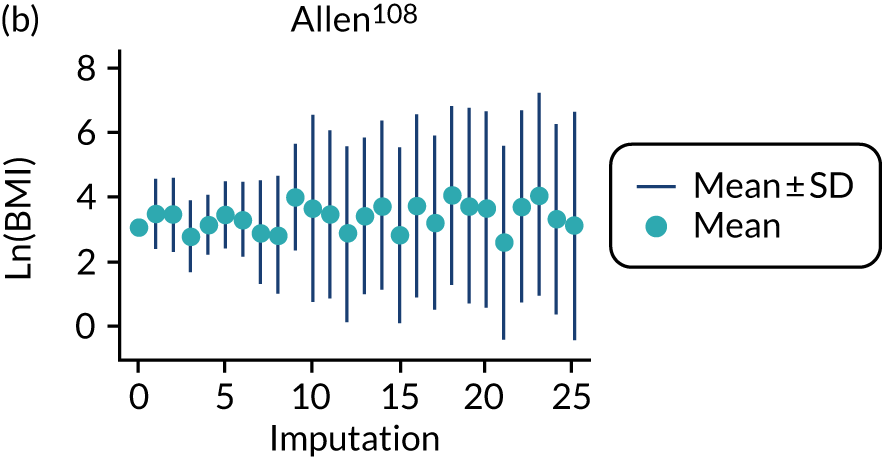
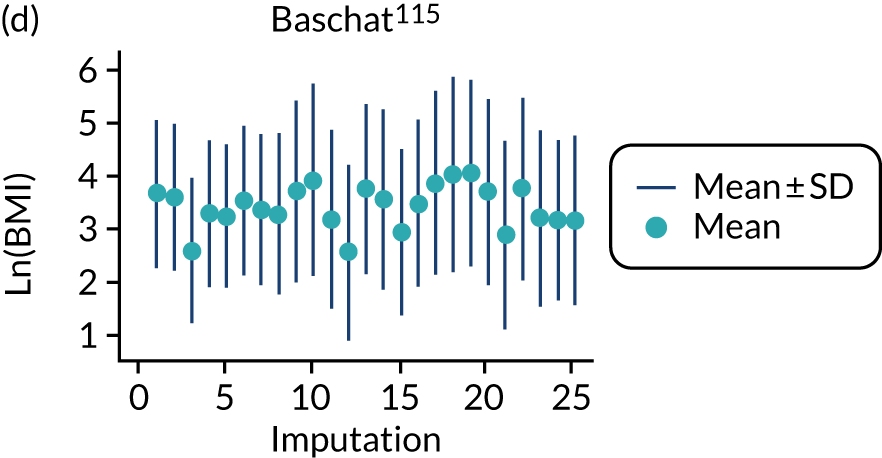
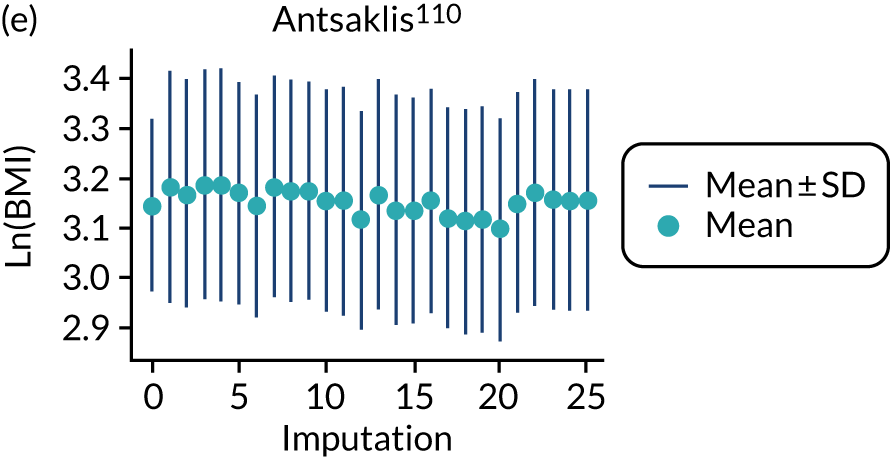
FIGURE 26.
Median and range of (BMI/10)–2 values in imputed data set for cohorts included in the development of models with trimester 2 clinical characteristics.
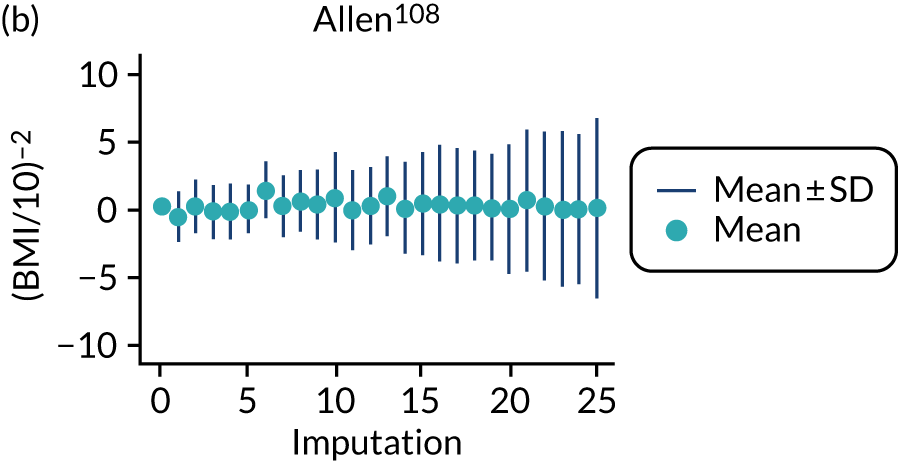
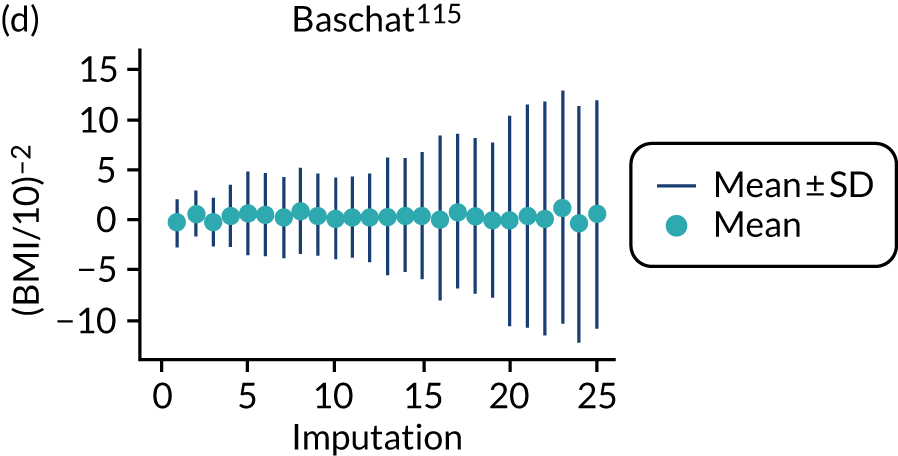
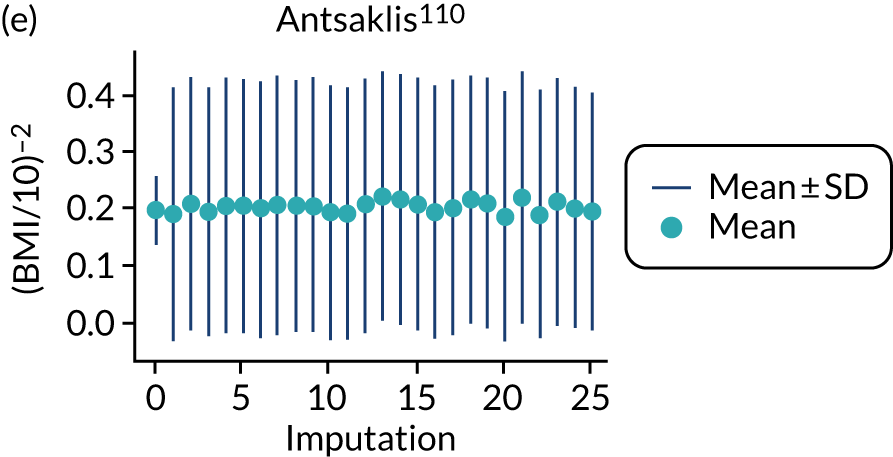
Imputation checking for clinical characteristics and biochemical markers models
Trimester 2 biomarkers
POUCH was excluded because of the unreliable imputation of PAPP-A, although the range of imputed values in POP was also concerning in later imputations.
FIGURE 27.
Median and range of ln(PAPP-A) values in imputed data set for cohorts included in the development of models with trimester 2 clinical characteristics.
Appendix 16 Comparison of clinical characteristics models in data imputed with BMI, ln(BMI) or BMI–2
| Variables | BMI | Ln(BMI) | BMI–2 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Beta coefficient (95% CI) | OR (95% CI) | p-value | Beta coefficient (95% CI) | OR (95% CI) | p-value | Beta coefficient (95% CI) | OR (95% CI) | p-value | |
| Age (years) | –0.016 (–0.028 to –0.005) | 0.984 (0.972 to 0.995) | 0.006 | –0.020 (–0.032 to –0.008) | 0.980 (0.969 to 0.992) | 0.001 | –0.025 (–0.038 to –0.012) | 0.975 (0.963 to 0.988) | < 0.001 |
| SBP | 0.017 (0.010 to 0.024) | 1.017 (1.010 to 1.025) | < 0.001 | –0.012 (–0.027 to 0.002) | 0.988 (0.974 to 1.002) | 0.100 | |||
| DBP | 0.033 (0.022 to 0.044) | 1.034 (1.022 to 1.045) | < 0.001 | 0.054 (0.033 to 0.075) | 1.055 (1.033 to 1.078) | < 0.001 | |||
| BMIa | 0.040 (0.027 to 0.054) | 1.041 (1.027 to 1.056) | < 0.001 | 0.510 (0.065 to 0.955) | 1.666 (1.068 to 2.599) | 0.026 | –0.51 (–0.794 to –0.226) | 0.601 (0.452 to 0.798) | 0.001 |
| Nulliparous | 1.015 (0.729 to 1.301) | 2.759 (2.073 to 3.673) | < 0.001 | 0.959 (0.668 to 1.250) | 2.609 (1.951 to 3.490) | < 0.001 | 0.917 (0.632 to 1.202) | 2.502 (1.882 to 3.326) | < 0.001 |
| Previous PE | 1.374 (0.990 to 1.758) | 3.952 (2.692 to 5.802) | < 0.001 | 1.39 (1.010 to 1.770) | 4.015 (2.745 to 5.873) | < 0.001 | 1.451 (1.067 to 1.835) | 4.268 (2.907 to 6.267) | < 0.001 |
| Renal disease | 1.641 (0.980 to 2.303) | 5.163 (2.665 to 10.002) | < 0.001 | 1.57 (0.905 to 2.236) | 4.809 (2.471 to 9.357) | < 0.001 | 1.541 (0.866 to 2.216) | 4.668 (2.377 to 9.168) | < 0.001 |
| Hypertension | 1.715 (1.478 to 1.951) | 5.555 (4.384 to 7.039) | < 0.001 | 1.713 (1.471 to 1.955) | 5.545 (4.352 to 7.065) | < 0.001 | 1.734 (1.485 to 1.983) | 5.663 (4.414 to 7.264) | < 0.001 |
| Diabetes | 0.391 (–0.125 to 0.907) | 1.479 (0.883 to 2.477) | 0.137 | 0.564 (0.058 to 1.069) | 1.757 (1.06 to 2.913) | 0.029 | 0.704 (0.196 to 1.213) | 2.022 (1.216 to 3.363) | 0.007 |
| Intercept | –6.868 (–7.904 to –5.831) | –7.711 (–9.300 to –6.123) | –5.875 (–7.306 to –4.443) | ||||||
| Model | C-statistic | Calibration slope | Calibration-in-the-large | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Estimate (95% CI) | τ2 (for logit C-statistic) | I2 (for logit C-statistic) | Estimate (95% CI) | τ2 | I 2 | Estimate (95% CI) | τ2 | I 2 | |
| Model with BMI | 0.677 (0.612 to 0.736) | 0.139 | 83.9 | 0.915 (0.603 to 1.226) | 0.155 | 86.4 | 0.012 (–0.630 to 0.654) | 0.881 | 98.7 |
| Model with ln(BMI) | 0.670 (0.589 to 0.742) | 0.228 | 88.7 | 0.874 (0.473 to 1.275) | 0.297 | 92.1 | 0.015 (–0.627 to 0.656) | 0.880 | 98.6 |
| Model with BMI-2 | 0.678 (0.597 to 0.750) | 0.229 | 87.3 | 0.872 (0.436 to 1.308) | 0.336 | 93.1 | 0.014 (–0.637 to 0.664) | 0.910 | 98.6 |
| Variables | BMI | Ln(BMI) | BMI–2 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Beta coefficient (95% CI) | OR (95% CI) | p-value | Beta coefficient (95% CI) | OR (95% CI) | p-value | Beta coefficient (95% CI) | OR (95% CI) | p-value | |
| Age (years) | –0.038 (–0.071 to –0.005) | 0.963 (0.932 to 0.995) | 0.025 | –0.036 (–0.068 to –0.004) | 0.965 (0.934 to 0.996) | 0.029 | |||
| SBP | 0.014 (–0.002 to 0.030) | 1.014 (0.998 to 1.031) | 0.089 | –0.018 (–0.041 to 0.006) | 0.983 (0.959 to 1.006) | 0.147 | |||
| DBP | 0.059 (0.031 to 0.088) | 1.061 (1.031 to 1.092) | < 0.001 | 0.075 (0.032 to 0.119) | 1.078 (1.032 to 1.126) | 0.001 | |||
| BMIa | 0.033 (–0.004 to 0.070) | 1.033 (0.996 to 1.072) | 0.080 | –0.593 (–1.311 to 0.125) | 0.553 (0.27 to 1.133) | 0.105 | |||
| Previous PE | 1.593 (0.964 to 2.222) | 4.918 (2.623 to 9.222) | < 0.001 | 1.682 (1.065 to 2.299) | 5.377 (2.902 to 9.961) | < 0.001 | 1.742 (1.127 to 2.358) | 5.710 (3.087 to 10.565) | < 0.001 |
| Renal disease | 2.538 (1.212 to 3.865) | 12.656 (3.360 to 47.681) | < 0.001 | 2.258 (0.910 to 3.607) | 9.567 (2.483 to 36.856) | 0.001 | 2.277 (0.930 to 3.625) | 9.749 (2.534 to 37.512) | 0.001 |
| Hypertension | 0.645 (0.071 to 1.219) | 1.906 (1.074 to 3.382) | 0.028 | 0.554 (–0.018 to 1.126) | 1.741 (0.982 to 3.085) | 0.058 | 0.548 (0.005 to 1.091) | 1.73 (1.005 to 2.976) | 0.048 |
| Diabetes | 1.298 (0.433 to 2.163) | 3.661 (1.541 to 8.695) | 0.003 | 1.603 (0.790 to 2.415) | 4.966 (2.204 to 11.190) | < 0.001 | 1.717 (0.896 to 2.538) | 5.567 (2.449 to 12.653) | < 0.001 |
| Intercept | –8.169 (–9.926 to –6.412) | –8.798 (–10.828 to –6.769) | –7.924 (–9.817 to –6.032) | ||||||
| Model | C-statistic | Calibration slope | Calibration-in-the-large | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Estimate (95% CI) | τ2 (for logit C-statistic) | I2 (for logit C-statistic) | Estimate (95% CI) | τ2 | I 2 | Estimate (95% CI) | τ2 | I 2 | |
| Model with BMI | 0.663 (0.504 to 0.791) | 0.469 | 71.2 | 1.032 (0.802 to 1.262) | 0.006 | 4.0 | 0.085 (–0.407 to 0.576) | 0.365 | 80.6 |
| Model with ln(BMI) | 0.711 (0.574 to 0.818) | 0.351 | 62.2 | 1.054 (0.828 to 1.279) | 0.000 | 0.0 | 0.089 (–0.374 to 0.552) | 0.311 | 77.7 |
| Model with BMI–2 | 0.719 (0.590 to 0.820) | 0.260 | 49.1 | 1.001 (0.795 to 1.206) | 0.000 | 0.0 | 0.090 (–0.377 to 0.557) | 0.318 | 77.9 |
| Variables | BMI | Ln(BMI) | BMI–2 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Beta coefficient (95% CI) | OR (95% CI) | p-value | Beta coefficient (95% CI) | OR (95% CI) | p-value | Beta coefficient (95% CI) | OR (95% CI) | p-value | |
| Age (years) | –0.015 (–0.028 to –0.003) | 0.985 (0.973 to 0.997) | 0.018 | –0.016 (–0.029 to –0.003) | 0.984 (0.972 to 0.997) | 0.013 | –0.022 (–0.035 to –0.009) | 0.978 (0.965 to 0.991) | 0.001 |
| SBP | 0.016 (0.008 to 0.025) | 1.016 (1.008 to 1.025) | < 0.001 | ||||||
| DBP | 0.027 (0.016 to 0.038) | 0.985 (0.973 to 0.997) | < 0.001 | 0.039 (0.023 to 0.055) | 1.039 (1.023 to 1.056) | < 0.001 | |||
| BMIa | 0.039 (0.023 to 0.054) | 1.039 (1.024 to 1.055) | < 0.001 | 0.622 (0.141 to 1.102) | 1.863 (1.152 to 3.012) | 0.013 | –0.471 (–0.784 to –0.157) | 0.625 (0.456 to 0.855) | 0.004 |
| Nulliparous | 1.128 (0.819 to 1.436) | 3.089 (2.268 to 4.206) | < 0.001 | 1.084 (0.771 to 1.397) | 2.957 (2.163 to 4.043) | < 0.001 | 1.030 (0.725 to 1.336) | 2.802 (2.065 to 3.803) | < 0.001 |
| Previous PE | 1.214 (0.790 to 1.638) | 3.368 (2.204 to 5.145) | < 0.001 | 1.244 (0.824 to 1.663) | 3.468 (2.280 to 5.275) | < 0.001 | 1.278 (0.857 to 1.700) | 3.591 (2.356 to 5.474) | < 0.001 |
| Renal disease | 1.359 (0.652 to 2.066) | 3.891 (1.919 to 7.892) | < 0.001 | 1.295 (0.585 to 2.006) | 3.652 (1.795 to 7.431) | < 0.001 | 1.267 (0.552 to 1.983) | 3.551 (1.736 to 7.263) | 0.001 |
| Hypertension | 1.835 (1.588 to 2.082) | 6.265 (4.895 to 8.018) | < 0.001 | 1.866 (1.618 to 2.114) | 6.460 (5.041 to 8.279) | < 0.001 | 1.846 (1.595 to 2.097) | 6.335 (4.928 to 8.143) | < 0.001 |
| Intercept | –7.052 (–8.173 to –5.931) | –8.042 (–9.834 to –6.250) | –6.552 (–7.824 to –5.280) | ||||||
| Model | C-statistic | Calibration slope | Calibration-in-the-large | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Estimate (95% CI) | τ2 (for logit C-statistic) | I2 (for logit C-statistic) | Estimate (95% CI) | τ2 | I 2 | Estimate (95% CI) | τ2 | I 2 | |
| Model with BMI | 0.677 (0.615 to 0.733) | 0.116 | 79.7 | 0.919 (0.621 to 1.217) | 0.131 | 82.0 | 0.016 (–0.705 to 0.736) | 1.105 | 98.8 |
| Model with ln(BMI) | 0.666 (0.590 to 0.733) | 0.181 | 84.5 | 0.845 (0.476 to 1.215) | 0.237 | 88.4 | 0.021 (–0.704 to 0.745) | 1.108 | 98.7 |
| Model with BMI–2 | 0.677 (0.599 to 0.746) | 0.202 | 85.5 | 0.856 (0.453 to 1.259) | 0.277 | 90.3 | 0.016 (–0.732 to 0.765) | 1.192 | 98.9 |
| Variables | BMI | Ln(BMI) | BMI–2 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Beta coefficient (95% CI) | OR (95% CI) | p-value | Beta coefficient (95% CI) | OR (95% CI) | p-value | Beta coefficient (95% CI) | OR (95% CI) | p-value | |
| Age (years) | –0.017 (–0.030 to –0.004) | 0.983 (0.970 to 0.996) | 0.013 | –0.011 (–0.025 to 0.002) | 0.989 (0.975 to 1.002) | 0.102 | –0.016 (–0.028 to –0.003) | 0.984 (0.972 to 0.997) | 0.014 |
| SBP | 0.011 (0.003 to 0.020) | 1.011 (1.003 to 1.020) | 0.010 | 0.017 (0.005 to 0.030) | 1.018 (1.005 to 1.030) | 0.008 | 0.019 (0.012 to 0.025) | 1.019 (1.012 to 1.026) | < 0.001 |
| DBP | 0.020 (0.007 to 0.033) | 1.021 (1.007 to 1.034) | 0.003 | 0.012 (0.004 to 0.021) | 1.013 (1.004 to 1.021) | 0.004 | |||
| BMIa | 0.063 (0.051 to 0.076) | 1.065 (1.052 to 1.079) | < 0.001 | 1.485 (1.119 to 1.851) | 4.414 (3.061 to 6.367) | < 0.001 | –1.317 (–2.029 to –0.604) | 0.268 (0.131 to 0.547) | < 0.001 |
| Nulliparous | 0.992 (0.644 to 1.340) | 2.697 (1.905 to 3.82) | < 0.001 | 1.015 (0.664 to 1.365) | 2.758 (1.943 to 3.916) | < 0.001 | 0.970 (0.628 to 1.313) | 2.638 (1.873 to 3.716) | < 0.001 |
| Previous PE | 1.452 (1.027 to 1.876) | 4.271 (2.794 to 6.528) | < 0.001 | 1.561 (1.112 to 2.010) | 4.765 (3.041 to 7.466) | < 0.001 | 1.575 (1.153 to 1.997) | 4.831 (3.167 to 7.368) | < 0.001 |
| Renal disease | 1.001 (0.280 to 1.722) | 2.721 (1.323 to 5.596) | 0.007 | 0.863 (0.224 to 1.502) | 2.371 (1.251 to 4.492) | 0.009 | 0.908 (0.276 to 1.540) | 2.480 (1.318 to 4.666) | 0.005 |
| Hypertension | 1.833 (1.581 to 2.085) | 6.253 (4.859 to 8.047) | < 0.001 | 1.906 (1.659 to 2.152) | 6.725 (5.255 to 8.606) | < 0.001 | 1.868 (1.606 to 2.130) | 6.476 (4.984 to 8.415) | < 0.001 |
| Intercept | –8.260 (–9.350 to –7.169) | –10.891 (–12.550 to –9.232) | –6.684 (–7.853 to –5.516) | ||||||
| Model | C-statistic | Calibration slope | Calibration-in-the-large | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Estimate (95% CI) | τ2 (for logit C-statistic) | I2 (for logit C-statistic) | Estimate (95% CI) | τ2 | I 2 | Estimate (95% CI) | τ2 | I 2 | |
| Model with BMI | 0.703 (0.648 to 0.753) | 0.076 | 79.3 | 0.969 (0.664 to 1.273) | 0.129 | 88.3 | 0.011 (–0.772 to 0.795) | 1.003 | 99.0 |
| Model with ln(BMI) | 0.690 (0.636 to 0.739) | 0.062 | 76.2 | 0.993 (0.625 to 1.361) | 0.181 | 89.9 | 0.010 (–0.789 to 0.809) | 1.044 | 99.0 |
| Model with BMI–2 | 0.684 (0.613 to 0.747) | 0.131 | 86.9 | 0.992 (0.556 to 1.429) | 0.291 | 93.3 | 0.011 (–0.888 to 0.910) | 1.336 | 99.2 |
| Variables | BMI | Ln(BMI) | BMI-2 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Beta coefficient (95% CI) | OR (95% CI) | p-value | Beta coefficient (95% CI) | OR (95% CI) | p-value | Beta coefficient (95% CI) | OR (95% CI) | p-value | |
| Age (years) | –0.025 (–0.060 to 0.009) | 0.975 (0.942 to 1.009) | 0.151 | ||||||
| SBP | 0.037 (0.007 to 0.066) | 1.037 (1.007 to 1.068) | 0.017 | 0.021 (0.007 to 0.036) | 1.022 (1.007 to 1.037) | 0.005 | |||
| DBP | 0.059 (0.018 to 0.099) | 1.060 (1.018 to 1.104) | 0.006 | 0.018 (0.005 to 0.030) | 1.018 (1.005 to 1.031) | 0.006 | |||
| Previous PE | 2.140 (1.521 to 2.758) | 8.497 (4.579 to 15.767) | < 0.001 | 2.142 (1.511 to 2.773) | 8.516 (4.532 to 16.003) | < 0.001 | 2.322 (1.718 to 2.926) | 10.195 (5.575 to 18.646) | < 0.001 |
| Renal disease | 1.351 (0.072 to 2.630) | 3.861 (1.074 to 13.878) | 0.039 | 1.444 (0.296 to 2.593) | 4.238 (1.344 to 13.363) | 0.014 | 1.422 (0.257 to 2.588) | 4.147 (1.293 to 13.300) | 0.017 |
| Autoimmune disease | 1.036 (–0.266 to 2.338) | 2.817 (0.766 to 10.359) | 0.118 | 1.084 (–0.338 to 2.505) | 2.955 (0.713 to 12.247) | 0.133 | 1.463 (0.190 to 2.737) | 4.321 (1.209 to 15.446) | 0.025 |
| Intercept | –9.641 (–12.515 to –6.768) | –9.766 (–13.178 to –6.355) | –8.631 (–10.526 to –6.735) | ||||||
| Model | C-statistic | Calibration slope | Calibration-in-the-large | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Estimate (95% CI) | τ2 (for logit C-statistic) | I2 (for logit C-statistic) | Estimate (95% CI) | τ2 | I 2 | Estimate (95% CI) | τ2 | I 2 | |
| Model with BMI | 0.723 (0.601 to 0.819) | 0.223 | 56.4 | 1.105 (0.868 to 1.341) | 0.000 | 0.0 | 0.037 (–0.465 to 0.539) | 0.287 | 76.4 |
| Model with ln(BMI) | 0.702 (0.631 to 0.764) | 0.000 | 0.0 | 1.169 (0.835 to 1.503) | 0.031 | 16.9 | 0.049 (–0.451 to 0.549) | 0.285 | 76.7 |
| Model with BMI–2 | 0.719 (0.650 to 0.779) | 0.006 | 3.7 | 1.126 (0.730 to 1.522) | 0.093 | 45.0 | 0.035 (–0.445 to 0.514) | 0.271 | 77.0 |
| Variables | BMI | Ln(BMI) | BMI–2 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Beta coefficient (95% CI) | OR (95% CI) | p-value | Beta coefficient (95% CI) | OR (95% CI) | p-value | Beta coefficient (95% CI) | OR (95% CI) | p-value | |
| Age (years) | –0.015 (–0.028 to –0.001) | 0.985 (0.972 to 0.999) | 0.034 | –0.011 (–0.026 to 0.003) | 0.989 (0.975 to 1.003) | 0.112 | –0.013 (–0.027 to 0.000) | 0.987 (0.974 to 1.000) | 0.052 |
| SBP | 0.011 (0.002 to 0.020) | 1.011 (1.002 to 1.020) | 0.012 | 0.009 (–0.003 to 0.021) | 1.009 (0.997 to 1.021) | 0.130 | 0.018 (0.010 to 0.026) | 1.018 (1.010 to 1.026) | < 0.001 |
| DBP | 0.013 (0.001 to 0.026) | 1.013 (1.001 to 1.026) | 0.035 | 0.010 (–0.002 to 0.021) | 1.010 (0.998 to 1.022) | 0.097 | 0.010 (0.001 to 0.019) | 1.010 (1.001 to 1.019) | 0.035 |
| BMIa | 0.070 (0.057 to 0.082) | 1.072 (1.059 to 1.086) | < 0.001 | 1.534 (1.186 to 1.882) | 4.636 (3.274 to 6.563) | < 0.001 | –1.566 (–2.371 to –0.760) | 0.209 (0.093 to 0.468) | < 0.001 |
| Nulliparous | 1.279 (0.904 to 1.654) | 3.592 (2.469 to 5.225) | < 0.001 | 1.212 (0.844 to 1.579) | 3.359 (2.325 to 4.852) | < 0.001 | 1.181 (0.798 to 1.563) | 3.257 (2.222 to 4.774) | < 0.001 |
| Previous PE | 1.253 (0.763 to 1.743) | 3.500 (2.144 to 5.713) | < 0.001 | 1.363 (0.859 to 1.866) | 3.906 (2.361 to 6.462) | < 0.001 | 1.346 (0.864 to 1.829) | 3.842 (2.371 to 6.226) | < 0.001 |
| Renal disease | 0.814 (0.096 to 1.531) | 2.257 (1.101 to 4.625) | 0.027 | 0.548 (–0.110 to 1.206) | 1.730 (0.896 to 3.341) | 0.102 | 0.753 (0.104 to 1.401) | 2.123 (1.110 to 4.060) | 0.023 |
| Hypertension | 2.046 (1.795 to 2.298) | 7.740 (6.020 to 9.953) | < 0.001 | 2.066 (1.801 to 2.330) | 7.890 (6.056 to 10.279) | < 0.001 | 2.093 (1.831 to 2.356) | 8.112 (6.240 to 10.545) | < 0.001 |
| Intercept | –8.475 (–9.645 to –7.306) | –11.157 (–12.740 to –9.574) | –6.850 (–8.166 to –5.535) | ||||||
| Model | C-statistic | Calibration slope | Calibration-in-the-large | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Estimate (95% CI) | τ2 (for logit C-statistic) | I2 (for logit C-statistic) | Estimate (95% CI) | τ2 | I 2 | Estimate (95% CI) | τ2 | I 2 | |
| Model with BMI | 0.705 (0.649 to 0.756) | 0.076 | 78.4 | 0.930 (0.656 to 1.204) | 0.096 | 84.2 | 0.014 (–0.897 to 0.925) | 1.358 | 99.1 |
| Model with ln(BMI) | 0.683 (0.613 to 0.746) | 0.107 | 83.8 | 0.888 (0.493 to 1.283) | 0.213 | 90.7 | 0.015 (–0.899 to 0.929) | 1.360 | 99.2 |
| Model with BMI–2 | 0.682 (0.613 to 0.743) | 0.119 | 84.7 | 0.883 (0.507 to 1.259) | 0.204 | 89.9 | 0.015 (–1.026 to 1.057) | 1.784 | 99.3 |
Appendix 17 Forest plots of predictive performance estimates in the individual data sets for the second-trimester model for any-onset pre-eclampsia
FIGURE 28.
Forest plot of logit C-statistics for the second-trimester model predicting any-onset pre-eclampsia in data sets used for model development and internal validation. Note: weights are from random-effects model.
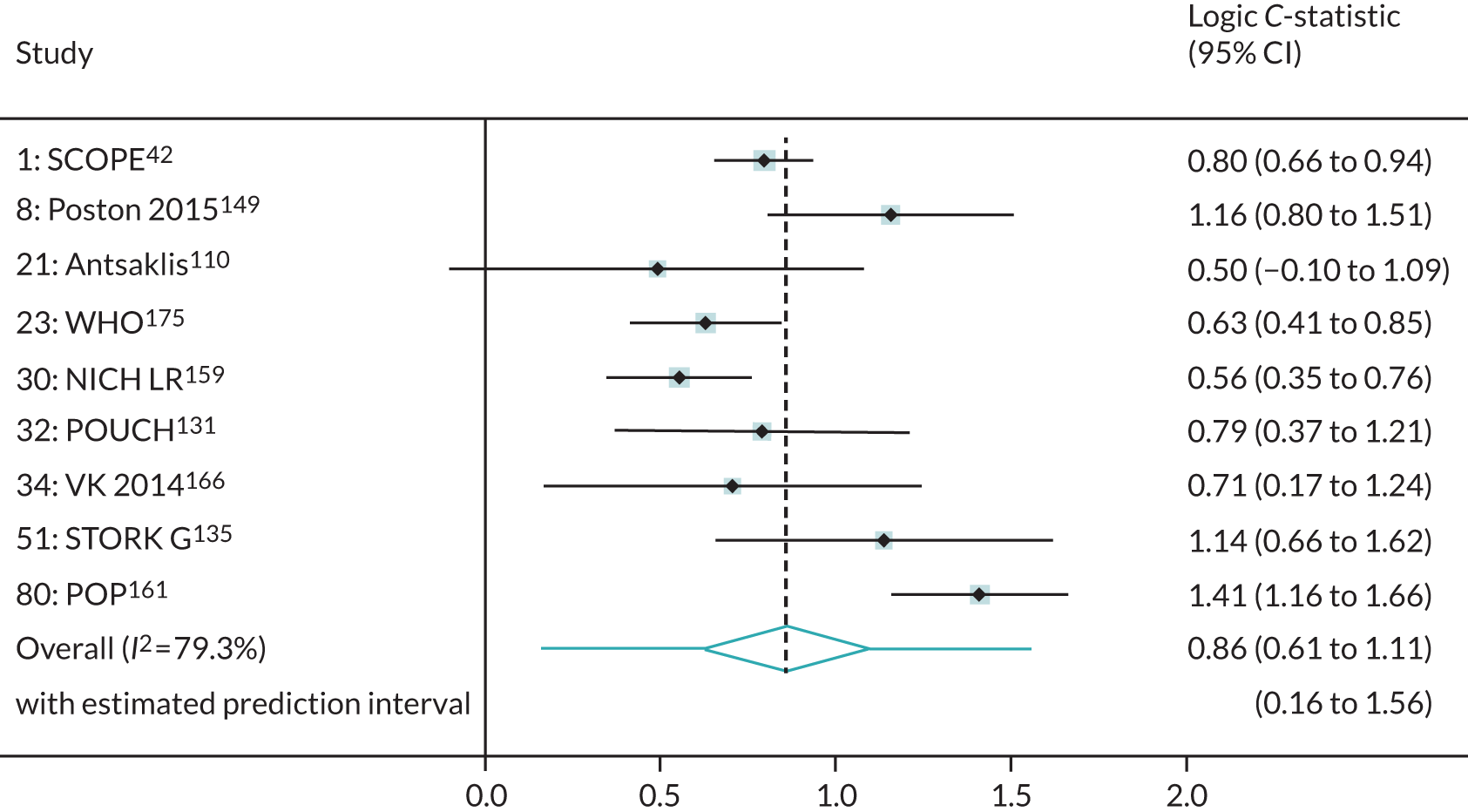
FIGURE 29.
Forest plot of calibration slope for the second-trimester model predicting any-onset pre-eclampsia in data sets used for model development and internal validation. Note: weights are from random-effects model.
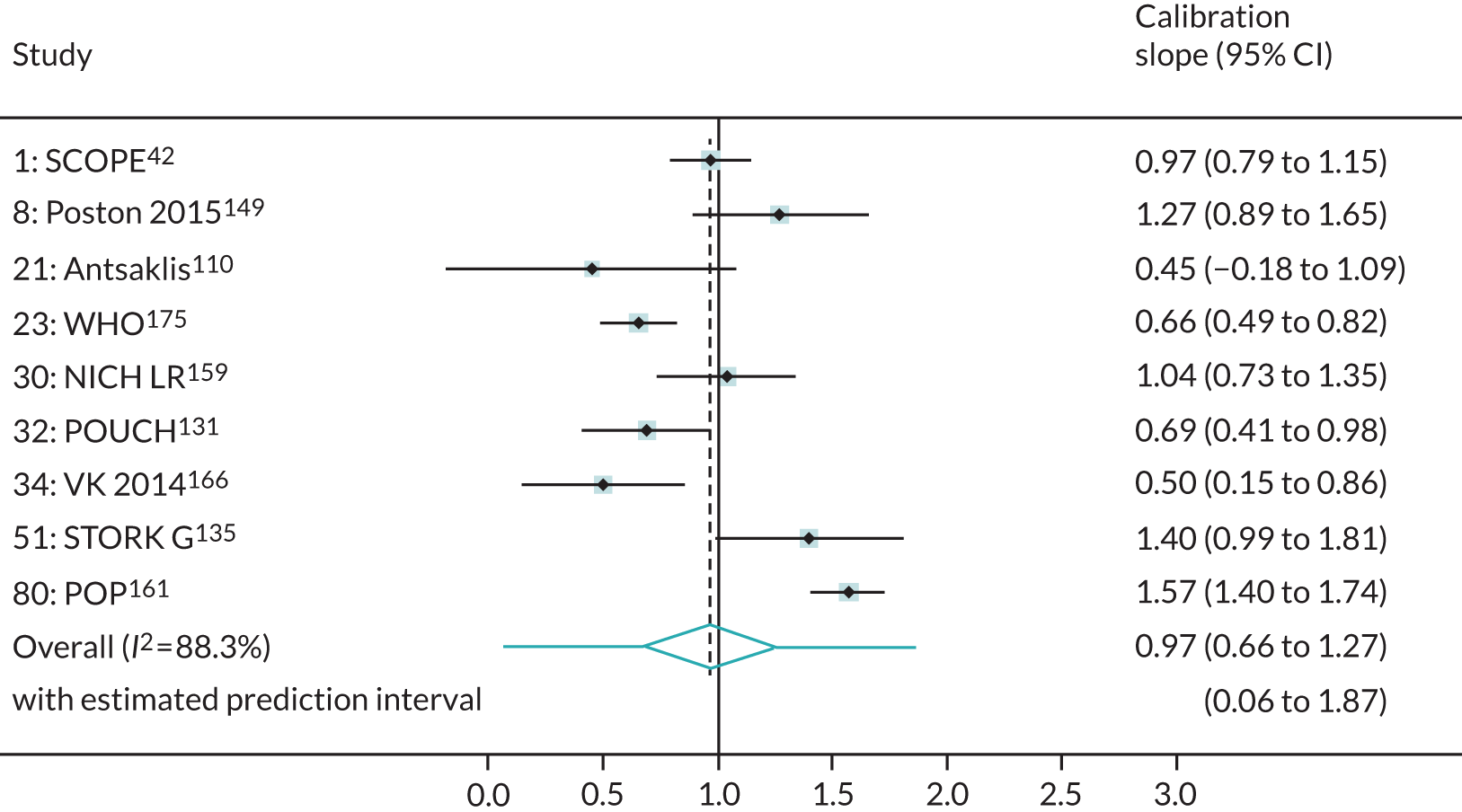
FIGURE 30.
Forest plot of calibration-in-the-large for the second-trimester model predicting any-onset pre-eclampsia in data sets used for model development and internal validation. Note: weights are from random-effects model.
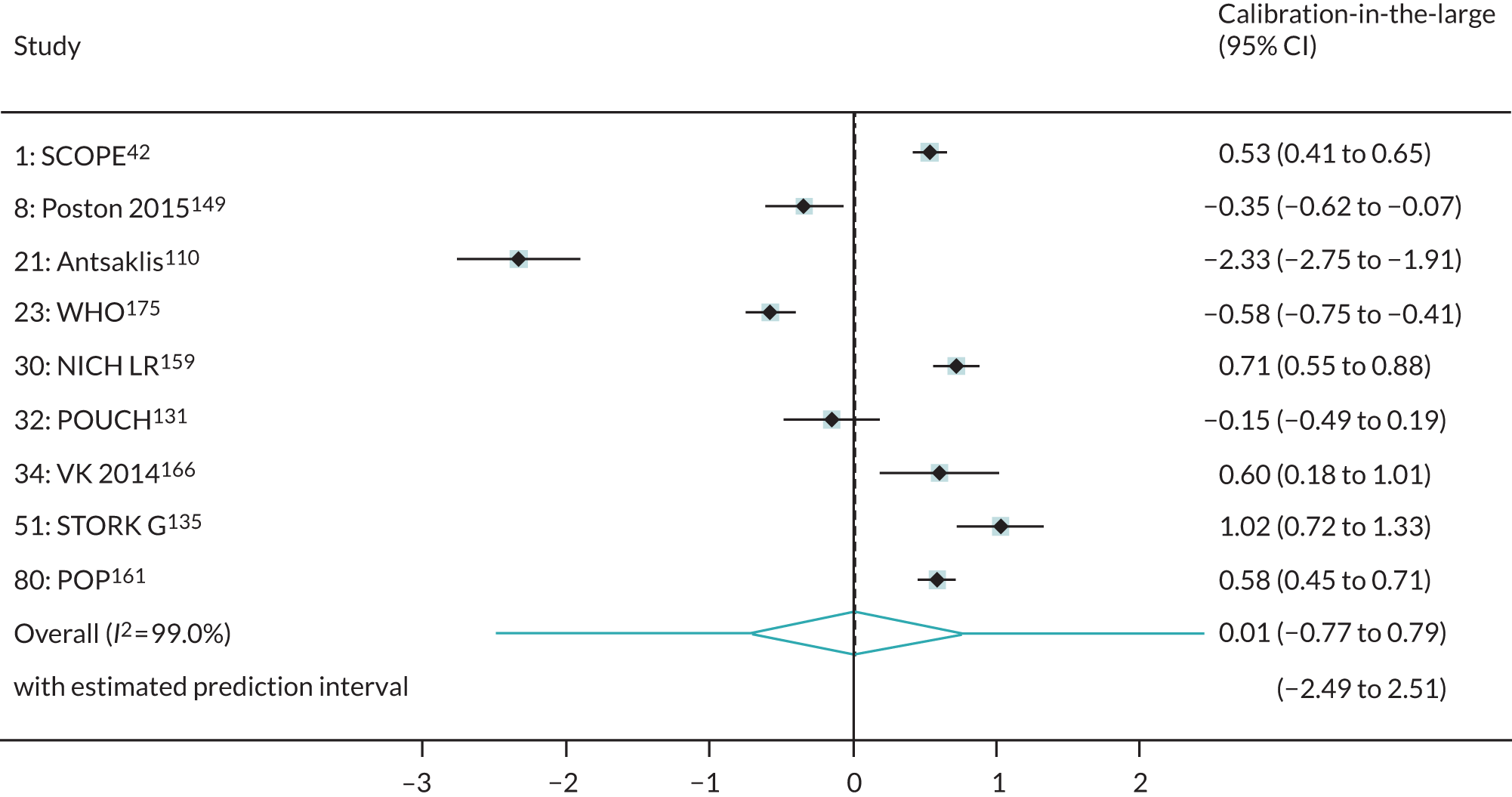
Appendix 18 Comparison of clinical characteristic and biochemical marker models in data imputed with original biochemical markers or natural log-transformed biochemical markers
| Variable | Original biochemical markers | Ln(biochemical markers) | ||||
|---|---|---|---|---|---|---|
| Beta coefficient (95% CI) | OR (95% CI) | p-value | Beta coefficient (95% CI) | OR (95% CI) | p-value | |
| Age (years) | –0.023 (–0.038 to –0.008) | 0.977 (0.962 to 0.992) | 0.003 | –0.023 (–0.039 to –0.007) | 0.977 (0.962 to 0.993) | 0.006 |
| SBP | 0.016 (0.004 to 0.028) | 1.016 (1.004 to 1.028) | 0.011 | 0.024 (0.014 to 0.034) | 1.024 (1.014 to 1.034) | < 0.001 |
| BMI | 0.041 (0.009 to 0.074) | 1.042 (1.009 to 1.077) | 0.015 | 0.036 (0.016 to 0.056) | 1.037 (1.016 to 1.057) | < 0.001 |
| Nulliparous | 1.136 (0.597 to 1.675) | 3.113 (1.816 to 5.337) | < 0.001 | 1.267 (0.703 to 1.831) | 3.551 (2.020 to 6.241) | < 0.001 |
| Previous PE | 1.350 (0.812 to 1.888) | 3.857 (2.252 to 6.607) | < 0.001 | 1.488 (0.901 to 2.076) | 4.430 (2.461 to 7.973) | < 0.001 |
| Renal disease | 0.957 (0.319 to 1.594) | 2.603 (1.376 to 4.925) | 0.004 | 1.058 (0.449 to 1.667) | 2.880 (1.566 to 5.296) | 0.001 |
| Hypertension | 2.241 (1.962 to 2.520) | 9.403 (7.113 to 12.430) | < 0.001 | 2.215 (1.949 to 2.482) | 9.165 (7.024 to 11.960) | < 0.001 |
| Diabetes | 0.021 (–0.915 to 0.957) | 1.021 (0.401 to 2.605) | 0.965 | 0.178 (–0.737 to 1.094) | 1.195 (0.479 to 2.985) | 0.702 |
| sFlt-1 | –2.42 × 10–4 (–4.3 × 10–4 to –5.5 × 10–5) | 0.9998 (0.9996 to 0.9999) | 0.012 | –0.052 (–0.109 to 0.005) | 0.949 (0.897 to 1.005) | 0.071 |
| PlGF | –0.005 (–0.008 to –0.002) | 0.995 (0.992 to 0.998) | 0.001 | –0.030 (–0.062 to 0.002) | 0.970 (0.940 to 1.002) | 0.064 |
| Intercept | –5.893 (–7.478 to –4.309) | –7.022 (–8.515 to –5.530) | ||||
| Model | C-statistic | Calibration slope | Calibration-in-the-large | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Estimate (95% CI) | τ2 (for logit C-statistic) | I2 (for logit C-statistic) | Estimate (95% CI) | τ2 | I 2 | Estimate (95% CI) | τ2 | I 2 | |
| Original biochemical markers | 0.696 (0.516 to 0.832) | 0.205 | 90.7 | 0.901 (0.355 to 1.447) | 0.094 | 86.7 | –0.004 (–0.939 to 0.931) | 0.333 | 97.9 |
| Ln(biochemical markers) | 0.693 (0.538 to 0.814) | 0.150 | 88.8 | 0.896 (0.302 to 1.491) | 0.120 | 90.5 | 0.008 (–0.874 to 0.889) | 0.291 | 97.1 |
| Variable | Original biochemical markers | Ln(biochemical markers) | ||||
|---|---|---|---|---|---|---|
| Beta coefficient (95% CI) | OR (95% CI) | p-value | Beta coefficient (95% CI) | OR (95% CI) | p-value | |
| Age (years) | –0.005 (–0.047 to 0.038) | 0.995 (0.954 to 1.039) | 0.831 | –0.013 (–0.054 to 0.028) | 0.987 (0.947 to 1.028) | 0.523 |
| SBP | 0.053 (0.025 to 0.082) | 1.055 (1.026 to 1.085) | < 0.001 | 0.024 (–0.007 to 0.054) | 1.024 (0.993 to 1.055) | 0.125 |
| DBP | –0.004 (–0.04 to 0.033) | 0.996 (0.960 to 1.033) | 0.836 | 0.042 (0.004 to 0.080) | 1.043 (1.004 to 1.083) | 0.031 |
| BMI | –0.007 (–0.069 to 0.056) | 0.993 (0.933 to 1.057) | 0.831 | 0.006 (–0.049 to 0.061) | 1.006 (0.952 to 1.062) | 0.837 |
| Previous PE | 1.488 (0.694 to 2.281) | 4.427 (2.002 to 9.789) | < 0.001 | 1.689 (0.909 to 2.469) | 5.412 (2.481 to 11.805) | < 0.001 |
| Renal disease | 1.481 (0.401 to 2.560) | 4.397 (1.494 to 12.942) | 0.007 | 1.414 (0.325 to 2.503) | 4.112 (1.384 to 12.216) | 0.011 |
| Hypertension | 0.833 (0.033 to 1.633) | 2.300 (1.034 to 5.117) | 0.041 | 0.978 (0.201 to 1.755) | 2.659 (1.222 to 5.786) | 0.014 |
| sFlt-1 | –5.15 × 10–4 (–1.1 × 10–3 to 6.1 × 10–5) | 0.9995 (0.9989 to 1.0001) | 0.079 | |||
| Intercept | –10.206 (–13.561 to –6.851) | –10.929 (–13.875 to –7.983) | ||||
| Model | C-statistic | Calibration slope | Calibration-in-the-large | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Estimate (95% CI) | τ2 (for logit C-statistic) | I2 (for logit C-statistic) | Estimate (95% CI) | τ2 | I 2 | Estimate (95% CI) | τ2 | I 2 | |
| Original biochemical markers | 0.762 (0.576 to 0.883) | 0.107 | 37.0 | 1.038 (0.666 to 1.410) | 0 | 0 | 0.036 (–1.269 to 1.342) | 0.610 | 91.7 |
| Ln(biochemical markers) | 0.745 (0.591 to 0.855) | 0.053 | 27.2 | 1.022 (0.695 to 1.350) | 0 | 0 | 0.014 (–1.240 to 1.268) | 0.552 | 90.2 |
| Variable | Original biochemical markers | Ln(biochemical markers) | ||||
|---|---|---|---|---|---|---|
| Beta coefficient (95% CI) | OR (95% CI) | p-value | Beta coefficient (95% CI) | OR (95% CI) | p-value | |
| Age (years) | –0.025 (–0.040 to –0.009) | 0.976 (0.961 to 0.991) | 0.002 | –0.024 (–0.042 to –0.006) | 0.976 (0.959 to 0.994) | 0.009 |
| SBP | 0.009 (–0.002 to 0.020) | 1.009 (0.998 to 1.020) | 0.113 | 0.020 (0.010 to 0.030) | 1.020 (1.010 to 1.031) | < 0.001 |
| BMI | 0.051 (0.024 to 0.077) | 1.052 (1.024 to 1.080) | < 0.001 | 0.040 (0.018 to 0.061) | 1.040 (1.019 to 1.063) | < 0.001 |
| Nulliparous | 1.409 (0.814 to 2.004) | 4.093 (2.258 to 7.418) | < 0.001 | 1.683 (1.035 to 2.330) | 5.379 (2.816 to 10.276) | < 0.001 |
| Previous PE | 1.197 (0.557 to 1.838) | 3.311 (1.745 to 6.283) | < 0.001 | 1.456 (0.774 to 2.138) | 4.289 (2.168 to 8.486) | < 0.001 |
| Renal disease | 0.758 (0.084 to 1.433) | 2.135 (1.088 to 4.190) | 0.028 | 0.886 (0.225 to 1.547) | 2.426 (1.253 to 4.698) | 0.009 |
| Hypertension | 2.374 (2.084 to 2.663) | 10.739 (8.039 to 14.346) | < 0.001 | 2.326 (2.049 to 2.604) | 10.238 (7.756 to 13.515) | < 0.001 |
| sFlt-1 | –2 × 10–4 (–3.8 × 10–4 to –2.4 × 10–5) | 0.9998 (0.9996 to 1.0000) | 0.026 | –0.06 (–0.119 to –0.001) | 0.942 (0.888 to 0.999) | 0.048 |
| PlGF | –0.006 (–0.009 to –0.003) | 0.994 (0.991 to 0.997) | 0.001 | –0.032 (–0.066 to 0.003) | 0.969 (0.936 to 1.003) | 0.072 |
| Intercept | –5.787 (–7.28 to –4.293) | –7.168 (–8.776 to –5.559) | ||||
| Model | C-statistic | Calibration slope | Calibration-in-the-large | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Estimate (95% CI) | τ2 (for logit C-statistic) | I2 (for logit C-statistic) | Estimate (95% CI) | τ2 | I 2 | Estimate (95% CI) | τ2 | I 2 | |
| Original biochemical markers | 0.688 (0.505 to 0.827) | 0.212 | 91.9 | 0.857 (0.338 to 1.376) | 0.085 | 84.4 | –0.005 (–0.932 to 0.922) | 0.322 | 97.5 |
| Ln(biochemical markers) | 0.685 (0.511 to 0.818) | 0.188 | 90.8 | 0.849 (0.277 to 1.422) | 0.112 | 88.6 | 0.010 (–0.856 to 0.876) | 0.276 | 96.5 |
| Variable | Original biochemical markers | Ln(biochemical markers) | ||||
|---|---|---|---|---|---|---|
| Beta coefficient (95% CI) | OR (95% CI) | p-value | Beta coefficient (95% CI) | OR (95% CI) | p-value | |
| Age (years) | –0.019 (–0.034 to –0.004) | 0.981 (0.966 to 0.996) | 0.012 | –0.026 (–0.041 to –0.011) | 0.974 (0.960 to 0.989) | 0.001 |
| SBP | 0.019 (0.009 to 0.029) | 1.019 (1.009 to 1.030) | < 0.001 | 0.005 (–0.005 to 0.015) | 1.005 (0.995 to 1.015) | 0.357 |
| DBP | –0.009 (–0.021 to 0.004) | 0.991 (0.979 to 1.004) | 0.167 | 0.020 (0.006 to 0.033) | 1.020 (1.006 to 1.034) | 0.004 |
| BMI | 0.069 (0.055 to 0.084) | 1.072 (1.057 to 1.088) | < 0.001 | 0.069 (0.055 to 0.084) | 1.072 (1.057 to 1.087) | < 0.001 |
| Nulliparous | 2.099 (1.263 to 2.935) | 8.156 (3.534 to 18.820) | < 0.001 | 2.144 (1.344 to 2.944) | 8.532 (3.833 to 18.993) | < 0.001 |
| Previous PE | 1.831 (1.013 to 2.648) | 6.237 (2.755 to 14.123) | < 0.001 | 1.947 (1.172 to 2.722) | 7.006 (3.227 to 15.209) | < 0.001 |
| Renal disease | 1.423 (0.609 to 2.237) | 4.150 (1.838 to 9.368) | 0.001 | 1.467 (0.658 to 2.275) | 4.335 (1.931 to 9.732) | < 0.001 |
| Hypertension | 2.401 (2.120 to 2.682) | 11.034 (8.334 to 14.608) | < 0.001 | 2.389 (2.112 to 2.666) | 10.905 (8.268 to 14.383) | < 0.001 |
| PlGF | –0.003 (–0.004 to –0.002) | 0.997 (0.996 to 0.998) | < 0.001 | –0.539 (–0.658 to –0.420) | 0.583 (0.518 to 0.657) | < 0.001 |
| Intercept | –7.895 (–9.314 to –6.476) | –6.070 (–7.559 to –4.581) | ||||
| Model | C-statistic | Calibration slope | Calibration-in-the-large | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Estimate (95% CI) | τ2 (for logit C-statistic) | I2 (for logit C-statistic) | Estimate (95% CI) | τ2 | I 2 | Estimate (95% CI) | τ2 | I 2 | |
| Original biochemical markers | 0.754 (0.526 to 0.894) | 0.151 | 92.4 | 0.961 (0.358 to 1.564) | 0.052 | 88.7 | 0.004 (–1.284 to 1.293) | 0.262 | 98.0 |
| Ln(biochemical markers) | 0.746 (0.521 to 0.888) | 0.150 | 93.6 | 0.945 (0.218 to 1.672) | 0.081 | 94.1 | 0.001 (–1.859 to 1.862) | 0.556 | 99.0 |
| Variable | Original biochemical markers | Ln(biochemical markers) | ||||
|---|---|---|---|---|---|---|
| Beta coefficient (95% CI) | OR (95% CI) | p-value | Beta coefficient (95% CI) | OR (95% CI) | p-value | |
| DBP | 0.071 (0.044 to 0.099) | 1.074 (1.045 to 1.104) | < 0.001 | 0.065 (0.039 to 0.092) | 1.068 (1.039 to 1.096) | < 0.001 |
| Previous PE | 1.998 (1.046 to 2.949) | 7.371 (2.847 to 19.084) | < 0.001 | 2.360 (1.529 to 3.191) | 10.594 (4.615 to 24.318) | < 0.001 |
| Renal disease | 2.291 (0.151 to 4.431) | 9.883 (1.163 to 83.975) | 0.036 | 3.148 (0.961 to 5.335) | 23.285 (2.613 to 207.474) | 0.005 |
| Autoimmune disease | 1.012 (–1.075 to 3.100) | 2.752 (0.341 to 22.193) | 0.342 | 1.154 (–0.924 to 3.231) | 3.169 (0.397 to 25.315) | 0.277 |
| sFlt-1 | 0.458 (0.096 to 0.820) | 1.581 (1.100 to 2.271) | 0.013 | |||
| PlGF | –0.003 (–0.005 to –0.001) | 0.997 (0.995 to 0.999) | 0.010 | –0.988 (–1.283 to –0.693) | 0.372 (0.277 to 0.500) | < 0.001 |
| Intercept | –10.116 (–12.119 to –8.114) | –9.343 (–12.661 to –6.025) | ||||
| Model | C-statistic | Calibration slope | Calibration-in-the-large | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Estimate (95% CI) | τ2 (for logit C-statistic) | I2 (for logit C-statistic) | Estimate (95% CI) | τ2 | I 2 | Estimate (95% CI) | τ2 | I 2 | |
| Original biochemical markers | 0.786 (0.389 to 0.955) | 0.304 | 61.2 | 1.054 (0.475 to 1.633) | 0 | 0 | 0.024 (–1.308 to 1.357) | 0.229 | 80.9 |
| Ln(biochemical markers) | 0.830 (0.629 to 0.934) | 0.041 | 17.4 | 1.079 (0.567 to 1.591) | 0.011 | 26.0 | 0.099 (–0.972 to 1.169) | 0.107 | 66.4 |
| Variables | Original biochemical markers | Ln(biochemical markers) | ||||
|---|---|---|---|---|---|---|
| Beta coefficient (95% CI) | OR (95% CI) | p-value | Beta coefficient (95% CI) | OR (95% CI) | p-value | |
| Age (years) | –0.020 (–0.036 to –0.004) | 0.980 (0.965 to 0.996) | 0.012 | –0.027 (–0.043 to –0.012) | 0.973 (0.958 to 0.989) | 0.001 |
| SBP | 0.018 (0.008 to 0.029) | 1.018 (1.008 to 1.029) | 0.001 | 0.002 (–0.009 to 0.014) | 1.002 (0.991 to 1.014) | 0.697 |
| DBP | –0.014 (–0.028 to –0.001) | 0.986 (0.973 to 0.999) | 0.031 | 0.018 (0.002 to 0.033) | 1.018 (1.002 to 1.034) | 0.026 |
| BMI | 0.073 (0.058 to 0.088) | 1.076 (1.060 to 1.093) | < 0.001 | 0.074 (0.059 to 0.089) | 1.077 (1.061 to 1.093) | < 0.001 |
| Nulliparous | 2.539 (1.564 to 3.515) | 12.673 (4.777 to 33.622) | < 0.001 | 2.567 (1.647 to 3.487) | 13.023 (5.190 to 32.681) | < 0.001 |
| Previous PE | 1.734 (0.799 to 2.669) | 5.664 (2.223 to 14.431) | < 0.001 | 1.839 (0.962 to 2.716) | 6.291 (2.617 to 15.120) | < 0.001 |
| Renal disease | 1.303 (0.463 to 2.143) | 3.680 (1.588 to 8.524) | 0.002 | 1.318 (0.487 to 2.148) | 3.735 (1.628 to 8.572) | 0.002 |
| Hypertension | 2.432 (2.144 to 2.720) | 11.380 (8.531 to 15.180) | < 0.001 | 2.426 (2.144 to 2.709) | 11.317 (8.531 to 15.012) | < 0.001 |
| PlGF | –0.003 (–0.004 to –0.002) | 0.997 (0.996 to 0.998) | < 0.001 | –0.424 (–0.545 to –0.302) | 0.655 (0.580 to 0.739) | < 0.001 |
| Intercept | –8.054 (–9.598 to –6.510) | –6.760 (–8.323 to –5.198) | ||||
| Model | C-statistic | Calibration slope | Calibration-in-the-large | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Estimate (95% CI) | τ2 (for logit C-statistic) | I2 (for logit C-statistic) | Estimate (95% CI) | τ2 | I 2 | Estimate (95% CI) | τ2 | I 2 | |
| Original biochemical markers | 0.746 (0.493 to 0.898) | 0.181 | 93.2 | 0.936 (0.355 to 1.517) | 0.047 | 86.5 | 0.004 (–1.295 to 1.303) | 0.266 | 97.8 |
| Ln(biochemical markers) | 0.730 (0.477 to 0.889) | 0.180 | 94.4 | 0.909 (0.186 to 1.632) | 0.078 | 92.8 | 0.001 (–1.697 to 1.700) | 0.461 | 98.7 |
Appendix 19 Predictive performance of final shrunken prediction models for any-, early- and late-onset pre-eclampsia in the individual data sets used for model development and validation
| Model | Data set | N | Events (n) | Performance statistic (95% CI) | ||
|---|---|---|---|---|---|---|
| C-statistic | Calibration slope | Calibration-in-the-large | ||||
| Any-onset pre-eclampsia | ||||||
| First-trimester clinical characteristics | SCOPE42 | 5628 | 278 | 0.597 (0.547 to 0.645) | 1.165 (0.722 to 1.608) | 0.458 (0.335 to 0.581) |
| Allen et al.108 | 1045 | 14 | 0.757 (0.633 to 0.849) | 1.253 (0.618 to 1.887) | –0.888 (–1.421 to –0.356) | |
| Poston et al. 2015149 | 1554 | 54 | 0.694 (0.565 to 0.799) | 1.340 (0.491 to 2.189) | –0.175 (–0.458 to 0.107) | |
| Baschat et al.115 | 1704 | 106 | 0.738 (0.688 to 0.783) | 0.747 (0.585 to 0.909) | 0.291 (0.079 to 0.503) | |
| Antsaklis et al.110 | 3328 | 32 | 0.585 (0.467 to 0.694) | 0.291 (–0.264 to 0.846) | –2.201 (–2.559 to –1.843) | |
| WHO175 | 7273 | 141 | 0.637 (0.577 to 0.693) | 0.808 (0.569 to 1.047) | –0.638 (–0.806 to –0.469) | |
| NICH LR159 | 3097 | 156 | 0.506 (0.425 to 0.587) | 0.024 (–0.858 to 0.906) | 0.411 (0.243 to 0.578) | |
| van Kuijk et al. 2014166 | 230 | 43 | 0.684 (0.581 to 0.772) | 0.705 (0.302 to 1.108) | 0.780 (0.413 to 1.147) | |
| STORK G135 | 812 | 46 | 0.723 (0.624 to 0.805) | 1.466 (0.957 to 1.975) | 1.064 (0.762 to 1.366) | |
| Vinter et al.173 | 304 | 19 | 0.679 (0.549 to 0.786) | 1.244 (0.260 to 2.227) | 0.442 (–0.026 to 0.910) | |
| POP161 | 4212 | 273 | 0.799 (0.765 to 0.829) | 1.744 (1.578 to 1.909) | 0.515 (0.389 to 0.642) | |
| First-trimester clinical characteristics and biochemical markers | SCOPE42 | 5628 | 278 | 0.583 (0.516 to 0.646) | 0.891 (0.423 to 1.359) | 0.354 (0.228 to 0.479) |
| WHO175 | 7273 | 141 | 0.687 (0.624 to 0.744) | 0.778 (0.565 to 0.992) | –0.835 (–1.007 to –0.664) | |
| POUCH131 | 3019 | 44 | 0.674 (0.553 to 0.775) | 0.737 (0.247 to 1.227) | 0.204 (–0.138 to 0.545) | |
| POP161 | 4212 | 273 | 0.807 (0.773 to 0.837) | 1.464 (1.328 to 1.600) | 0.282 (0.154 to 0.411) | |
| Second-trimester clinical characteristics | SCOPE42 | 5628 | 278 | 0.689 (0.658 to 0.719) | 1.004 (0.819 to 1.190) | 0.514 (0.391 to 0.636) |
| Poston et al. 2015149 | 1554 | 54 | 0.761 (0.691 to 0.819) | 1.310 (0.914 to 1.706) | –0.342 (–0.617 to –0.067) | |
| Antsaklis et al.110 | 3328 | 32 | 0.621 (0.475 to 0.748) | 0.468 (–0.191 to 1.128) | –2.293 (–2.707 to –1.879) | |
| WHO175 | 7273 | 141 | 0.653 (0.602 to 0.700) | 0.677 (0.506 to 0.847) | –0.585 (–0.756 to –0.414) | |
| NICH LR159 | 3097 | 156 | 0.635 (0.586 to 0.682) | 1.076 (0.755 to 1.396) | 0.693 (0.531 to 0.856) | |
| POUCH131 | 3019 | 44 | 0.688 (0.592 to 0.770) | 0.714 (0.419 to 1.008) | –0.15 (–0.487 to 0.186) | |
| van Kuijk et al. 2014166 | 230 | 43 | 0.67 (0.543 to 0.776) | 0.519 (0.151 to 0.887) | 0.632 (0.222 to 1.041) | |
| STORK G135 | 812 | 46 | 0.757 (0.659 to 0.835) | 1.443 (1.019 to 1.867) | 0.998 (0.693 to 1.303) | |
| POP161 | 4212 | 273 | 0.804 (0.762 to 0.840) | 1.621 (1.446 to 1.795) | 0.573 (0.444 to 0.703) | |
| Second-trimester clinical characteristics and biochemical markers | SCOPE42 | 5628 | 278 | 0.670 (0.637 to 0.701) | 0.927 (0.743 to 1.110) | 0.063 (–0.060 to 0.185) |
| WHO175 | 7273 | 141 | 0.768 (0.697 to 0.826) | 0.782 (0.580 to 0.984) | –0.549 (–0.726 to –0.371) | |
| POP161 | 4212 | 273 | 0.814 (0.781 to 0.844) | 1.264 (1.145 to 1.383) | 0.482 (0.345 to 0.620) | |
| Early-onset pre-eclampsia | ||||||
| First-trimester clinical characteristics | SCOPE42 | 5628 | 44 | 0.783 (0.455 to 0.940) | 1.680 (–0.026 to 3.387) | 0.986 (0.673 to 1.300) |
| Allen et al.108 | 1045 | 1 | NE | 0.808 (–0.391 to 2.008) | –1.566 (–3.551 to 0.420) | |
| Poston et al. 2015149 | 1554 | 5 | 0.736 (0.419 to 0.915) | 0.934 (–0.251 to 2.118) | –0.462 (–1.374 to 0.451) | |
| Baschat et al.115 | 1704 | 21 | 0.846 (0.716 to 0.923) | 1.018 (0.752 to 1.284) | 0.681 (0.226 to 1.135) | |
| Antsaklis et al.110 | 3328 | 13 | 0.459 (0.272 to 0.659) | –0.233 (–1.342 to 0.876) | –0.019 (–0.539 to 0.500) | |
| WHO175 | 7273 | 18 | 0.811 (0.659 to 0.905) | 1.079 (0.728 to 1.430) | –0.646 (–1.112 to –0.181) | |
| NICH LR159 | 3097 | 12 | 0.536 (0.283 to 0.771) | 0.189 (–1.446 to 1.824) | 0.162 (–0.413 to 0.737) | |
| van Kuijk et al. 2014166 | 230 | 15 | 0.672 (0.446 to 0.839) | 0.620 (–0.301 to 1.541) | 0.778 (0.221 to 1.335) | |
| STORK G135 | 812 | 1 | NE | –1.513 (–7.175 to 4.150) | –1.125 (–3.088 to 0.837) | |
| Vinter et al.173 | 304 | 2 | 0.775 (0.261 to 0.971) | 2.114 (–0.318 to 4.547) | –0.033 (–1.425 to 1.359) | |
| POP161 | 4212 | 10 | 0.779 (0.568 to 0.904) | 1.168 (0.676 to 1.660) | –0.235 (–0.858 to 0.388) | |
| First-trimester clinical characteristics and biochemical markers | SCOPE42 | 5628 | 44 | 0.664 (0.509 to 0.790) | 0.934 (0.165 to 1.703) | 0.899 (0.591 to 1.207) |
| WHO175 | 7273 | 18 | 0.879 (0.727 to 0.952) | 1.010 (0.687 to 1.333) | –0.714 (–1.188 to –0.241) | |
| POUCH131 | 3019 | 12 | 0.773 (0.620 to 0.877) | 0.889 (0.475 to 1.302) | 0.479 (–0.049 to 1.008) | |
| POP161 | 4212 | 10 | 0.743 (0.496 to 0.895) | 1.195 (0.654 to 1.737) | –0.703 (–1.326 to –0.081) | |
| Second-trimester clinical characteristics | SCOPE42 | 5628 | 44 | 0.674 (0.583 to 0.754) | 1.268 (0.748 to 1.789) | 0.978 (0.681 to 1.275) |
| Poston 2015149 | 1554 | 5 | 0.882 (0.703 to 0.959) | 1.155 (0.497 to 1.814) | –0.701 (–1.587 to 0.185) | |
| Antsaklis110 | 3328 | 13 | 0.701 (0.331 to 0.917) | 0.863 (–0.93 to 2.655) | –0.258 (–1.581 to 1.064) | |
| WHO175 | 7273 | 18 | 0.838 (0.671 to 0.930) | 1.036 (0.744 to 1.327) | –0.634 (–1.101 to –0.167) | |
| NICH LR159 | 3097 | 12 | 0.556 (0.379 to 0.720) | 0.422 (–0.594 to 1.437) | 0.557 (–0.017 to 1.132) | |
| POUCH131 | 3019 | 12 | 0.832 (0.691 to 0.917) | 0.965 (0.637 to 1.294) | 0.377 (–0.211 to 0.965) | |
| van Kuijk et al. 2014166 | 230 | 15 | 0.672 (0.428 to 0.848) | 0.659 (–0.270 to 1.588) | 0.293 (–0.308 to 0.894) | |
| STORK G135 | 812 | 1 | NE | 0.311 (–3.395 to 4.018) | –0.970 (–2.932 to 0.991) | |
| POP161 | 4212 | 10 | 0.558 (0.311 to 0.779) | 0.404 (–0.806 to 1.613) | –0.286 (–0.911 to 0.339) | |
| Second-trimester clinical characteristics and biochemical markers | SCOPE42 | 5628 | 44 | 0.805 (0.728 to 0.865) | 1.175 (0.891 to 1.458) | –0.159 (–0.458 to 0.141) |
| WHO175 | 7273 | 18 | 0.912 (0.778 to 0.968) | 0.876 (0.598 to 1.153) | –0.128 (–0.61 to 0.354) | |
| POP161 | 4212 | 10 | 0.810 (0.575 to 0.931) | 0.905 (0.453 to 1.356) | 0.805 (0.156 to 1.455) | |
| Late-onset pre-eclampsia | ||||||
| First-trimester clinical characteristics | SCOPE42 | 5628 | 234 | 0.630 (0.578 to 0.679) | 1.266 (0.844 to 1.688) | 0.432 (0.298 to 0.565) |
| Allen et al.108 | 1045 | 13 | 0.738 (0.599 to 0.842) | 1.258 (0.596 to 1.921) | –0.757 (–1.308 to –0.205) | |
| Poston et al. 2015149 | 1554 | 49 | 0.682 (0.548 to 0.791) | 1.208 (0.388 to 2.027) | –0.048 (–0.342 to 0.246) | |
| Baschat et al.115 | 1704 | 85 | 0.701 (0.646 to 0.751) | 0.601 (0.417 to 0.785) | 0.281 (0.050 to 0.513) | |
| Antsaklis et al.110 | 3328 | 19 | 0.676 (0.527 to 0.796) | 0.781 (–0.075 to 1.637) | –2.669 (–3.128 to –2.211) | |
| WHO175 | 7273 | 123 | 0.606 (0.541 to 0.668) | 0.672 (0.374 to 0.970) | –0.588 (–0.768 to –0.409) | |
| NICH LR159 | 3097 | 144 | 0.506 (0.42 to 0.591) | 0.025 (–0.948 to 0.998) | 0.495 (0.323 to 0.668) | |
| van Kuijk et al. 2014166 | 230 | 28 | 0.686 (0.574 to 0.780) | 0.674 (0.242 to 1.106) | 0.536 (0.110 to 0.961) | |
| STORK G135 | 812 | 45 | 0.733 (0.634 to 0.813) | 1.442 (0.947 to 1.937) | 1.226 (0.921 to 1.532) | |
| Vinter et al.173 | 304 | 17 | 0.659 (0.521 to 0.774) | 0.954 (0.026 to 1.883) | 0.532 (0.039 to 1.026) | |
| POP161 | 4212 | 263 | 0.798 (0.763 to 0.828) | 1.667 (1.508 to 1.827) | 0.621 (0.493 to 0.750) | |
| First-trimester clinical characteristics and biochemical markers | SCOPE42 | 5628 | 234 | 0.576 (0.521 to 0.630) | 0.854 (0.435 to 1.274) | 0.304 (0.169 to 0.438) |
| WHO175 | 7273 | 123 | 0.679 (0.612 to 0.739) | 0.810 (0.540 to 1.08) | –0.804 (–0.985 to –0.622) | |
| POUCH131 | 3019 | 32 | 0.662 (0.555 to 0.755) | 0.717 (0.224 to 1.211) | 0.109 (–0.314 to 0.531) | |
| POP161 | 4212 | 263 | 0.805 (0.771 to 0.835) | 1.468 (1.331 to 1.606) | 0.396 (0.265 to 0.526) | |
| Second-trimester clinical characteristics | SCOPE42 | 5628 | 234 | 0.678 (0.644 to 0.711) | 0.961 (0.760 to 1.161) | 0.473 (0.340 to 0.606) |
| Poston et al. 2015149 | 1554 | 49 | 0.737 (0.667 to 0.797) | 1.153 (0.746 to 1.560) | –0.182 (–0.468 to 0.105) | |
| Antsaklis et al.110 | 3328 | 19 | 0.721 (0.555 to 0.843) | 1.023 (0.041 to 2.005) | –2.720 (–3.195 to –2.245) | |
| WHO175 | 7273 | 123 | 0.638 (0.586 to 0.688) | 0.624 (0.408 to 0.839) | –0.513 (–0.694 to –0.332) | |
| NICH LR159 | 3097 | 144 | 0.636 (0.584 to 0.685) | 1.107 (0.764 to 1.451) | 0.752 (0.583 to 0.921) | |
| POUCH131 | 3019 | 32 | 0.701 (0.604 to 0.784) | 0.741 (0.422 to 1.060) | –0.235 (–0.624 to 0.155) | |
| van Kuijk et al. 2014166 | 230 | 28 | 0.655 (0.505 to 0.779) | 0.457 (0.050 to 0.865) | 0.611 (0.154 to 1.067) | |
| STORK G135 | 812 | 45 | 0.761 (0.662 to 0.838) | 1.346 (0.938 to 1.754) | 1.195 (0.886 to 1.503) | |
| POP161 | 4212 | 263 | 0.805 (0.765 to 0.840) | 1.554 (1.397 to 1.710) | 0.648 (0.517 to 0.780) | |
| Second-trimester clinical characteristics and biochemical markers | SCOPE42 | 5628 | 234 | 0.654 (0.617 to 0.688) | 0.887 (0.686 to 1.087) | 0.003 (–0.129 to 0.136) |
| WHO175 | 7273 | 123 | 0.754 (0.681 to 0.815) | 0.812 (0.585 to 1.040) | –0.531 (–0.717 to –0.345) | |
| POP161 | 4212 | 263 | 0.815 (0.782 to 0.845) | 1.262 (1.141 to 1.383) | 0.524 (0.384 to 0.663) | |
Appendix 20 Calibration plots for final shrunken prediction models for any- and late-onset pre-eclampsia
Appendix 21 Decision curve analysis for developed models in each data set
Early-onset pre-eclampsia models
FIGURE 38.
Decision curves for the final (shrunken) model predicting early-onset pre-eclampsia using first-trimester clinical characteristics, in data sets used in the development and validation of the model. (a) SCOPE;42 (b) Allen et al.;108 (c) Poston et al. 2015;149 (d) Baschat et al.;115 (e) Antsaklis;110 (f) WHO;175 (g) NICH LR;159 (h) Van Kuijk et al. 2014;166 (i) STORK G;135 (j) Vinter et al.;173 and (k) POP. 161
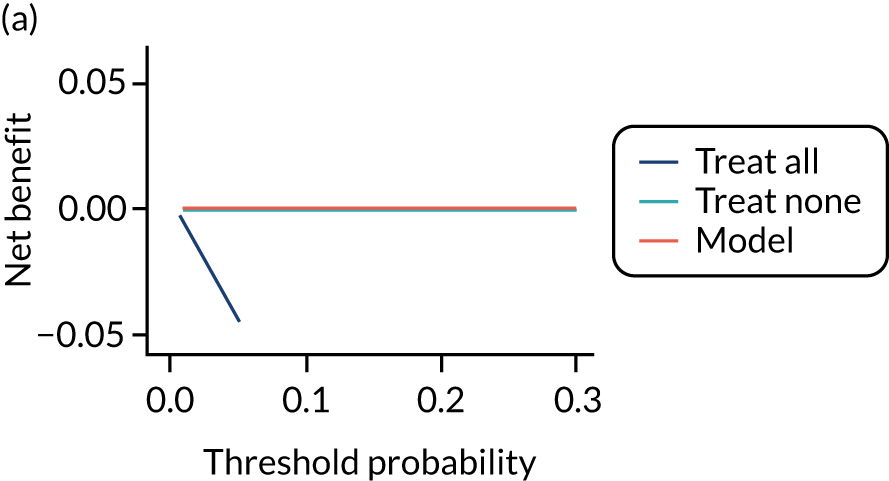
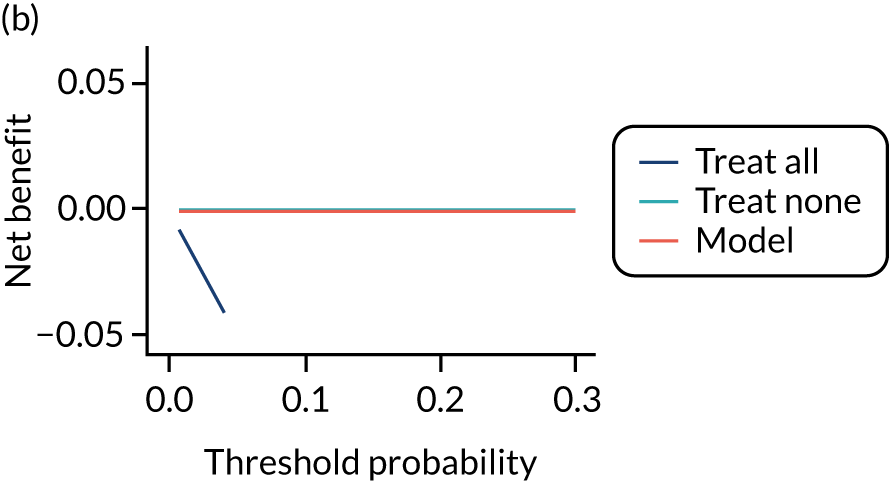
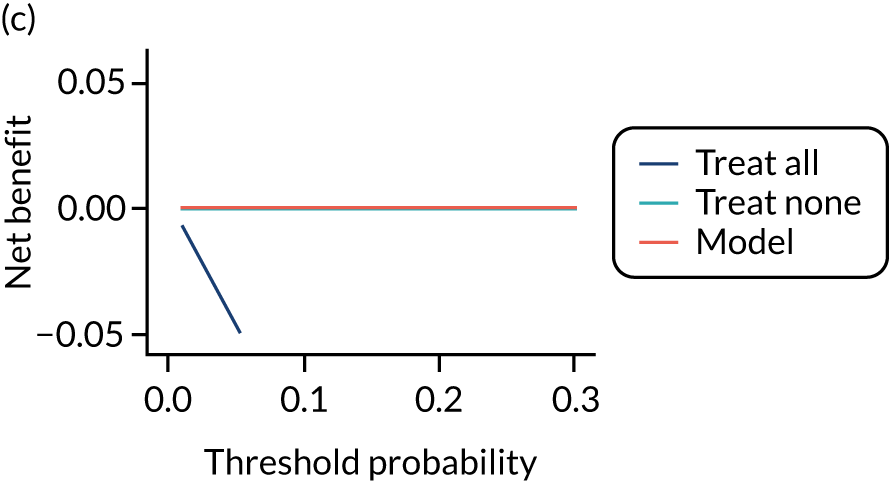
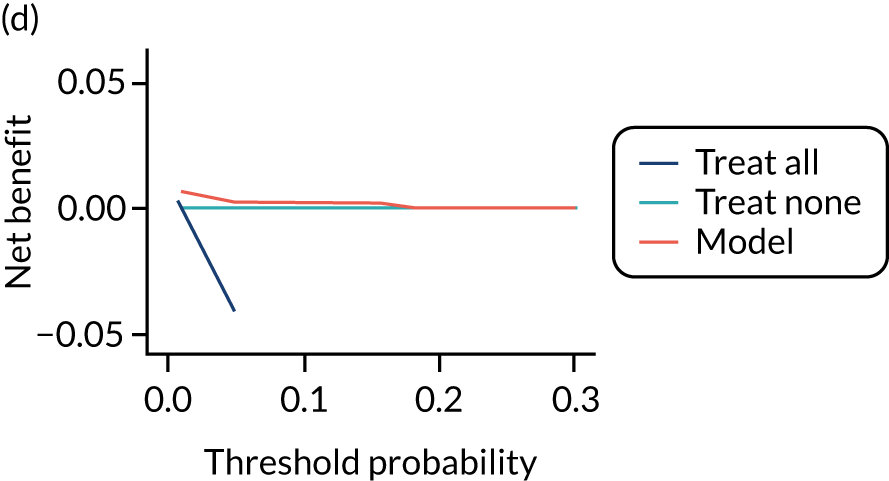
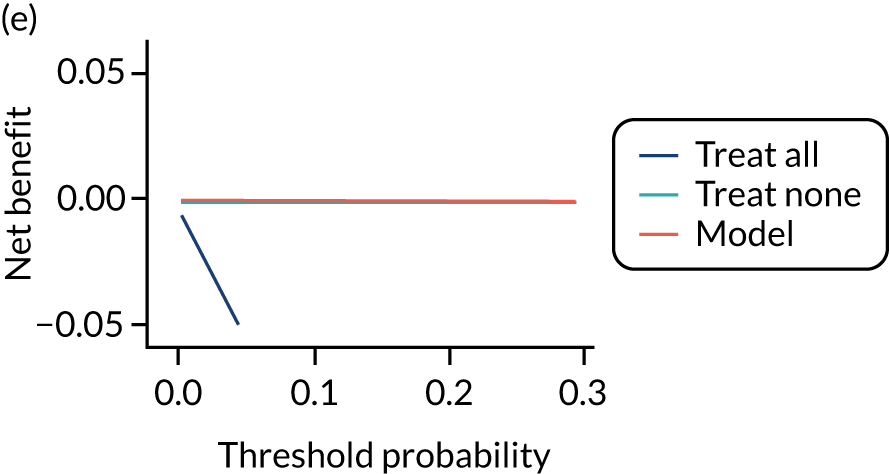
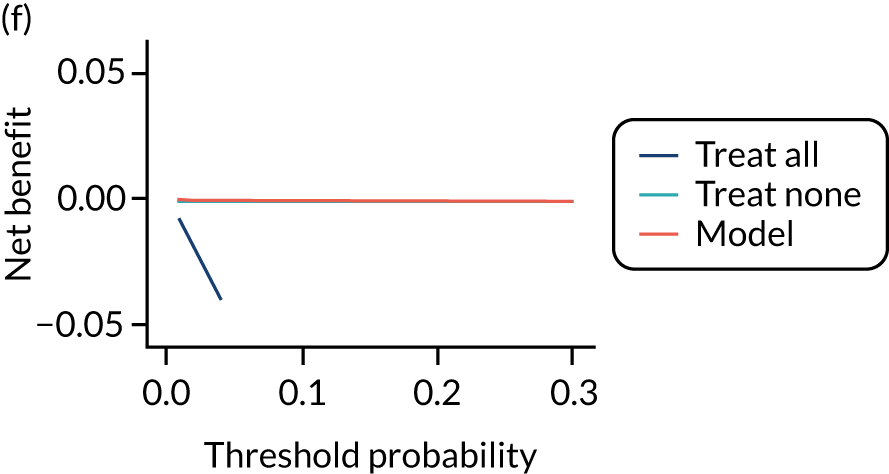
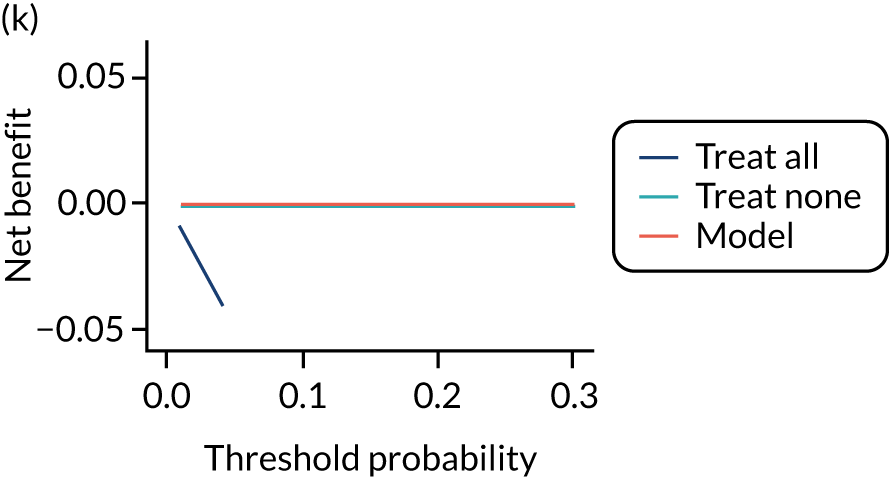
FIGURE 39.
Decision curves for the final (shrunken) model predicting early-onset pre-eclampsia using second-trimester clinical characteristics, in data sets used in the development and validation of the model. (a) SCOPE;42 (b) Poston et al. 2015;149 (c) Antsaklis et al.;110 (d) WHO;175 (e) NICH LR;159 (f) POUCH;131 (g) Van Kuijk et al. 2014;166 (h) STORK G;135 and (i) POP. 161
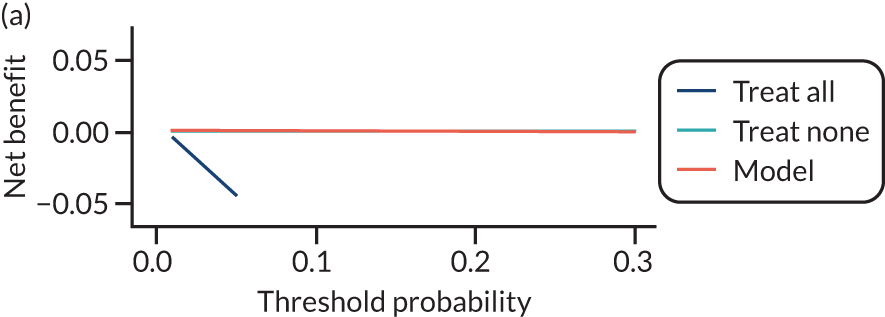
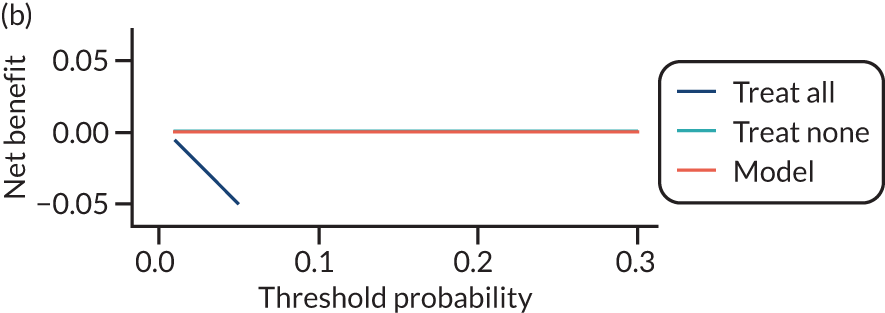
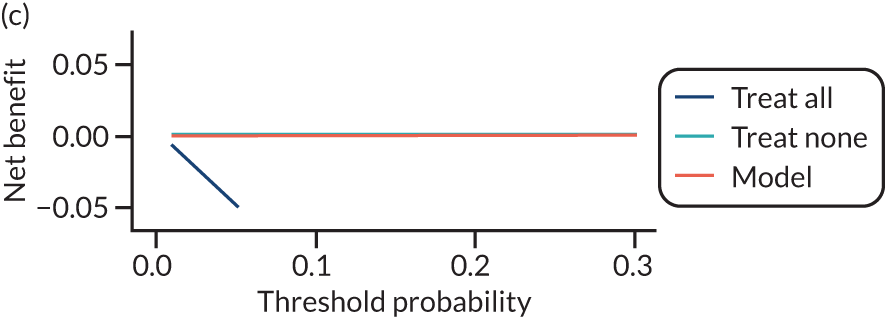
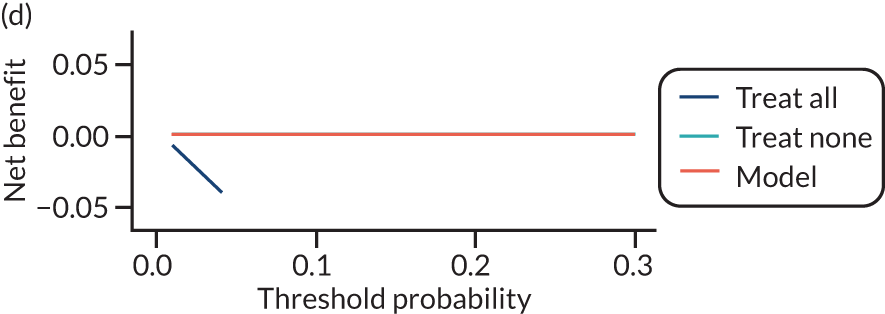
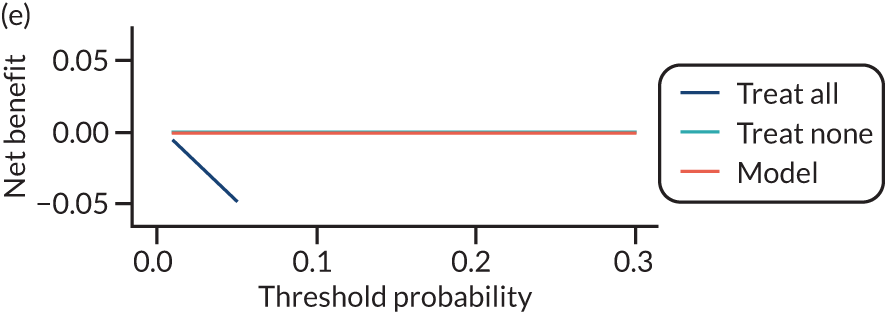
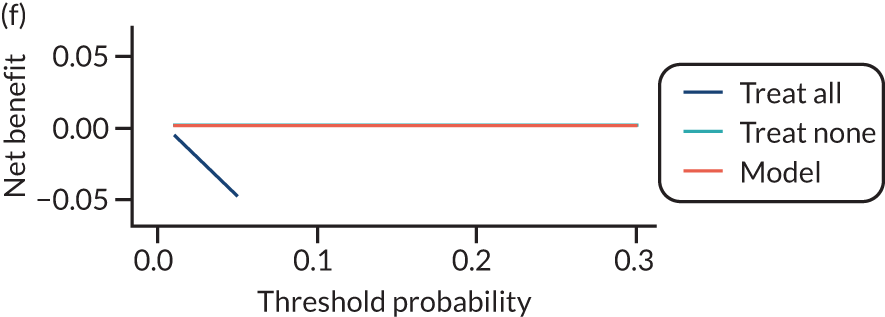
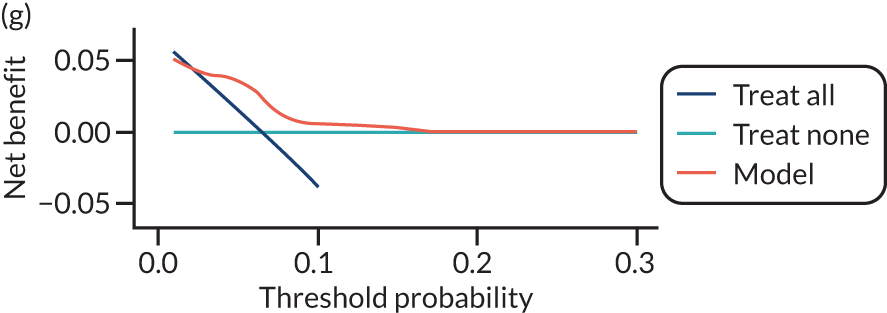
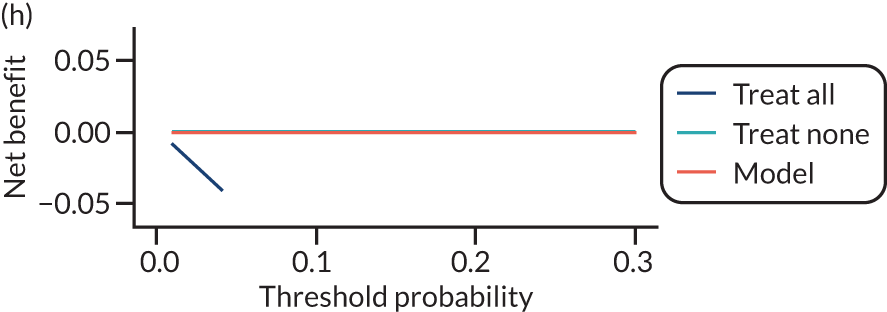
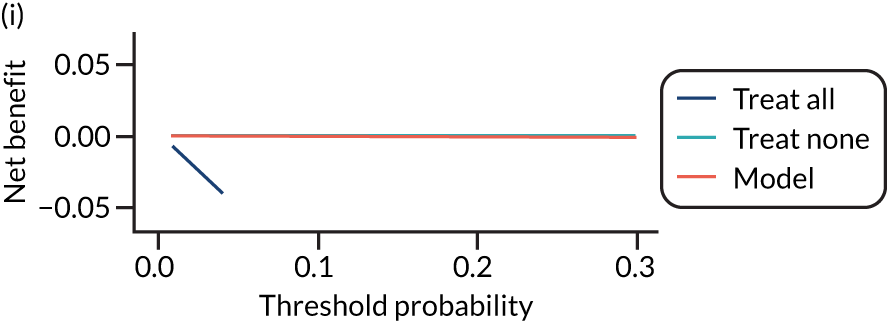
Late-onset pre-eclampsia models
FIGURE 42.
Decision curves for the final (shrunken) model predicting late-onset pre-eclampsia using first-trimester clinical characteristics, in data sets used in the development and validation of the model. (a) SCOPE;42 (b) Allen et al.;108 (c) Poston et al. 2015;149 (d) Baschat et al.;115 (e) Antsaklis;110 (f) WHO;175 (g) NICH LR;159 (h) Van Kuijk et al. 2014;166 (i) STORK G;135 (j) Vinter et al.;173 and (k) POP. 161
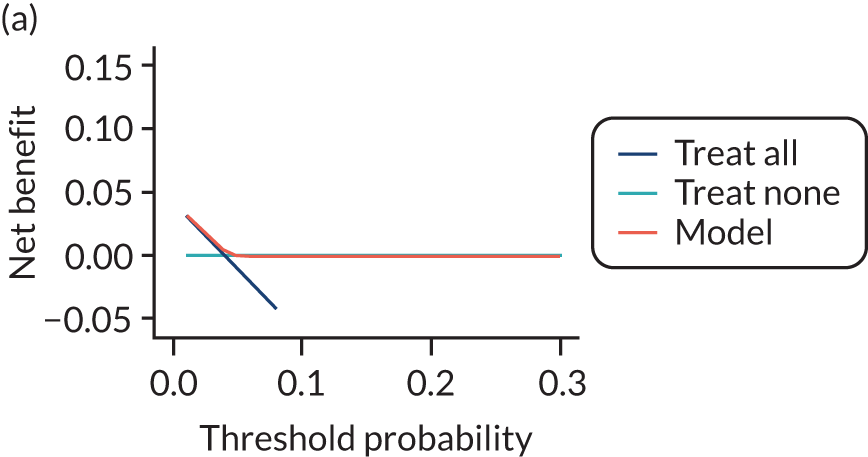
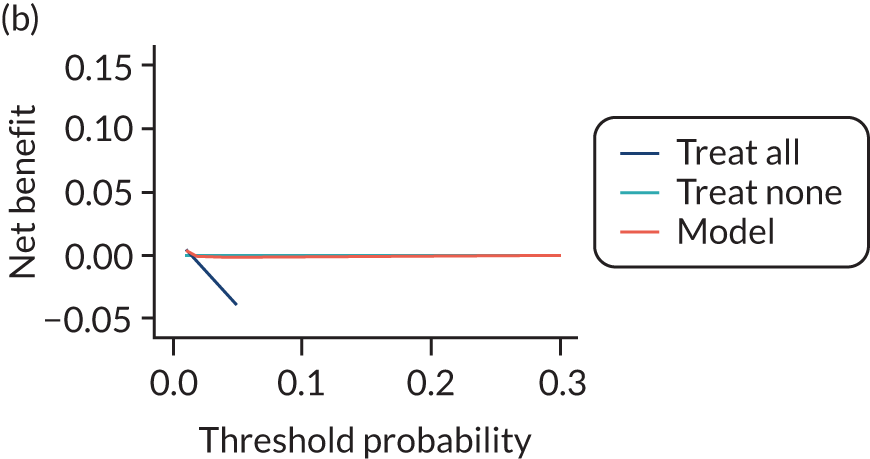
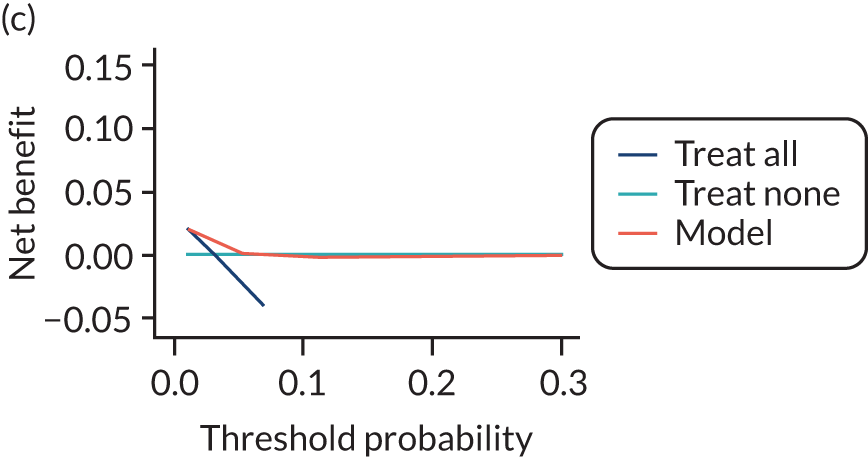
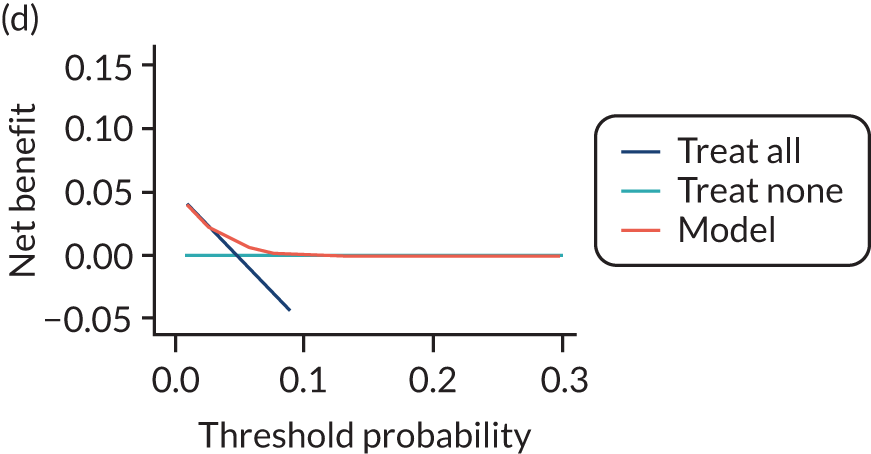
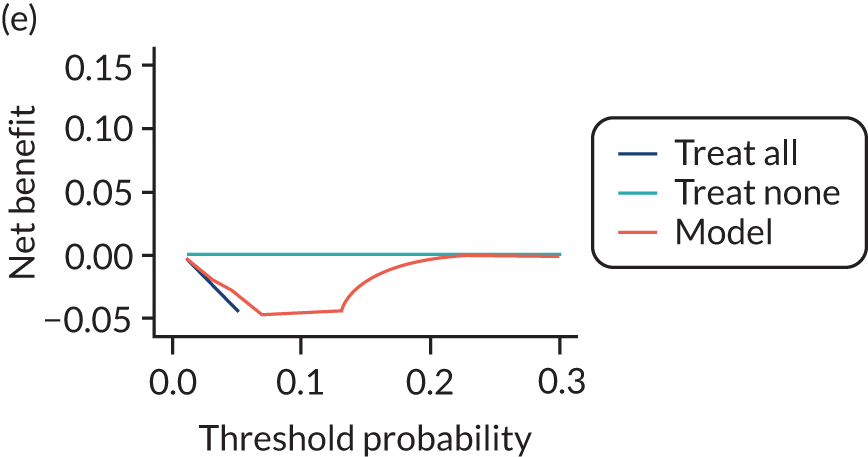
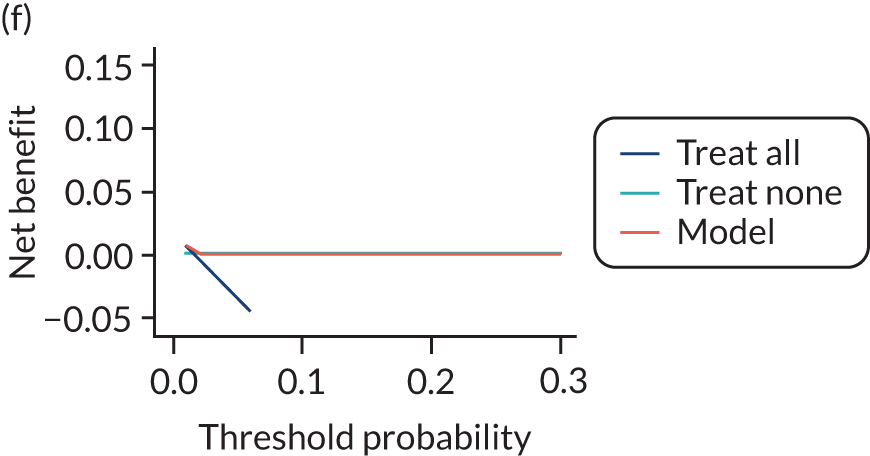
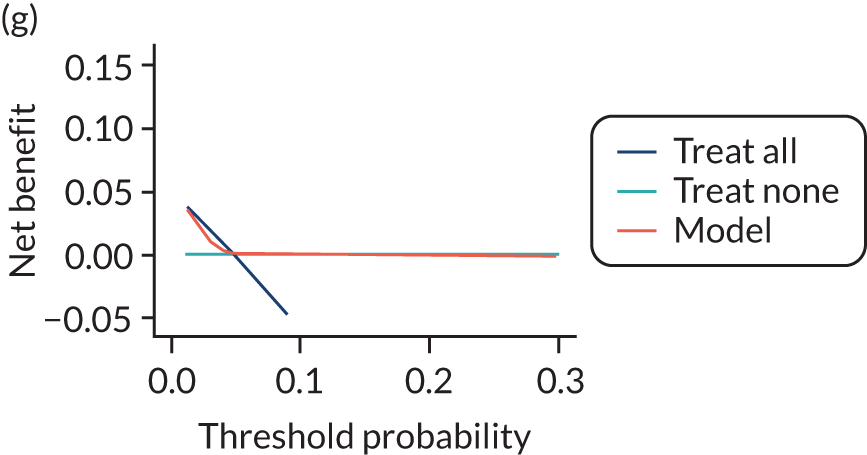
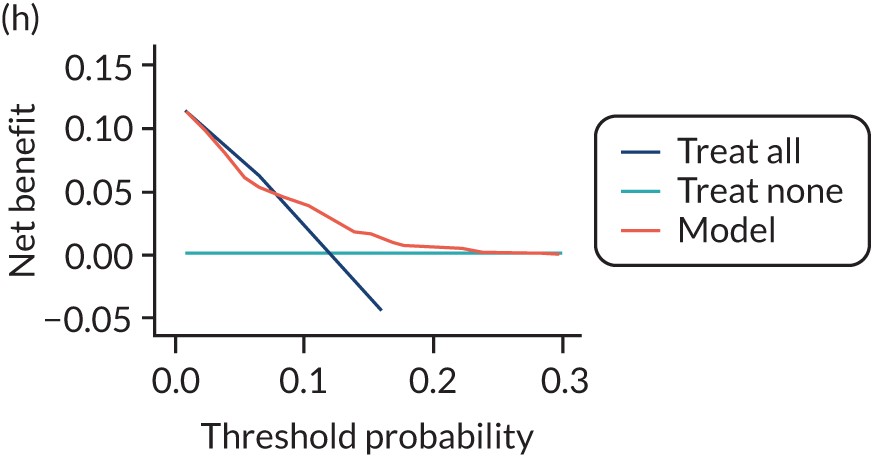
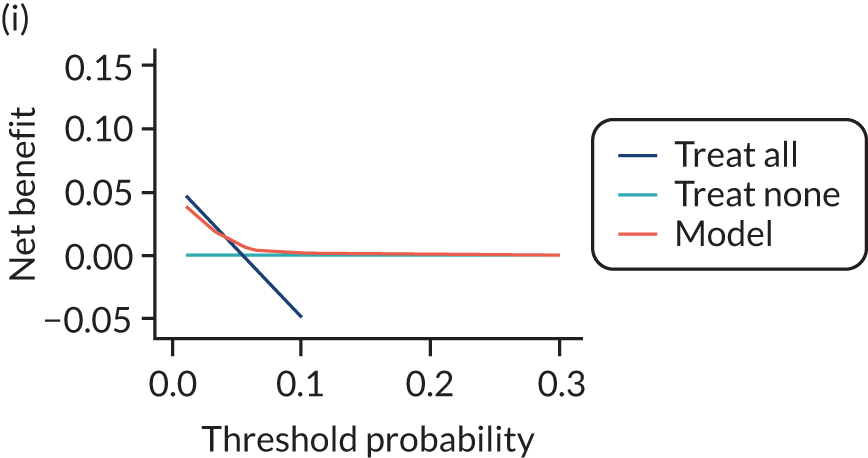
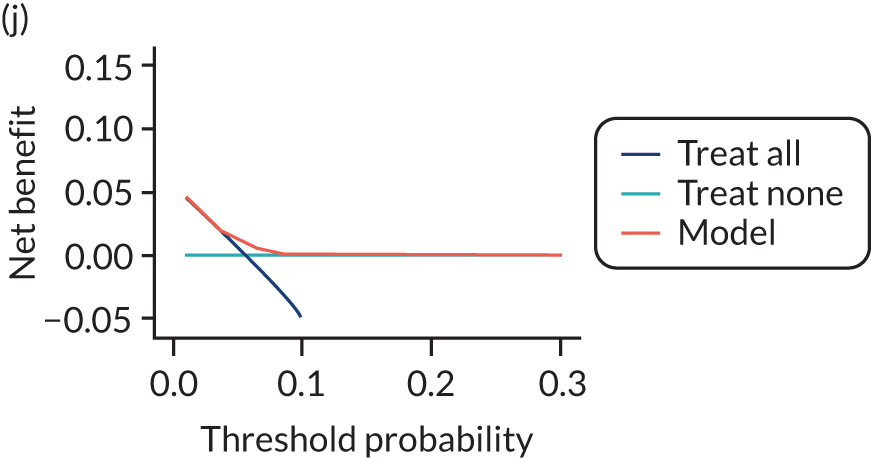
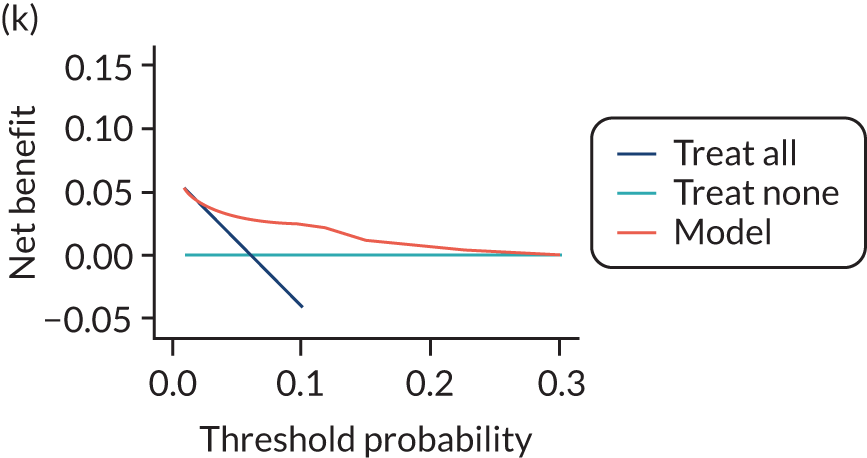
FIGURE 43.
Decision curves for the final (shrunken) model predicting late-onset pre-eclampsia using second-trimester clinical characteristics, in data sets used in the development and validation of the model. (a) SCOPE;42 (b) Poston et al. 2015;149 (c) Antsaklis;110 (d) WHO;175 (e) NICH LR;159 (f) POUCH;131 (g) Van Kuijk et al. 2014;166 (h) STORK G;135 and (i) POP. 161
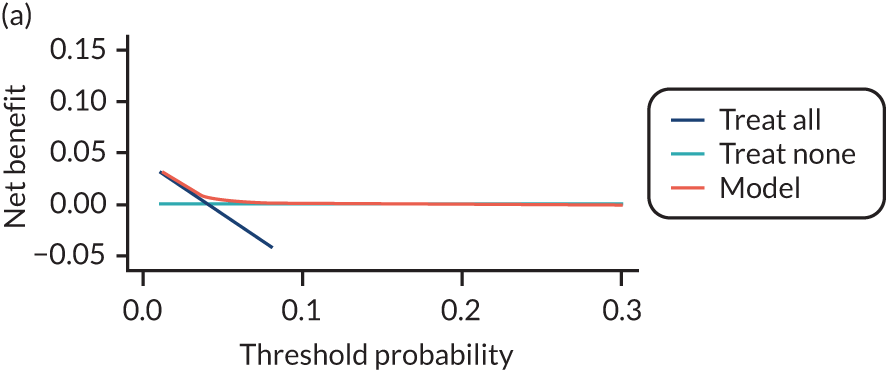
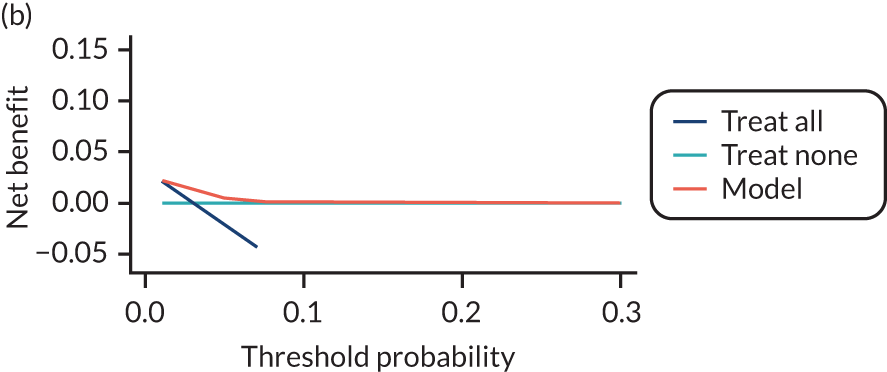
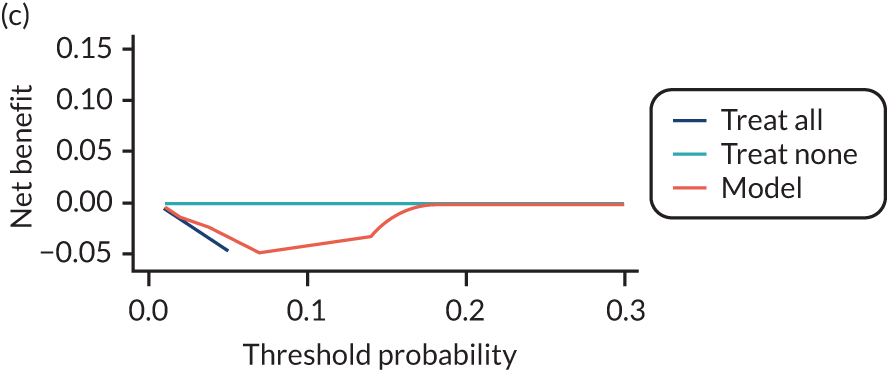
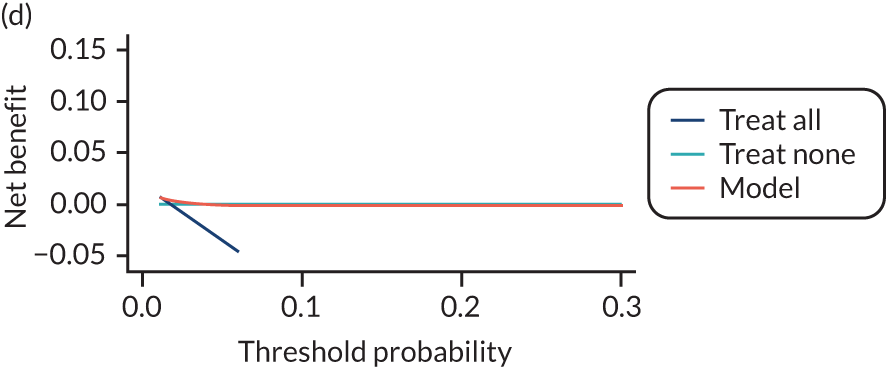
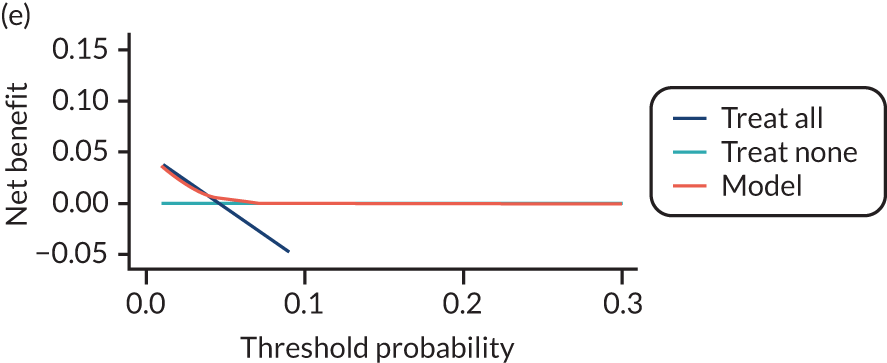
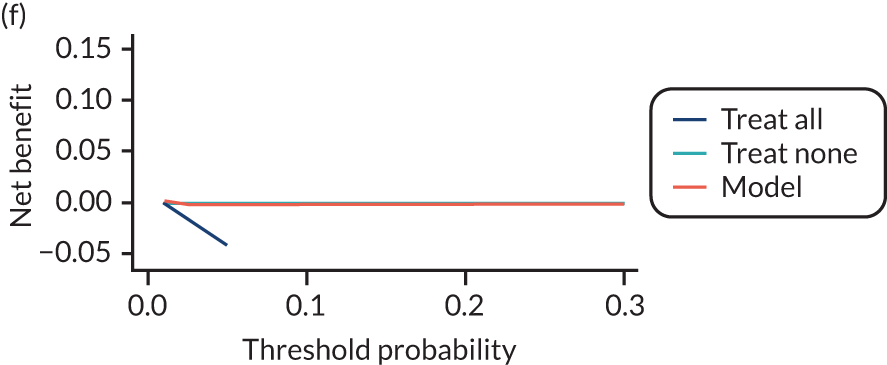
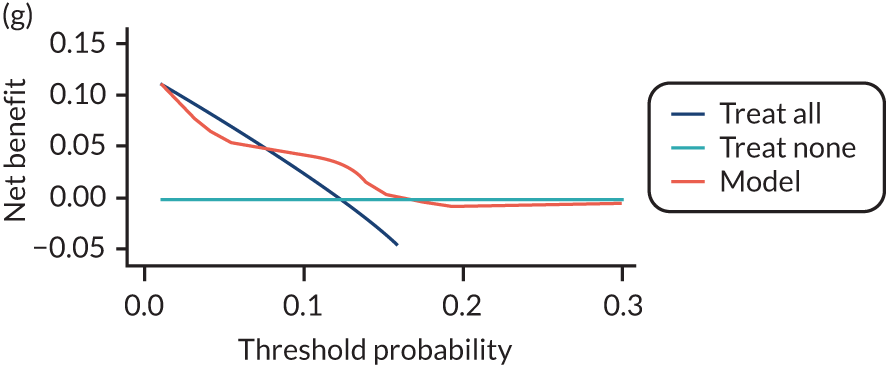
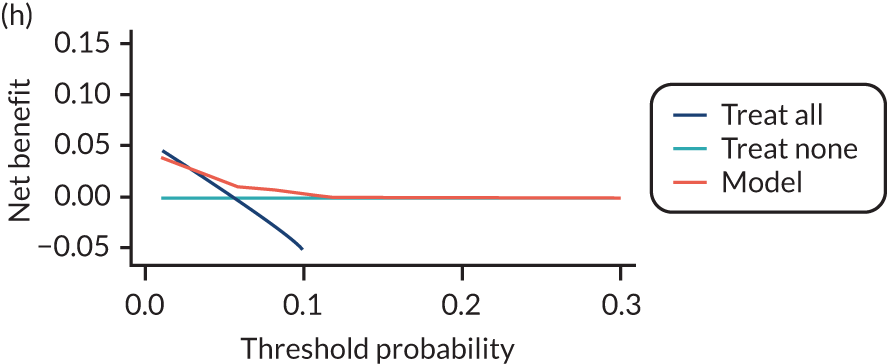
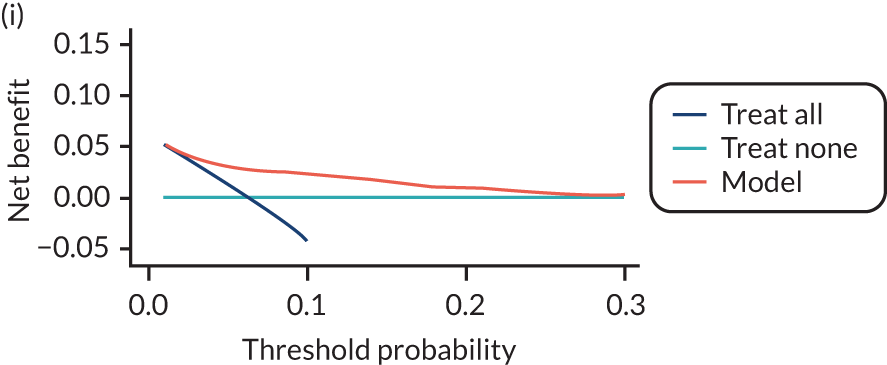
List of abbreviations
- AMND
- Aberdeen Maternity and Neonatal Databank
- BMI
- body mass index
- CI
- confidence interval
- CRP
- C-reactive protein
- DBP
- diastolic blood pressure
- HELLP
- haemolysis, elevated liver enzymes and low platelet count
- IPD
- individual participant data
- IPPIC
- International Prediction of Pregnancy Complications
- IQR
- interquartile range
- LP
- linear predictor
- MAP
- mean arterial blood pressure
- OR
- odds ratio
- PAPP-A
- pregnancy-associated plasma protein A
- PCR
- protein–creatinine ratio
- PlGF
- placental growth factor
- POP
- Pregnancy Outcome Prediction
- PROBAST
- Prediction study Risk of Bias Assessment Tool
- SBP
- systolic blood pressure
- SCOPE
- Screening for Pregnancy Endpoints
- sFlt-1
- soluble fms-like tyrosine kinase-1
- WHO
- World Health Organization
Notes
Supplementary material can be found on the NIHR Journals Library report page (https://doi.org/10.3310/hta24720).
Supplementary material has been provided by the authors to support the report and any files provided at submission will have been seen by peer reviewers, but not extensively reviewed. Any supplementary material provided at a later stage in the process may not have been peer reviewed.